Abstract
Previous research has found that milk is associated with a decreased risk of colorectal cancer (CRC). However, it is unclear whether the milk digestion by the enzyme lactase-phlorizin hydrolase (LPH) plays a role in CRC susceptibility. Our study aims to investigate the direct causal relationship of CRC risk with LPH levels by applying a two-sample Mendelian Randomization (MR) strategy. Genetic instruments for LPH were derived from the Fenland Study, and CRC-associated summary statistics for these instruments were extracted from the FinnGen Study, PLCO Atlas Project, and Pan-UK Biobank. Primary MR analyses focused on a cis-variant (rs4988235) for LPH levels, with results integrated via meta-analysis. MR analyses using all variants were also undertaken. This analytical approach was further extended to assess CRC subtypes (colon and rectal). Meta-analysis across the three datasets illustrated an inverse association between genetically predicted LPH levels and CRC risk (OR: 0.92 [95% CI, 0.89–0.95]). Subtype analyses revealed associations of elevated LPH levels with reduced risks for both colon (OR: 0.92 [95% CI, 0.89–0.96]) and rectal cancer (OR: 0.92 [95% CI, 0.87, 0.98]). Consistency was observed across varied analytical methods and datasets. Further exploration is warranted to unveil the underlying mechanisms and validate LPH’s potential role in CRC prevention.
1. Introduction
Colorectal cancer (CRC) is one of the most common forms of cancer in the digestive system. There are estimated to be over 1.9 million incident cases and 93,500 CRC-related deaths in 2020, making CRC the third most common cancer and the second leading cause of cancer-related death worldwide [1]. The complex etiology of CRC points to a confluence of genetic, dietary, and lifestyle determinants of risk [2,3,4].
Lactase-phlorizin hydrolase (LPH) is a pivotal enzyme in the human body that helps hydrolyze lactose, the main carbohydrate in milk, into glucose and galactose [5,6]. The reduced expression or activity of LPH, known as lactase non-persistence (LNP), leads to a clinical condition called lactose intolerance, in which milk and other dairy products cannot be properly digested. Individuals with lactose intolerance experience symptoms such as abdominal pain, bloating, diarrhea, nausea, and vomiting after consumption of milk and other dairy products [5,6]. Genetically, LPH is encoded by the lactase gene (LCT) on chromosome 2. Genetic expression of LCT has been found to be regulated by single nucleotide polymorphisms (SNPs) located on the gene MCM6, a regulatory region 14 kb upstream from the LCT gene [6,7,8]. Specifically, the SNP rs4988235 on MCM6 confers the LNP phenotype.
Diminished LPH levels or activity, leading to lactose maldigestion, are linked to decreased calcium [9] and vitamin D intake [10], along with a reduced abundance of beneficial gut bacteria, Bifidobacterium [11]. Observational studies have reported that reduced calcium [12,13] and vitamin D [14,15] intake are associated with increased CRC risk, suggesting protective roles of calcium and vitamin D in CRC development. In addition, clinical studies have shown that dietary intake of Bifidobacterium modulates gut microbiota towards CRC prevention [16]. Given LPH’s pivotal role in milk digestion and its downstream influence on crucial nutrient absorption and gut microbiota composition, it may also have a significant impact on CRC susceptibility. In addition, LPH could potentially serve as a potential candidate biomarker for CRC risk stratification or a druggable target for CRC treatment, as several other circulating proteins associated with CRC risk have been implemented for these purposes [17,18,19,20]. Yet, the specific role of LPH in the development of CRC remains unclear, highlighting the need for detailed studies exploring this potential association.
There has been no research directly studying the relationship between LPH levels and CRC risk in the medical literature. Instead, previous epidemiologic studies have investigated this relationship using LNP status, LPH-related SNPs, and dietary milk intake as proxies for LPH levels [21,22,23,24,25,26,27]. However, these studies have several limitations, including exposure misclassification, residual confounding, and reverse causality. For instance, lactase persistence/non-persistence status was often binarily defined by individual genotype. However, the negative impacts of lactose maldigestion among lactase-non-persistent individuals are actually determined by continuous residual LPH expression levels [6,28,29]. In addition, CRC patients undergoing adjuvant 5-fluorouracil chemotherapy can develop secondary lactose intolerance due to gastrointestinal damage [30,31], disrupting small intestine enzyme and transporter functions [32]. Consequently, the potential for reverse causation (i.e., CRC leading to reduced LPH levels and thus milk intake) remains plausible.
