Inventory and Evolution of Mitochondrion-localized Family A DNA Polymerases in Euglenozoa
Abstract
:1. Introduction
2. Results
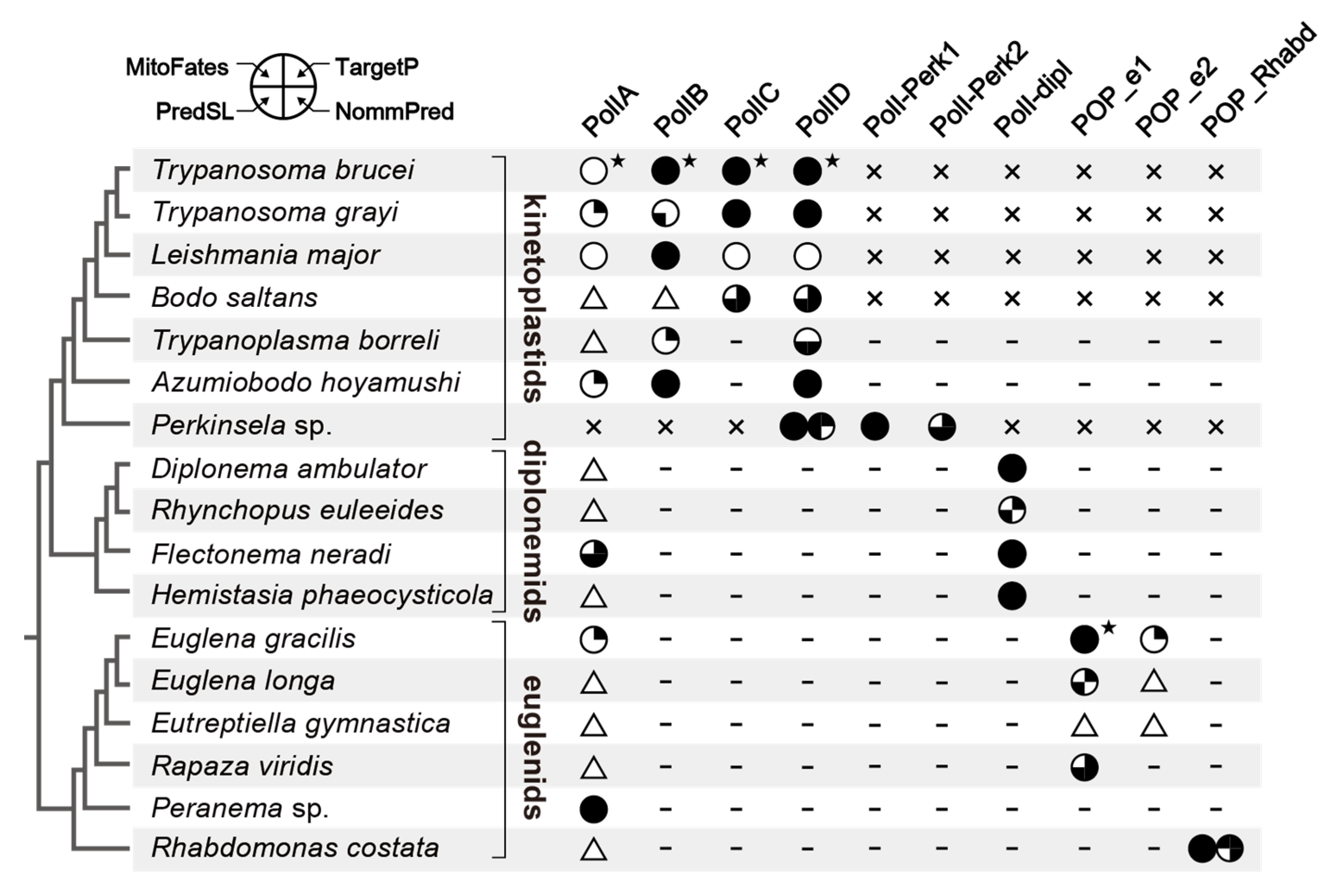
2.1. PolIA is Ubiquitous in Euglenida, Diplonemea, and Kinetoplastea
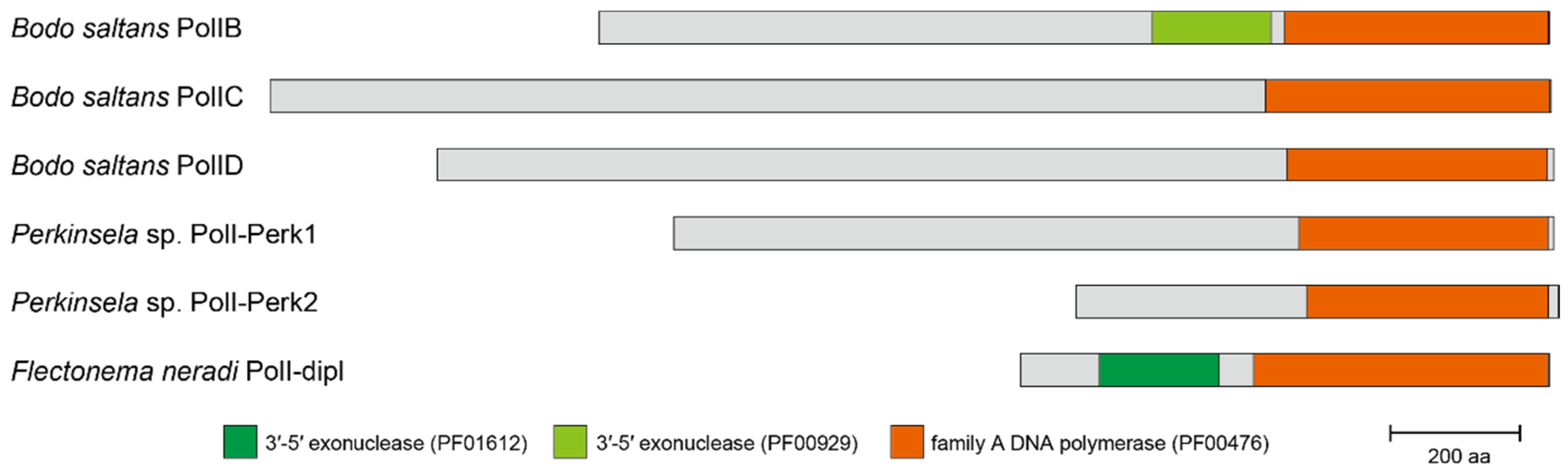
2.2. PolIB, C, D, and “PolIBCD-Related” DNA Polymerases in Diplonemea and Kinetoplastea
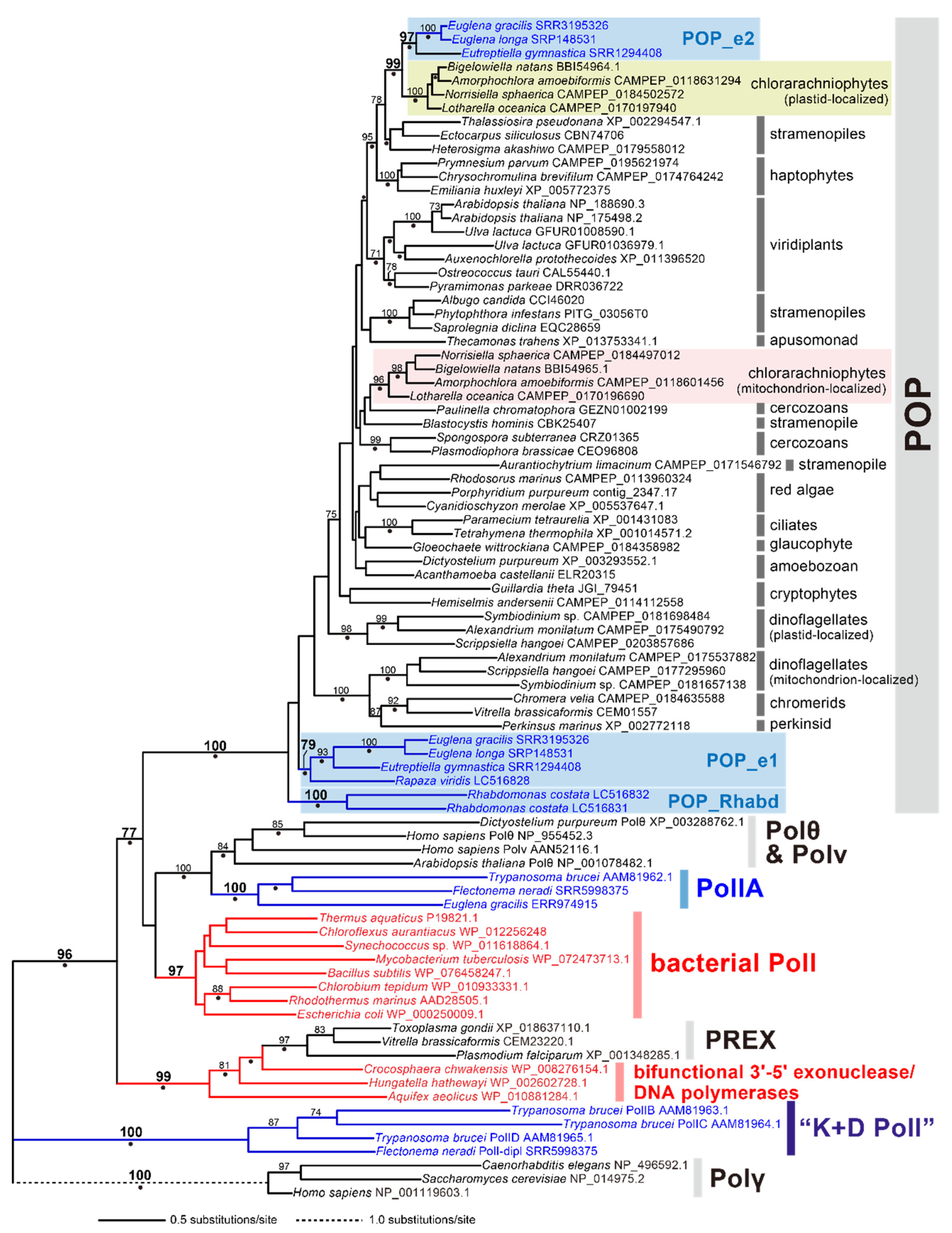
2.3. POP in Euglenida
3. Discussion
4. Materials and Methods
4.1. Sequence Data Preparation
4.2. Phylogenetic Analysis of family A DNA Polymerases
4.3. Phylogenetic Analysis of POP
4.4. In silico Prediction of Subcellular Localization and Functional Domains of Family A DNA Polymerases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barrett, M.P.; Burchmore, R.J.S.; Stich, A.; Lazzari, J.O.; Frasch, A.C.; Cazzulo, J.J.; Krishna, S. The trypanosomiases. Lancet 2003, 362, 1469–1480. [Google Scholar] [CrossRef]
- Verner, Z.; Basu, S.; Benz, C.; Dixit, S.; Dobáková, E.; Faktorová, D.; Hashimi, H.; Horáková, E.; Huang, Z.; Paris, Z.; et al. Malleable mitochondrion of Trypanosoma brucei. In International Review of Cell and Molecular Biology; Academic Press: Cambridge, MA, USA, 2015; Volume 315, pp. 73–151. [Google Scholar]
- Klingbeil, M.M.; Motyka, S.A.; Englund, P.T. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell 2002, 10, 175–186. [Google Scholar] [CrossRef]
- Krasich, R.; Copeland, W.C. DNA polymerases in the mitochondria: A critical review of the evidence. Physiol. Behav. 2017, 22, 692–709. [Google Scholar]
- Saxowsky, T.T.; Choudhary, G.; Klingbeil, M.M.; Englund, P.T. Trypanosoma brucei has two distinct mitochondrial DNA polymerase β enzymes. J. Biol. Chem. 2003, 278, 49095–49101. [Google Scholar] [CrossRef] [Green Version]
- Rajão, M.A.; Passos-Silva, D.G.; DaRocha, W.D.; Franco, G.R.; Macedo, A.M.; Pena, S.D.J.; Teixeira, S.M.; Machado, C.R. DNA polymerase kappa from Trypanosoma cruzi localizes to the mitochondria, bypasses 8-oxoguanine lesions and performs DNA synthesis in a recombination intermediate. Mol. Microbiol. 2009, 71, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Graziewicz, M.A.; Longley, M.J.; Copeland, W.C. DNA polymerase γ in mitochondrial DNA replication and repair. Chem. Rev. 2006, 106, 383–405. