The Emerging Concern and Interest SARS-CoV-2 Variants
Abstract
1. Introduction
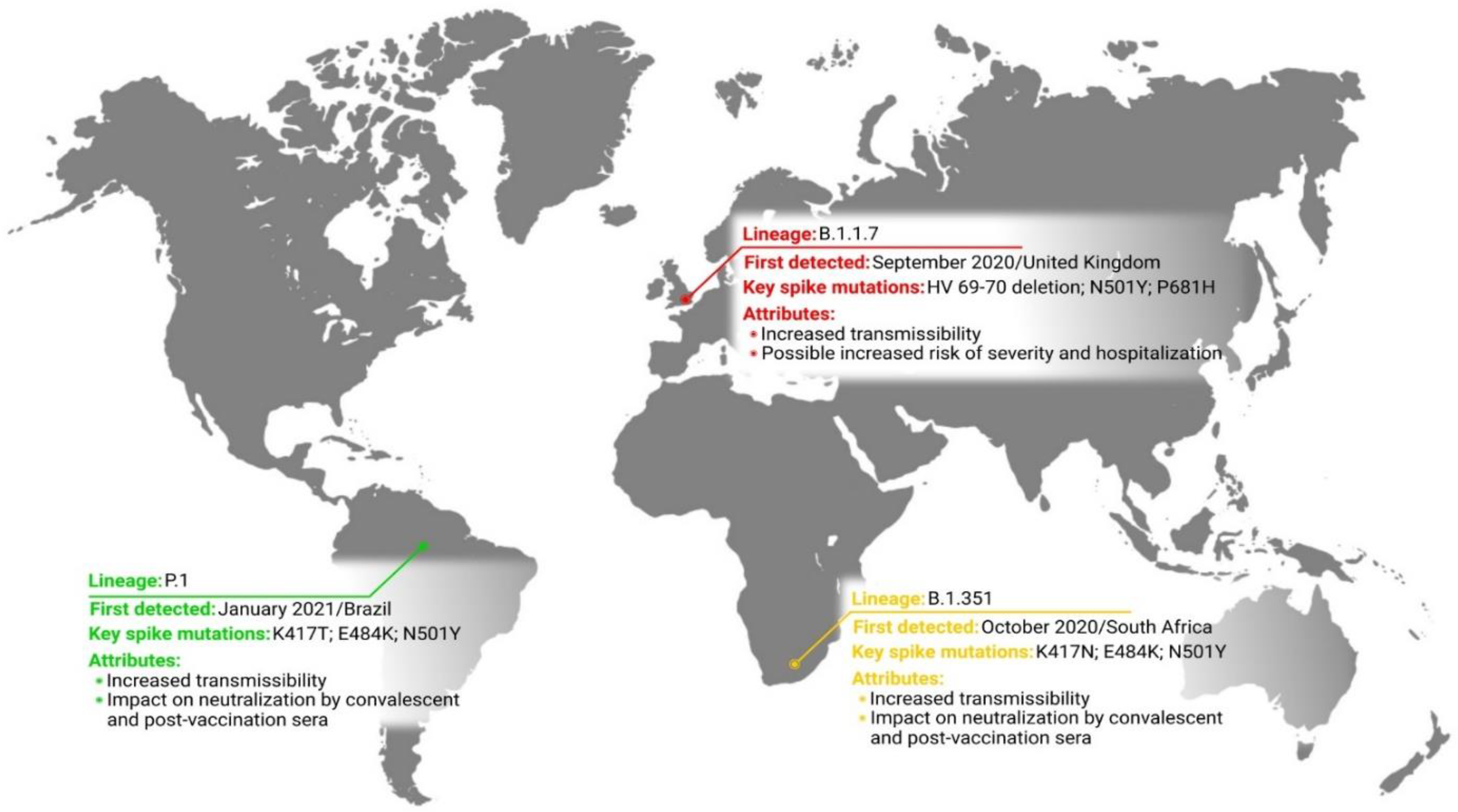
2. Variant of Concern
2.1. Lineage B.1.1.7
2.2. Lineage B.1.351
2.3. Lineage P.1
3. Variant of Interest
4. Variant of High Consequence
5. Mutations Impact on SARS-CoV-2 Diagnostics/Detection Protocols
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. 2020. Available online: https://covid19.who.int/ (accessed on 3 March 2021).