To circumvent these challenges, we utilize Mendelian Randomization (MR) analysis, an innovative method that employs genetic variants as instrumental variables (IVs) for LPH levels [33]. The random assignment of these variants during meiosis helps mitigate confounding bias and reverse causality issues, offering a robust means to explore potential causality [33,34,35]. While conventional genome-wide MR studies encompass both cis-variants (i.e., located near the gene of interest) and trans-variants (i.e., often located on different chromosomes), there is a rising trend in cis-MR studies that exclusively use cis-variants as IVs, especially in contexts where protein expression is a key consideration [36,37,38,39,40]. The appeal of cis-MR studies has grown due to their potential for drug target identification and validation [38,40]. In our study, we focus on continuous LPH levels as the exposure, selecting both cis- and trans-variants associated with LPH levels from a large-scale genome-wide association study (GWAS). We then use sets of (1) only cis-variants and (2) combined cis- and trans-variants as separate IVs in our MR analyses.
This study leverages MR to probe the potential causal influence of genetically determined elevated LPH levels on the risk of CRC and its subtypes, namely colon and rectal cancer. Utilizing publicly accessible summary-level GWAS data from three large-scale, independent cohorts of European ancestry, we seek to enhance our understanding of the genetic underpinnings of CRC and inform future preventive strategies.
2. Materials and Methods
2.1. Study Design
Our study utilized a two-sample MR approach, using genetic variants as IVs, to investigate whether there is a causal relationship between elevated LPH levels and the risk of CRC. The MR analyses rest on three fundamental assumptions: (1) the Relevance assumption establishes that the genetic IVs are associated with the exposure (e.g., LPH levels); (2) the Independence assumption states that the genetic IVs have no correlation with potential confounders; and (3) the Exclusion restriction assumption dictates that the genetic IVs could only affect the outcome of interest (e.g., CRC) via the exposure (i.e., no horizontal pleiotropy where genetic IVs can affect multiple outcomes) [41].
The schematic overview of our study design is presented in Figure 1. Our process commenced with the selection of genetic instruments for LPH levels from the GWAS Catalog [42], followed by the extraction of summary statistics of these selected genetic instruments from prior GWAS of CRC risk performed in three independent cohorts: the FinnGen Study, the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) Atlas Project, and the Pan-UK Biobank. Each cohort had prior ethical approvals, negating the need for additional approvals for this study.
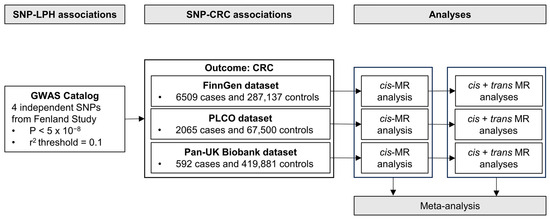
Figure 1.
Schematic overview of the study design for the primary Mendelian Randomization analyses. Abbreviations: SNP, single nucleotide polymorphism; LPH, lactase-phlorizin hydrolase; CRC, colorectal cancer; GWAS, genome-wide association study; MR, Mendelian Randomization.
To assess the causal effect of elevated LPH levels on CRC risk, we primarily conducted two-sample MR analyses in each cohort using a cis-variant for LPH levels. The results from the three cohorts were subsequently integrated using meta-analysis. For validation, MR analyses incorporating all variants (cis- + trans-) were also performed. Further, this identical workflow was used for the analysis of CRC subtypes (i.e., colon and rectal cancer). Our study followed the Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization (STROBE-MR) reporting guidelines [43].
2.2. Genetic Instruments
Genetic instruments for LPH levels were retrieved from the NHGRI-EBI GWAS Catalog, a large-scale open GWAS database collaboratively developed by the European Bioinformatics Institute (EBI) and the Human Genome Research Institute (NHGRI) with over 24,000 traits (www.ebi.ac.uk/gwas, accessed on 2 May 2023) [42]. Our focus was on the Fenland Study data from the GWAS Catalog, which offers the largest and most recent GWAS of LPH levels (GWAS Catalog accession ID: GCST90248315). Summaries of the study are listed in Table 1. The Fenland Study consisted of 10,708 genotyped participants of European ancestry who were recruited from general practice surgeries in the Cambridgeshire region of the UK from 2005 to 2015 [44]. Genotyping was conducted using three different arrays (Affymetrix UK Biobank Axiom array [Affymetrix, Santa Clara, CA, USA], Illumina Infinium Core Exome 24v1 [Illumina, San Diego, CA, USA], and Affymetrix SNP5.0 [Affymetrix, Santa Clara, CA, USA]), and levels for each protein target were measured using the rank-based inverse normal-transformed aptamer abundance method [44]. GWAS analysis was then performed using the transformed protein levels, with the residuals used as input for the genetic association analyses [45]. The beta coefficients for each protein target, representing one standard deviation (SD) change in normalized plasma abundance of protein per effect allele of the SNPs, were estimated, adjusting for age, sex, sample collection site, and the first ten principal components [44].