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.C.; Lyznik, A.; Mohammed, S.; Elowsky, C.G.; Elo, A.; Yule, R.; Mackenzie, S.A. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 2005, 17, 2805–2816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyama, T.; Sato, N. Enzymes involved in organellar DNA replication in photosynthetic eukaryotes. Front. Plant Sci. 2014, 5, 480:1–480:12. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, Y.; Watanabe, A. Organellar DNA polymerases in complex plastid-bearing algae. Biomolecules 2019, 9, 140. [Google Scholar] [CrossRef] [Green Version]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [Green Version]
- Lukeš, J.; Guilbride, D.L.; Votýpka, J.; Zíková, A.; Benne, R.; Englund, P.T. Kinetoplast DNA network: Evolution of an improbable structure. Eukayot. Cell 2002, 1, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, V.; Flegontov, P.; Gerasimov, E.; Tanifuji, G.; Hashimi, H.; Logacheva, M.D.; Maruyama, S.; Onodera, N.T.; Gray, M.W.; Archibald, J.M.; et al. Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, an endosymbiotic kinetoplastid. MBio 2015, 6, e01498-15:1–e01498-15:12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yabuki, A.; Tanifuji, G.; Kusaka, C.; Takishita, K.; Fujikura, K. Hyper-eccentric structural genes in the mitochondrial genome of the algal parasite Hemistasia phaeocysticola. Genome Biol. Evol. 2016, 8, 2870–2878. [Google Scholar] [PubMed] [Green Version]
- Burger, G.; Valach, M. Perfection of eccentricity: Mitochondrial genomes of diplonemids. IUBMB Life 2018, 70, 1197–1206. [Google Scholar] [CrossRef] [Green Version]
- Kaur, B.; Záhonová, K.; Valach, M.; Faktorová, D.; Prokopchuk, G.; Burger, G.; Lukeš, J. Gene fragmentation and RNA editing without borders: Eccentric mitochondrial genomes of diplonemids. Nucleic Acids Res. 2020, 48, 2694–2708. [Google Scholar] [CrossRef]
- Roy, J.; Faktorová, D.; Lukeš, J.; Burger, G. Unusual mitochondrial genome structures throughout the Euglenozoa. Protist 2007, 158, 385–396. [Google Scholar] [CrossRef]