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, 325, 529. [Google Scholar] [CrossRef]
- Almubaid, Z.; Al-Mubaid, H. Analysis and comparison of genetic variants and mutations of the novel coronavirus SARS-CoV-2. Gene Rep. 2021, 23, 101064. [Google Scholar] [CrossRef]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Petrone, M.E.; Holmes, E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020, 5, 529–530. [Google Scholar] [CrossRef]
- Ferron, F.; Subissi, L.; De Morais, A.T.S.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA 2018, 115, E162–E171. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Walensky, R.P.; Walke, H.T.; Fauci, A.S. SARS-CoV-2 Variants of Concern in the United States—Challenges and Opportunities. JAMA 2021, 325, 1037. [Google Scholar] [CrossRef]
- Carrasco-Hernandez, R.; Jácome, R.; Vidal, Y.L.; De León, S.P. Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review. ILAR J. 2017, 58, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem. Biophys. Res. Commun. 2021, 534, 374–380. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- World Health Organization. An Update on SARS-CoV-2 Virus Mutations & Variants. Available online: https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update47-sars-cov-2-variants.pdf?sfvrsn=f2180835_4 (accessed on 25 March 2021).
- Centers for Disease Control and Prevention. About Variants of the Virus that Causes COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html (accessed on 25 March 2021).
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462. [Google Scholar] [CrossRef]
- Centers for Disease and Control Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html#Interest (accessed on 24 March 2021).
- Lineages, P. Global Report Investigating Novel Coronavirus Haplotypes. 2021 Copyright© 2021 Massachusetts Medical Society. Available online: https://cov-lineages.org/global_report (accessed on 24 March 2021).
- World Health Organization. Weekly Epidemiological Update on COVID-19—23 March 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---23-march-2021 (accessed on 24 March 2021).
- Huang, S.-W.; Miller, S.O.; Yen, C.-H.; Wang, S.-F. Impact of Genetic Variability in ACE2 Expression on the Evolutionary Dynamics of SARS-CoV-2 Spike D614G Mutation. Genes 2020, 12, 16. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Flores, M.; Cardozo, T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020, 74, 13525. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. SARS-CoV-2 Genomic Sequencing for Public Health Goals. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-genomic_sequencing-2021.1 (accessed on 10 May 2021).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; Du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- World Health Organization. COVID-19 Weekly Epidemiological Update. Special Edition: Proposed Working Definitions of SARS-CoV-2 Variants of Interest and Variants of Concern. Available online: https://www.who.int/Publications/m/item/covid-19-weekly-epidemiological-update (accessed on 24 March 2021).
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372. [Google Scholar] [CrossRef]
- Grint, D.J.; Wing, K.; Williamson, E.; McDonald, H.I.; Bhaskaran, K.; Evans, D.; Evans, S.J.; Walker, A.J.; Hickman, G.; Nightingale, E.; et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Eurosurveillance 2021, 26, 2100256. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. US COVID-19 Cases Caused by Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html (accessed on 22 March 2021).
- Global Report Investigating Novel Coronavirus Haplotypes. B.1.1.7. Available online: https://cov-lineages.org/global_report_B.1.1.7.html#table2link (accessed on 23 March 2021).
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertson, D.L.; Volz, E. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 18 March 2021).