Table 1.
Summary of GWAS datasets used for LPH levels and CRC.
Our study selected SNPs associated with LPH levels at the genome-wide significant threshold of p < 5 × 10−8 [46]. Correlated SNPs were excluded according to measures of linkage disequilibrium (LD) r2 < 0.1 and minor allele frequency (MAF) > 0.01 based on the European populations from the 1000 Genomes phase 3 reference panel using the SNPclip online tool (https://ldlink.nih.gov/, accessed on 4 May 2023) [47].
Following exclusions, our analysis included four variants, one cis-variant (rs4988235) and three trans-variants (rs516246, rs532436, and rs641476), that were used as genetic instruments to genetically predict LPH levels. The characteristics of the genetic instruments for elevated LPH levels included in our study are presented in Table 2. Four independent SNPs associated with MCM6 (rs4988235), FUT2 (rs516246), ABO (rs532436), and GAREM1 (rs641476) were selected based on the genome-wide significance level (p < 5 × 10−8) and LD-based pruning (r2 < 0.1). Overall, the four selected SNPs accounted for 36.42% of the observed variance in elevated LPH levels, with the cis-variant rs4988235 contributing the majority of the variance.

Table 2.
Characteristics of genetic instruments for elevated LPH levels from the GWAS identified in the GWAS Catalog.
To assess the strength of the genetic instruments selected, we calculated R2 (the percent variation in LPH levels explained by the genetic instrument) and the Cragg–Donald F-statistics (the strength of the association between the genetic instrument and LPH levels) for each LPH-associated SNP using the formula: R2 = β2 × 2 × EAF × (1 − EAF) and F = R2 × (N − 2)/(1 − R2), where EAF denotes the effect allele frequency of the SNP and N represents the sample size of the exposure GWAS [48,49]. A F-statistic greater than 10 indicates strong genetic instruments for the MR analyses [50]. The F-statistics for the four SNPs ranged from 87.01 to 5340.06, underscoring their strength as genetic instruments for MR analyses.
2.3. Outcome Data Sources
A summary of the GWAS datasets for CRC is presented in Table 1. Summary-level data pertaining to the association of SNPs with CRC were obtained from three publicly available GWAS: (1) the FinnGen Study (available at https://www.finngen.fi/en/access_results, accessed on 2 May 2023); (2) the PLCO Atlas Project (available at https://exploregwas.cancer.gov/plco-atlas/#/gwas/summary, accessed on 2 May 2023); and (3) the Pan-UK Biobank (available at https://pan.ukbb.broadinstitute.org/, accessed on 2 May 2023). Detailed information for these studies was reported in the original publications [51,52,53]. CRC cases were identified by: (1) ICD-10 codes C18–C20 in the FinnGen Study; (2) ICD-O-2 codes 180, 182–189, 199, 209, 212, and 218 in the PLCO Atlas; and (3) self-report through verbal interview with a trained nurse in the Pan-UK Biobank [51,52,53]. To minimize population stratification bias, only GWAS results from individuals of European ancestry were included.
All genetic association estimates between the SNPs and CRC were calculated using logistic regression comparing cases and controls, adjusting for age, sex, and genetic principal components (the first ten in the FinnGen consortium and Pan-UK Biobank, and the first twenty in the PLCO Atlas). In addition, some studies also included study-relevant covariates in their logistic regression models, such as age2 (in the Pan-UK Biobank), study center (in the PLCO Atlas), and genotyping batch (in the FinnGen Study).
We extracted estimates (e.g., effective alleles, beta coefficients, standard errors, and p-values) for the associations between the selected genetic instruments and the risk of CRC and CRC subtypes (colon and rectal cancer) from the FinnGen, PLCO Atlas, and Pan-UK Biobank GWAS. For SNPs not available in these GWAS, we identified proxy SNPs in linkage disequilibrium (r2 > 0.7 within a ±500,000 base pairs window) based on the European populations from the 1000 Genomes phase 3 reference panel utilizing the LDProxy online tool (https://ldlink.nih.gov/, accessed on 4 May 2023) [47]. All four genetic instruments were found in the PLCO and Pan-UK Biobank datasets. Rs532436 was not available in the FinnGen dataset, and thus we used the proxy SNP rs635634, which was in high linkage disequilibrium with rs532436 (r2 = 0.99). Details of the genetic association between the SNPs and the risk of CRC are presented in Table 3.

Table 3.
Summary of four genetic instruments and their proxies (where necessary) from the FinnGen, PLCO, and Pan-UK Biobank GWAS on CRC.