- Spencer, D.F.; Gray, M.W. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: Fragmented genes in a seemingly fragmented genome. Mol. Genet. Genom. 2011, 285, 19–31. [Google Scholar] [CrossRef]
- Dobáková, E.; Flegontov, P.; Skalický, T.; Lukeš, J. Unexpectedly streamlined mitochondrial genome of the euglenozoan Euglena gracilis. Genome Biol. Evol. 2015, 7, 3358–3367. [Google Scholar] [CrossRef] [Green Version]
- Simpson, A.G.B.; Hoff, J.V.D.; Bernard, C.; Burton, H.R.; Patterson, D.J. The ultrastructure and systematic position of the euglenozoon Postgaardi mariagerensis. Arch. Protistenkd. 1997, 147, 213–225. [Google Scholar] [CrossRef]
- Yubuki, N.; Edgcomb, V.P.; Bernhard, J.M.; Leander, B.S. Ultrastructure and molecular phylogeny of Calkinsia aureus: Cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiol. 2009, 9, 16:1–16:22. [Google Scholar] [CrossRef] [Green Version]
- Breglia, S.A.; Yubuki, N.; Hoppenrath, M.; Leander, B.S. Ultrastructure and molecular phylogenetic position of a novel euglenozoan with extrusive episymbiotic bacteria: Bihospites bacati n. gen. et sp. (Symbiontida). BMC Microbiol. 2010, 10, 145:1–145:21. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, T.; Terasawa, K.; Fujiwara, M.; Sato, N. Purification and characterization of organellar DNA polymerases in the red alga Cyanidioschyzon merolae. FEBS J. 2008, 275, 2899–2918. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.; Vandoros, A.V.; Mozeleski, B.; Klingbeil, M.M. Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes. Eukaryot. Cell 2008, 7, 2141–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruhn, D.F.; Mozeleski, B.; Falkin, L.; Klingbeil, M.M. Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Mol. Microbiol. 2010, 75, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Bruhn, D.F.; Sammartino, M.P.; Klingbeil, M.M. Three mitochondrial DNA polymerases are essential for kinetoplast DNA replication and survival of bloodstream form Trypanosoma brucei. Eukaryot. Cell 2011, 10, 734–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panigrahi, A.K.; Ogata, Y.; Zíková, A.; Anupama, A.; Dalley, R.A.; Acestor, N.; Myler, P.J.; Stuart, K.D. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 2009, 9, 434–450. [Google Scholar] [CrossRef] [Green Version]