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nat. Cell Biol. 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.-Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef]
- Santos, J.C.; Passos, G.A. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Pereira, F. SARS-CoV-2 variants combining spike mutations and the absence of ORF8 may be more transmissible and require close monitoring. Biochem. Biophys. Res. Commun. 2021, 550, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tang, H.; McDanal, C.; Wagh, K.; Fischer, W.; Theiler, J.; Yoon, H.; Li, D.; Haynes, B.F.; Sanders, K.O.; et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021, 29, 529–539.e3. [Google Scholar] [CrossRef]
- Graham, M.S.; Sudre, C.H.; May, A.; Antonelli, M.; Murray, B.; Varsavsky, T.; Kläser, K.; Canas, L.S.; Molteni, E.; Modat, M.; et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: An ecological study. Lancet Public Health 2021, 6, e335–e345. [Google Scholar] [CrossRef]
- Davies, N.G.; Jarvis, C.I.; Edmunds, W.J.; Jewell, N.P.; Diaz-Ordaz, K.; Keogh, R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nat. Cell Biol. 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Calistri, P.; Amato, L.; Puglia, I.; Cito, F.; Di Giuseppe, A.; Danzetta, M.L.; Morelli, D.; Di Domenico, M.; Caporale, M.; Scialabba, S.; et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021, 105, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; De Marco, A.; Ferreira, I.A.T.M.; Meng, B.; Datir, R.P.; Walls, A.C.; Bassi, J.; Pinto, D.; Fregni, C.S. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nat. Cell Biol. 2021, 593, 136–141. [Google Scholar] [CrossRef]
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. BioRxiv 2021. [Google Scholar] [CrossRef]
- Muik, A.; Wallisch, A.-K.; Sänger, B.; Swanson, K.A.; Mühl, J.; Chen, W.; Cai, H.; Maurus, D.; Sarkar, R.; Türeci, Ö.; et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science 2021, 371, 1152–1153. [Google Scholar] [CrossRef]
- Mwenda, M.; Saasa, N.; Sinyange, N.; Busby, G.; Chipimo, P.J.; Hendry, J.; Kapona, O.; Yingst, S.; Hines, J.Z.; Minchella, P.; et al. Detection of B.1.351 SARS-CoV-2 Variant Strain—Zambia, December 2020. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 280–282. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Science Brief: Emerging SARS-CoV-2 Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html (accessed on 22 March 2021).
- European Centre for Disease Prevention and Control. SARS-CoV-2 Increased Circulation of Variants of Concern and Vaccine Rollout in the EU/EEA, 14th Update. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-covid-19-14th-update-15-feb-2021.pdf (accessed on 22 March 2021).
- Global Report Investigating Novel Coronavirus Haplotypes. B.1.35. Available online: https://cov-lineages.org/global_report_B.1.351.html (accessed on 23 March 2021).
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.-H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; Wilkinson, E.; Naidoo, Y.; et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nat. Cell Biol. 2021, 593, 142–146. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nat. Cell Biol. 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Lopez-Rincon, A.; Perez-Romero, C.A.; Tonda, A.; Mendoza-Maldonado, L.; Claassen, E.; Garssen, J.; Kraneveld, A.D. Design of Specific Primer Sets for the Detection of B.1.1.7, B.1.351 and P.1 SARS-CoV-2 Variants using Deep Learning. bioRxiv 2021. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476.e6. [Google Scholar] [CrossRef]
- Huang, S.-W.; Wang, S.-F. SARS-CoV-2 Entry Related Viral and Host Genetic Variations: Implications on COVID-19 Severity, Immune Escape, and Infectivity. Int. J. Mol. Sci. 2021, 22, 3060. [Google Scholar] [CrossRef]
- Faria, N.R.; Claro, I.M.; Candido, D.; Moyses Franco, L.A.; Andrade, P.S.; Coletti, T.M.; Silva, C.A.; Sales, F.C.; Manuli, E.R.; Aguiar, R.S. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings. J. Virol. 2021, 12. [Google Scholar] [CrossRef]
- Fujino, T.; Nomoto, H.; Kutsuna, S.; Ujiie, M.; Suzuki, T.; Sato, R.; Fujimoto, T.; Kuroda, M.; Wakita, T.; Ohmagari, N. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg. Infect. Dis. 2021, 27, 1243–1245. [Google Scholar] [CrossRef]
- Firestone, M.J.; Lorentz, A.J.; Meyer, S.; Wang, X.; Como-Sabetti, K.; Vetter, S.; Smith, K.; Holzbauer, S.; Beaudoin, A.; Garfin, J.; et al. First Identified Cases of SARS-CoV-2 Variant P.1 in the United States—Minnesota, January 2021. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 346–347. [Google Scholar] [CrossRef] [PubMed]
- Global Report Investigating Novel Coronavirus Haplotypes. P.1. Available online: https://cov-lineages.org/global_report_P.1.html (accessed on 23 March 2021).