2.4. Statistical Power Calculation
The statistical power of our MR analyses was calculated using an online tool (https://sb452.shinyapps.io/power/, accessed on 15 May 2023) with several parameters, including the total sample size, the percent variance in the exposure explained by the genetic instruments (R2), and the ratio of cases to controls [54]. Calculations were performed separately for each cohort. The significance level for the power calculation was set at α = 0.05. Results from the power calculation indicated that our study has an 80% power to detect a 6% change in the odds of CRC per SD increase in normalized plasma LPH levels.
2.5. Statistical Analysis
Effect alleles were defined for each SNP as the allele contributing to increased LPH levels. We performed strand alignment to harmonize the relationships between genetic instruments and CRC, as well as between LPH levels and CRC for the same allele. We primarily performed the Wald ratio two-sample cis-MR using rs4988235 as the genetic instrument. For validation, we then employed the inverse-variance weighted (IVW) two-sample MR across all four genetic instruments. The IVW method assumes that all SNPs are valid instruments and that horizontal pleiotropic effects are absent or balanced, constraining the intercepts to zero [55]. The Cochran’s Q statistic and I2 index were used to test for the presence of heterogeneity, which is an indicator of whether the IVW estimates on LPH levels and CRC risk are different across different genetic variants [56].
Further enhancing the robustness of our investigation, we performed a series of sensitivity MR analyses, including penalized IVW, robust IVW, penalized robust IVW, MR–Egger, weighted median, mode-based estimation, and MR–Lasso. The robust IVW method uses robust regression to downweight outliers, while the penalized IVW method improves the robustness of the estimates by penalizing the weights of genetic instruments with heterogeneous causal estimates for the outcome [57,58]. The penalized robust IVW method further provides robustness both to outliers and to data points with high leverage through robust regression [57]. The MR–Egger method allows the inclusion of horizontal pleiotropic SNPs and provides a bias-corrected exposure-outcome effect estimate, with a deviating intercept indicating mean pleiotropic effects [59]. Despite relaxing the exclusion restriction assumption, MR–Egger mandates the InSIDE (Instrument Strength Independent of Direct Effect) assumption, which requires that the associations of the genetic instruments with the exposure and the direct effects of the genetic instruments on the outcome are independent [60]. Consequently, we also incorporated MR analyses that do not require the InSIDE assumption (e.g., weighted median and the mode-based estimation) [59,61]. To assess the distortions of the IVW estimate from any heterogeneity or horizontal pleiotropy, MR–Lasso was used to detect and remove pleiotropic outliers [62].
The effect estimates of genetically predicted LPH on CRC and its subtypes were reported as odds ratios (ORs), along with their 95% confidence intervals (CIs), per one SD increase in normalized plasma abundance of LPH. Each SNP’s association was plotted against its corresponding effect on CRC risk. To evaluate the potential influence of a single SNP on MR results, iterative leave-one-out analyses were executed [60].
All of the primary and sensitivity MR analyses were conducted separately within each of the three outcome data sources (i.e., FinnGenn, PLCO Atlas, and Pan-UK Biobank). For comparison and consolidation of effect estimates from varying data sources, we utilized meta-analysis with fixed effects models to integrate the IVW estimates across the three cohorts. The degree of heterogeneity between the IVW estimates was quantified using the I2 index and Cochran Q statistics [63].
All statistical tests were two-sided, with the level of significance predetermined at p < 0.05. We performed all analyses using R version 4.1.2 (The R Foundation for Statistical Computing) [64]. We used the “MendelianRandomization” package [65] for MR analyses and the “meta” package for meta-analyses [66].
3. Results
3.1. FinnGen Dataset
The FinnGen GWAS summary statistics on CRC consisted of 6509 CRC cases and 287,137 controls. Using only the cis-variant rs4988235 as the genetic instrument, the FinnGen dataset showed that genetically determined higher levels of LPH were associated with decreased odds of CRC (OR per SD higher normalized plasma abundance of LPH: 0.91 [95% CI, 0.88–0.95], p < 0.001) (Table S1). The IVW estimate from the MR analysis using all LPH-associated genetic variants showed similar results as the cis-MR analysis (OR: 0.92 [95% CI, 0.88–0.95], p < 0.001) (Table S1, Figure S1A). Results for sensitivity analyses were presented in Table S1 and Figures S1–S3. Little heterogeneity across SNPs was evidenced by Cochran’s Q statistics (Q = 2.5, p = 0.482), and sensitivity analyses produced consistent results. There was no evidence of horizontal pleiotropy according to the MR–Egger results (PEgger-intercept = 0.552). Based on the leave-one-out analysis (Figure S3A), the primary influence on the effect came from the SNP rs4988235 on MCM6, which is the most well-characterized SNP responsible for LPH synthesis and the only cis-variant selected in the GWAS for LPH levels [4,6].