- Hammond, M.J.; Nenarokova, A.; Butenko, A.; Zoltner, M.; Dobáková, E.L.; Field, M.C.; Lukeš, J. A uniquely complex mitochondrial proteome from Euglena gracilis. Mol. Biol. Evol. 2020. Epub ahead of print. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Heijne, G.V. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Kume, K.; Amagasa, T.; Hashimoto, T.; Kitagawa, H. NommPred: Prediction of mitochondrial and mitochondrion-related organelle proteins of nonmodel organisms. Evol. Bioinform. 2018, 14, 1–12. [Google Scholar] [CrossRef]
- Petsalaki, E.I.; Bagos, P.G.; Litou, Z.I.; Hamodrakas, S.J. PredSL: A tool for the N-terminal sequence-based prediction of protein subcellular localization. Genom. Proteom. Bioinf. 2006, 4, 48–55. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bicudo, C.E.d.M.; Menezes, M. Phylogeny and classification of Euglenophyceae: A brief review. Front. Ecol. Evol. 2016, 4, 17:1–17:15. [Google Scholar] [CrossRef] [Green Version]
- Tashyreva, D.; Prokopchuk, G.; Yabuki, A.; Kaur, B.; Faktorová, D.; Votýpka, J.; Kusaka, C.; Fujikura, K.; Shiratori, T.; Ishida, K.I.; et al. Phylogeny and morphology of new diplonemids from Japan. Protist 2018, 169, 158–179. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, E.; Ishikawa, S.A.; Kume, K.; Kumagai, A.; Kamaishi, T.; Tanifuji, G.; Hashimoto, T.; Inagaki, Y. Global kinetoplastea phylogeny inferred from a large-scale multigene alignment including parasitic species for better understanding transitions from a free-living to a parasitic lifestyle. Genes Genet. Syst. 2017, 92, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosal, L.; Comendador, M.A.; Sierra, L.M. The mus308 locus of Drosophila melanogaster is implicated in the bypass of ENU-induced O-alkylpyrimidine adducts. Mol. Gen. Genet. 2000, 263, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Concepción-Acevedo, J.; Miller, J.C.; Boucher, M.J.; Klingbeil, M.M. Cell cycle localization dynamics of mitochondrial DNA polymerase IC in African trypanosomes. Mol. Biol. Cell 2018, 29, 2540–2552. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Sakai, A.; Takechi, K.; Takio, S.; Takusagawa, M.; Takano, H. NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007, 48, 1679–1692. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, T.; Tajima, N.; Sekine, K.; Sato, N. Localization and phylogenetic analysis of enzymes related to organellar genome replication in the unicellular rhodophyte Cyanidioschyzon merolae. Genome Biol. Evol. 2014, 6, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Bendtsen, J.D.; Nielsen, H.; Heijne, G.V.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Heijne, G.V.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Novák Vanclová, A.M.G.; Zoltner, M.; Kelly, S.; Soukal, P.; Záhonová, K.; Füssy, Z.; Ebenezer, T.E.; Lacová Dobáková, E.; Eliáš, M.; Lukeš, J.; et al. Metabolic quirks and the colourful history of the Euglena gracilis secondary plastid. New Phytol. 2020, 225, 1578–1592. [Google Scholar] [CrossRef] [PubMed]
- Tanifuji, G.; Cenci, U.; Moog, D.; Dean, S.; Nakayama, T.; David, V.; Fiala, I.; Curtis, B.A.; Sibbald, S.J.; Onodera, N.T.; et al. Genome sequencing reveals metabolic and cellular interdependence in an amoeba-kinetoplastid symbiosis. Sci. Rep. 2017, 7, 11688:1–11688:13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turmel, M.; Gagnon, M.C.; O’Kelly, C.J.; Otis, C.; Lemieux, C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 2009, 26, 631–648. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Hirakawa, Y.; Kofuji, R.; Sugita, M.; Ishida, K. Plastid genome sequences of Gymnochlora stellata, Lotharella vacuolata, and Partenskyella glossopodia reveal remarkable structural conservation among chlorarachniophyte species. J. Plant Res. 2016, 129, 581–590. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Brister, J.R.; Bolton, E.E.; Canese, K.; Comeau, D.C.; Funk, K.; Ketter, A.; Kim, S.; Kimchi, A.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48, D9–D16. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421:1–421:9. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, S.R. HMMER: Biosequence Analysis Using Profile Hidden Markov Models. Available online: http://hmmer.org/ (accessed on 10 December 2019).