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, eabh2644. [Google Scholar] [CrossRef]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A., Jr.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased Resistance of SARS-CoV-2 Variant P.1 to Antibody Neutralization. bioRxiv 2021. [Google Scholar] [CrossRef]
- De Souza, W.M.; Amorim, M.R.; Sesti-Costa, R.; Coimbra, L.D.; de Toledo-Teixeira, D.A.; Parise, P.L.; Barbosa, P.P.; Bispo-Dos-Santos, K.; Mofatto, L.S.; Simeoni, C.L.; et al. Levels of SARS-CoV-2 Lineage P.1 Neutralization by Antibodies Elicited after Natural Infection and Vaccination. SSRN Electron. J. 2021. [Google Scholar] [CrossRef]
- Naveca, F.; da Costa, C.; Nascimento, V.; Souza, V.; Corado, A.; Nascimento, F.; Costa, Á.; Duarte, D.; Silva, G.; Mejía, M. SARS-CoV-2 Reinfection by the New Variant of Concern (VOC) P.1 in Amazonas, Brazil. Virological. Org. Preprint. Available online: https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/5962021 (accessed on 24 March 2021).
- PANGO Lineages. Lineage B.1.526. Available online: https://cov-lineages.org/lineages/lineage_B.1.526.html (accessed on 24 March 2021).
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv 2021. [Google Scholar] [CrossRef]
- Böger, B.; Fachi, M.M.; Vilhena, R.O.; Cobre, A.F.; Tonin, F.S.; Pontarolo, R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021, 49, 21–29. [Google Scholar] [CrossRef]
- Wollschlaeger, P.; Gerlitz, N.; Todt, D.; Pfaender, S.; Bollinger, T.; Sing, A.; Dangel, A.; Ackermann, N.; Korn, K.; Ensser, A.; et al. SARS-CoV-2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a widely used commercial multiplex PCR assay. medRxiv 2021. [Google Scholar] [CrossRef]
- Artesi, M.; Bontems, S.; Göbbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.-P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58, e01598-20. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Sundararaju, S.; Manickam, C.; Mirza, F.; Al-Hail, H.; Lorenz, S.; Tang, P. A Novel Point Mutation in the N Gene of SARS-CoV-2 May Affect the Detection of the Virus by Reverse Transcription-Quantitative PCR. J. Clin. Microbiol. 2021, 59, e03278-20. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, K.; Steininger, P.; Ziegler, R.; Steinmann, J.; Korn, K.; Ensser, A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Eurosurveillance 2020, 25, 2001650. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, C.; Brancaccio, G.; Brazzale, A.R.; Lavezzo, E.; Onelia, F.; Franchin, E.; Manuto, L.; Bianca, F.; Cianci, V.; Cattelan, A.; et al. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv 2021. [Google Scholar] [CrossRef]
- The U.S. Food and Drug Administration. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests (accessed on 12 May 2021).
- Zuckerman, N.S.; Fleishon, S.; Bucris, E.; Bar-Ilan, D.; Linial, M.; Bar-Or, I.; Indenbaum, V.; Weil, M.; Mendelson, E.; Mandelboim, M.; et al. A unique SARS-CoV-2 spike protein P681H strain detected in Israel. medRxiv 2021. [Google Scholar] [CrossRef]
- Statista. Rate of COVID-19 Vaccination Worldwide as of 31 March 2021, by Country or Territory*. Available online: https://www.statista.com/statistics/1194939/rate-covid-vaccination-by-county-worldwide/ (accessed on 1 April 2021).