3.2. PLCO Dataset
The PLCO GWAS dataset included 2065 CRC participants and 67,500 controls. The PLCO dataset illustrated a non-significant association between genetically determined elevated LPH levels and CRC risk in the cis-MR (OR: 0.92 [95% CI, 0.85–1.00], p = 0.063) (Table S1). Similar results were found in the MR analysis including all genetic instruments (OR: 0.94 [95% CI, 0.85–1.03], p = 0.170), whereas the confidence interval was slightly wider than that in the cis-MR (Table S1, Figure S1B). Table S1 and Figures S1–S3 show the results from the sensitivity analyses. With penalized robust IVW, the association became significant (OR: 0.94 [95% CI, 0.90–0.98], p = 0.002), indicating the presence of potential outliers. Results from the MR–Egger, weighted median, and mode-based estimation analyses did not provide strong evidence for horizontal pleiotropic effects among the SNPs (Table S1). The leave-one-out analysis plot suggested that the MR IVW estimates were largely influenced by rs4988235, which was consistent with results in the FinnGen dataset (Figure S3B).
3.3. Pan-UK Biobank Dataset
There were 592 CRC cases and 419,881 controls in the Pan-UK Biobank. The cis-MR Wald ratio did not provide evidence supporting the effect of genetically determined elevated LPH levels on CRC risk in the Pan-UK Biobank dataset (OR: 1.00 [95% CI, 0.87–1.14], p = 0.971), and this result was similar with the IVW estimate including both cis- and trans-variants (OR: 1.03 [95% CI, 0.83–1.27], p = 0.812) (Table S1, Figure S1C). In addition, the intercept for the MR–Egger analysis was not significantly different from zero (PEgger-intercept = 0.712), indicating little evidence of horizontal pleiotropic effects in the selected genetic instruments. Sensitivity analyses mirrored the IVW estimate, with the leave-one-out analysis affirming rs4988235’s substantial impact (Figure S3C).
3.4. Meta-Analysis Combining FinnGen, PLCO, and Pan-UK Biobank Results
Meta-analysis combining the cis-MR estimates from FinnGen, PLCO, and Pan-UK Biobank showed an inverse association between genetically predicted elevated LPH and CRC risk (OR: 0.92 [95% CI, 0.89–0.95], p < 0.001), with no discernible heterogeneity in the effect across the three datasets (I2 = 0%, Pcochran-Q = 0.470) (Figure 2). Similarly, the combined IVW estimate for MR studies utilizing all four genetic variants showed a sightly attenuated association (OR: 0.93 [95% CI, 0.89–0.96], p < 0.001). We did not find strong evidence indicating heterogeneity across the three datasets (I2 = 0%, Pcochran-Q = 0.554) (Figure 2).
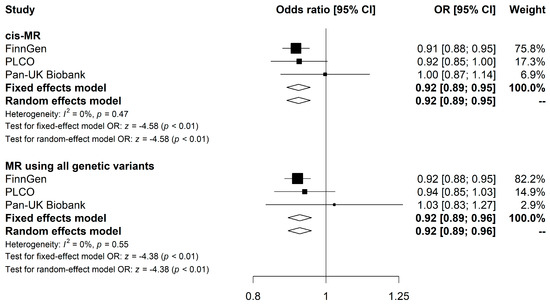
Figure 2.
Meta-analysis results for the association of LPH levels with CRC risk using MR analyses. Forest plots show results from cis-MR and MR using all genetic variants. Squares represent study-specific MR estimates. Diamonds represent meta-analyzed MR estimates using fixed and random effects models. Detailed results from the MR analyses and sensitivities analyses for each CRC GWAS study are presented in Table S1. Abbreviations: LPH, lactase-phlorizin hydrolase; CRC, colorectal cancer; MR, Mendelian Randomization.
3.5. CRC Subtype-Specific MR Analyses
CRC subtype-specific MR analyses are reported in Tables S2–S4, and sensitivity analysis results are presented in Figures S4–S6 and S8–S10. The cis-MR analysis of the FinnGen dataset revealed an inverse association between genetically predicted elevated LPH levels and colon cancer risk (OR: 0.92 [95% CI, 0.87–0.97], p = 0.001, Table S2). Although similar Wald ratio estimates were observed in the PLCO and Pan-UK Biobank datasets, statistical significance was not reached, potentially due to small sample sizes (PLCO: OR 0.93 [95% CI, 0.85–1.02], p = 0.144; Pan-UK Biobank: OR 0.95 [95% CI, 0.86–1.05], p = 0.285, Table S2). Combining the cis-MR results from the three datasets, the meta-analyzed estimate (Table S4, Figure S7) suggested a significant association between genetically predicted higher LPH levels and decreased risk of colon cancer (OR: 0.92 [95% CI, 0.89–0.96], p < 0.001). Results from the MR analyses utilizing all four genetic instruments further confirmed the association with similar estimates but wider confidence intervals (meta-analyzed OR: 0.93 [95% CI, 0.89–0.97], p < 0.001) (Tables S2 and S4, Figure S7).