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- López-García, P.; Duperron, S.; Philippot, P.; Foriel, J.; Susini, J.; Moreira, D. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 2003, 5, 961–976. [Google Scholar] [CrossRef] [Green Version]
- Scheckenbach, F.; Hausmann, K.; Wylezich, C.; Weitere, M.; Arndt, H. Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc. Natl. Acad. Sci. USA 2010, 107, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Marquardt, M.; Vader, A.; Stübner, E.I.; Reigstad, M.; Gabrielsen, T.M. Strong seasonality of marine microbial eukaryotes in a high-Arctic fjord (Isfjorden, in West Spitsbergen, Norway). Appl. Environ. Microbiol. 2016, 82, 1868–1880. [Google Scholar] [CrossRef] [Green Version]
- Gawryluk, R.M.R.; del Campo, J.; Okamoto, N.; Strassert, J.F.H.; Lukeš, J.; Richards, T.A.; Worden, A.Z.; Santoro, A.E.; Keeling, P.J. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 2016, 26, 3053–3059. [Google Scholar] [CrossRef] [Green Version]
- Wideman, J.G.; Lax, G.; Leonard, G.; Milner, D.S.; Rodríguez-Martínez, R.; Simpson, A.G.B.; Richards, T.A. A single-cell genome reveals diplonemid-like ancestry of kinetoplastid mitochondrial gene structure. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190100. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harada, R.; Hirakawa, Y.; Yabuki, A.; Kashiyama, Y.; Maruyama, M.; Onuma, R.; Soukal, P.; Miyagishima, S.; Hampl, V.; Tanifuji, G.; et al. Inventory and Evolution of Mitochondrion-localized Family A DNA Polymerases in Euglenozoa. Pathogens 2020, 9, 257. https://doi.org/10.3390/pathogens9040257
Harada R, Hirakawa Y, Yabuki A, Kashiyama Y, Maruyama M, Onuma R, Soukal P, Miyagishima S, Hampl V, Tanifuji G, et al. Inventory and Evolution of Mitochondrion-localized Family A DNA Polymerases in Euglenozoa. Pathogens. 2020; 9(4):257. https://doi.org/10.3390/pathogens9040257
Chicago/Turabian StyleHarada, Ryo, Yoshihisa Hirakawa, Akinori Yabuki, Yuichiro Kashiyama, Moe Maruyama, Ryo Onuma, Petr Soukal, Shinya Miyagishima, Vladimír Hampl, Goro Tanifuji, and et al. 2020. "Inventory and Evolution of Mitochondrion-localized Family A DNA Polymerases in Euglenozoa" Pathogens 9, no. 4: 257. https://doi.org/10.3390/pathogens9040257
APA StyleHarada, R., Hirakawa, Y., Yabuki, A., Kashiyama, Y., Maruyama, M., Onuma, R., Soukal, P., Miyagishima, S., Hampl, V., Tanifuji, G., & Inagaki, Y. (2020). Inventory and Evolution of Mitochondrion-localized Family A DNA Polymerases in Euglenozoa. Pathogens, 9(4), 257. https://doi.org/10.3390/pathogens9040257