| Gene | Nucleotide | Amino Acid |
|---|---|---|
| ORF1ab | C3267T | T1001I |
| C5388A | A1708D | |
| T6954C | I2230T | |
| 11288–11296 deletion | SGF 3675–3677 deletion | |
| Spike | 21765–21770 deletion | HV 69–70 deletion (mutation of concern) |
| 21991–21993 deletion | Y144 deletion | |
| A23063T | N501Y (mutation of concern) | |
| C23271A | A570D | |
| C23604A | P681H (mutation of concern) | |
| C23709T | T716I | |
| T24506G | S982A | |
| G24914C | D1118H | |
| ORF8 | C27972T | Q27stop |
| G28048T | R52I | |
| A28111G | Y73C | |
| N | 28280 GAT- > CTA | D3L |
| C28977T | S235F |
| Gene | Nucleotide | Amino Acid |
|---|---|---|
| ORF1ab | C1059T | T265I |
| G5230T | K1655N | |
| C8660T | H2799Y | |
| C8964T | S2900L | |
| A10323G | K3353R | |
| G13843T | D4527Y | |
| C17999T | T5912I | |
| Spike | C21614T | L18F |
| A21801C | D80A | |
| A2206G | D215G | |
| 22286-22294 deletion | L242_244L deletion | |
| G22299T | R246I | |
| G22813T | K417N | |
| G23012A | E484K | |
| A23063T | N501Y | |
| C23664T | A701V | |
| ORF3a | G25563T | Q57H |
| C25904T | S71L | |
| E | C26456T | P71L |
| N | C28887T | T205I |
| Characteristic | B.1.1.7 | B.1.351 | P.1 |
|---|---|---|---|
| Characteristic spike mutations | HV 69/70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | L18F, D80A, D215G, L242_244L deletion, R246I, K417N, E484K, N501Y, D614G, A701V | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G H655Y, T1027I, V1176F |
| Transmissibility | Increased transmissibility (43% to 90% more transmissible than previously circulating variants). | Suggested increased transmissibility (50% more transmissible than the previously circulating variants). | Increased, 1.7–2.4-fold more transmissible than previous circulating variants |
| Severity | Possible increased risk of severity and mortality of illness. | No significant evidence of an impact on COVID-19 course severity or mortality. | No significant evidence of an impact on COVID-19 course severity or mortality. |
| Immune susceptibility | Refractory or moderately resistant to neutralization by monoclonal antibodies (mAbs). Not resistant to neutralization by convalescent plasma and vaccine sera. | Potential increased risk of reinfection. Resistant to neutralization by mAbs. Markedly resistant to neutralization by convalescent plasma and vaccine sera. | Potential increased risk of reinfection. Resistant to neutralization by mAbs. Notable loss of neutralizing activity by convalescent plasma and vaccine sera. |
| References | [32,33,34,39,42,43,44] | [45,46,47,51] | [47,60,62,63,64] |
| Name (PANGO Lineage) | Next Strain Clade | First Detected | Key Spike Mutations | Characteristic |
|---|---|---|---|---|
| B.1.526 | 20C | United States (November 2020) | T95I, D253G, L5F, S477N, E484K, D614G, A701V | Potential depletion in neutralization by convalescent and post-vaccination sera or monoclonal antibody treatments |
| B.1.525 | 20C | United States (December 2020) | H69-V70 deletion, Y144 deletion, Q52R; E484K, Q677H; D614G, F888L | Potential depletion in neutralization by convalescent and post-vaccination sera or monoclonal antibody treatments |
| B.1.427/B.1.429 | 20C/S:452R | United States (June 2020) | L452R, W152C, S13I, D614G | Decrease in neutralization using convalescent and post-vaccination sera, ~20% increased transmissibility |
| P.2 | 20J | Brazil (April 2020) | L18F, T20N, P26S, F157L, E484K, D614G, S929I, V1176F | Potential depletion in neutralization by convalescent and post-vaccination sera or monoclonal antibody treatments |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janik, E.; Niemcewicz, M.; Podogrocki, M.; Majsterek, I.; Bijak, M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens 2021, 10, 633. https://doi.org/10.3390/pathogens10060633
Janik E, Niemcewicz M, Podogrocki M, Majsterek I, Bijak M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens. 2021; 10(6):633. https://doi.org/10.3390/pathogens10060633
Chicago/Turabian StyleJanik, Edyta, Marcin Niemcewicz, Marcin Podogrocki, Ireneusz Majsterek, and Michal Bijak. 2021. "The Emerging Concern and Interest SARS-CoV-2 Variants" Pathogens 10, no. 6: 633. https://doi.org/10.3390/pathogens10060633
APA StyleJanik, E., Niemcewicz, M., Podogrocki, M., Majsterek, I., & Bijak, M. (2021). The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens, 10(6), 633. https://doi.org/10.3390/pathogens10060633