With respect to rectal cancer, the FinnGen dataset indicated an inverse association between genetically predicted elevated LPH levels and rectal cancer susceptibility when using the single cis-variant rs4988235 (OR: 0.91 [95% CI, 0.85–0.97], p = 0.005, Table S3). The PLCO dataset suggested a negative but non-significant estimate (OR: 0.86 [95% CI, 0.70–1.06], p = 0.172, Table S3). Results from the Pan-UK Biobank dataset, however, demonstrated an incongruous positive, albeit non-significant, estimate (OR: 1.13 [95% CI, 0.91–1.40], p = 0.267, Table S3). The subsequent meta-analysis (Table S4, Figure S11) suggested an inverse association between elevated LPH levels and rectal cancer risk (OR: 0.92 [95% CI, 0.87, 0.98], p = 0.0083), and moderate heterogeneity was observed across the datasets (I2 = 50%, Pcochran-Q = 0.136). Further MR analyses including both cis- and trans-variants showed consistent results (Tables S3 and S4, Figure S11), indicating the robustness of our cis-MR estimates.
4. Discussion
In this study, we leveraged summary-level statistics from three large-scale GWAS of European ancestry and employed a two-sample MR framework to investigate the potential causal relationship between LPH levels and CRC risk using both cis-variants and all genetic instruments (cis- + trans-). The results from the cis-MR analysis provided genetic evidence suggesting an inverse causal association between elevated LPH levels and CRC risk. This finding was consistent and validated by MR analyses using both cis- and trans-variants. Further MR analyses by CRC subtypes indicated that this causal relationship seemed applicable to both colon cancer and rectal cancer.
While the FinnGen dataset showed a significant inverse association between genetically predicted elevated LPH levels and CRC risk, the findings from the PLCO and Pan-UK Biobank datasets were not statistically significant, likely due to insufficient statistical power attributed to smaller sample sizes and lower case-to-control ratios. We confirmed this hypothesis through power calculations, revealing 85% power in the FinnGen dataset to detect a 6% change in the odds of CRC, compared with just 39% and 15% power in the PLCO and Pan-UK Biobank datasets, respectively. Therefore, to bolster statistical power, we conducted a meta-analysis of the separate MR analyses within each of the three cohorts. Subgroup analyses for colon and rectal cancer revealed similar trends. With a relatively small number of rectal cancer cases in both the PLCO (320 cases) and Pan-UK Biobank (301 cases) datasets, these analyses were likely hindered by limited statistical power.
It is worth noting that the Pan-UK Biobank dataset showed a higher number of colon cancer cases compared to overall CRC cases. This discrepancy might be explained by the case identification method in the Pan-UK Biobank, which is reliant on self-reported cancer diagnoses and therefore subject to potential measurement error. Although more accurate cancer case ascertainment methods might be employed in individual-level UK Biobank datasets, such information was not available in the publicly accessible summary statistic data that we utilized.
The potential underlying biological mechanisms linking elevated LPH levels with reduced CRC risk warrant further exploration. Evidence suggests that lactase persistent individuals typically consume more milk than their lactase-non-persistent counterparts [67,68]. Given the known impact of milk consumption on CRC risk [26,27,69,70,71,72,73,74], it is plausible that the protective effect of LPH on CRC risk is partially mediated through catalyzed products of milk [75,76,77] and key milk components, namely calcium [12,13,78] and vitamin D [14,79]. Other milk-derived compounds, such as butyric acid, conjugated linoleic acid, sphingolipids, and lactoferrin [80,81,82], also contribute to the protective effect of LPH. Moreover, the effect of LPH on gut microbiota diversity could also play a role in modifying CRC risk [11].
Calcium and vitamin D, abundant components of milk, have been recognized for their multifaceted roles in CRC prevention. Calcium’s protective effects can be attributed to its capacity to bind secondary bile acids and ionized fatty acids, thereby reducing their toxicity on colonocytes and inhibiting mucosal proliferation [6]. In addition, it may activate certain signaling pathways via the calcium-sensing receptor (CaSR), including E-cadherin expression promotion, beta-catenin/T cell factor activation suppression, and p38 mitogen-activated protein kinase cascade activation [78]. There is also evidence linking calcium to a lower risk of mutations in the KRAS gene, a significant determinant in the carcinogenesis of CRC [6]. Vitamin D modulates molecular pathways relevant to CRC development, including the downregulation of the COX-2 gene and the upregulation of 15-hydroxyprostaglandin dehydrogenase (15-PDGH), leading to a reduction in local prostaglandin levels and hence inhibiting cancer cell survival [14]. Moreover, it interferes with β-catenin-mediated gene transcription, primarily by promoting Vitamin D receptor (VDR) binding to β-catenin, emphasizing its suppressive role on tumor growth [79].
Other milk compounds, such as butyric acid, conjugated linoleic acid, and lactoferrin, may also contribute to CRC prevention [80,81,82]. These components have shown various anti-carcinogenic effects in in vitro and animal studies, ranging from suppressing proliferation to enhancing immune function [80,81,82,83,84,85]. Additionally, LPH levels might impact CRC risk by modifying the gut microbiota. For instance, studies have linked increased LPH levels to a greater abundance of Bifidobacterium [11], which is known for augmenting antitumor immunity and facilitating the efficacy of immunotherapy [86]. In this context, our MR findings provide genetic support for this biological rationale, underscoring the relevance of LPH metabolism in CRC prevention.
While no study has directly investigated the effects of LPH on CRC risk, our findings are comparable to prior epidemiologic studies investigating CRC risk associated with LNP status or genetic instruments for milk consumption. Two studies conducted in Finnish and Hungarian populations observed a statistically significant increased risk of CRC risk among LNP individuals, with ORs reported at 1.40 and 4.04, respectively [22,24]. Although other studies conducted in British, Spanish, and Italian populations observed no association between LNP and CRC, these had limited statistical power due to small sample sizes (44-283 CRC cases) [22,25]. Furthermore, two other studies using rs4988235 as a genetic instrument for milk consumption found that genetically predicted milk intake was associated with a reduced risk of CRC (reported ORs of 0.89 and 0.95) [26,27]. This is similar to the effect size observed in our current analysis for genetically predicted LPH levels and CRC risk (OR 0.92) using the same cis-variant (rs4988235).
Our findings on the protective effect of LPH against CRC development highlight its potential role in CRC prevention and treatment. Specifically, LNP individuals identified through screening methods, such as lactose breath tests or genetic testing of the rs4988235 polymorphism, could benefit from specific dietary recommendations (e.g., calcium or vitamin D supplements) to mitigate CRC risk. Such targeted interventions could not only enhance individual health outcomes, but also contribute to more personalized and potentially cost-effective approaches to CRC risk management. Furthermore, LPH can perhaps serve as a novel therapeutic target for CRC, providing potential avenues for CRC treatment strategies.
Our study has several notable strengths. We implemented a cis-MR approach as our primary analysis, which not only mitigates biases such as residual confounding and reverse causation that typically complicate observational studies, but also minimizes potential horizontal pleiotropy. The use of the cis-variant (rs4988235), located within the MCM6 gene and in close proximity of the LPH-encoded gene LCT, ensures that the observed effects on CRC can be attributed solely to variations in LPH expression, given the regulatory role of rs4988235 [6,7,8]. This study’s findings suggest the potential therapeutic role of LPH for CRC, underscoring its clinical significance. Furthermore, the utilization of all genetic variants (cis- + trans-) served as a validation of the cis-MR approach and allowed for a series of sensitivity analyses. These included various MR methods, such as weighted median, mode-based estimation, and MR–Egger, which helped to examine the potential effects of horizontal pleiotropy from selected genetic instruments. Previous studies may have also been subject to several limitations, such as binary definitions of lactase persistence status and potential violations of the relevance assumption of MR [21,22,23,24,25,26,27]. Our study addressed these issues by using genetically predicted continuous LPH levels as the exposure and selecting genetic instruments directly associated with LPH levels from large-scale GWAS datasets. By our calculations, the SNPs selected in our study explained 36.43% of the variance in LPH levels, with rs4988235 displaying a strong association with LPH levels (variance explained: 33.28%). In addition, by using distinct GWAS datasets for LPH levels (exposure) and CRC (outcome) in our two sample MR analyses, we also reduced the potential inflation of bias associated with weak instrument variables [87]. Furthermore, we accounted for heterogeneity introduced by specific SNPs with outlier causal estimates by employing penalized IVW and MR–Lasso estimations. The application of leave-one-out analyses also helped us verify the consistency of estimates across genetic instruments and determine whether specific SNPs substantially influenced our causal estimates. We further integrated three large-scale, independent GWAS datasets into our MR analyses and meta-analyses, ensuring sufficient sample sizes for the outcome. Lastly, by conducting MR analyses across different CRC subtypes, we offered a comprehensive view of LPH’s potential biological role in various tumor locations.
However, our study has some limitations. We acknowledge that the limited number of CRC cases in the Pan-UK Biobank, and especially the smaller number of rectal cancer cases across all three cohorts, could have constrained our study’s statistical power. To mitigate this limitation, we employed meta-analysis techniques, maximizing data utilization to yield more robust results and inferences. A further limitation of our study lies in our exclusive inclusion of individuals of European descent. It is worth noting that the prevalence of lactase non-persistence significantly varies across populations; it is highest in East Asians (for example, 85% in Chinese and 100% in South Koreans) and lowest in individuals of Northern European descent (for instance, 8% in Finns and 7.8% in Swedes) [8]. Consequently, these variations in LPH levels among different populations restrict the generalizability of our findings to individuals of European ancestry. Future research should include other populations and delve into sex-specific causal estimates for a more nuanced understanding of LPH and CRC.
This study, to our knowledge, is the first to explore the causal relationships between LPH levels and the risk of CRC using MR analyses with large-scale GWAS datasets. The findings underscore the importance of LPH and its downstream effects in influencing CRC risk. Moreover, it may provide new insights into preventive strategies and a potential drug target for interventions aimed at reducing the burden of CRC. Further studies are necessary to better delineate these mechanisms and validate the potential of LPH as a biomarker for CRC risk.
5. Conclusions
Our study suggests that there is an inverse causal relationship between LPH levels and CRC risk. These findings, consistent across cohorts for both colon and rectal cancers, highlight a potential causal role for LPH as a preventative biomarker. Further study is needed to clarify the mechanisms and extend these findings to other populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu16060808/s1. Table S1. Associations of genetically predicted elevated LPH Levels and CRC in the FinnGen, PLCO, and Pan-UK Biobank datasets; Table S2. Associations of genetically predicted elevated LPH levels and colon cancer in the FinnGen, PLCO, and Pan-UK Biobank datasets; Table S3. Associations of genetically predicted elevated LPH levels and rectal cancer in the FinnGen, PLCO, and Pan-UK Biobank datasets; Table S4. Meta-analysis results for the association between elevated LPH levels and CRC, colon cancer, and rectal cancer; Figure S1. Scatter plots of the IVW and MR–Egger methods investigating the effect of elevated LPH levels on CRC in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S2. Forest plots of the IVW estimates on the association between genetically predicted LPH levels and CRC risk for each genetic instrument in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S3. Leave-one-out analyses for the MR analysis on LPH levels and CRC risk in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S4. Scatter plots of the IVW and MR–Egger methods investigating the effect of elevated LPH levels on colon cancer in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S5. Forest plots of the IVW estimates on the association between genetically predicted LPH levels and colon cancer risk for each genetic instrument in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S6. Leave-one-out analyses for the MR analysis on LPH levels and colon cancer risk in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S7. Meta-analysis results for the association of elevated LPH levels with colon cancer risk using cis-MR and MR using all genetic variants; Figure S8. Scatter plots of the IVW and MR-Egger methods investigating the effect of elevated LPH levels on rectal cancer in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S9. Forest plots of the IVW estimate on the association between genetically predicted LPH levels and rectal cancer risk for each genetic instrument in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S10. Leave-one-out analyses for the MR analysis on LPH levels and rectal cancer risk in the FinnGen, PLCO, and Pan-UK Biobank datasets; Figure S11. Meta-analysis results for the association of elevated LPH levels with rectal cancer risk using cis-MR and MR using all genetic variants.
Author Contributions
Conceptualization, S.H.; methodology, S.H., J.Y., H.Y., S.A.S. and B.Z.H.; software, S.H., J.Y. and B.Z.H.; validation, S.H., J.Y. and B.Z.H.; formal analysis, S.H., J.Y. and B.Z.H.; investigation, S.H., J.Y. and B.Z.H.; resources, S.H. and B.Z.H.; data curation, S.H., J.Y. and B.Z.H.; writing—original draft preparation, S.H.; writing—review and editing, S.H., J.Y., H.Y., S.A.S., J.R., R.A.N., Z.Z. and B.Z.H.; visualization, S.H. and B.Z.H.; supervision, Z.Z. and B.Z.H.; project administration, S.H. and B.Z.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH/NCI, grant number R00CA256525 (Brian Z. Huang); NIH/NIGMS, grant number R25GM143298 (Brian Z. Huang); and NIH/NCI, grant number T32CA229110 (Samantha A. Streicher).
Institutional Review Board Statement
Ethical review and approval were waived for this study because our analyses used publicly available GWAS summary statistics from FinnGen, PLCO, and Pan-UK Biobank. All original studies have been approved by the corresponding ethical review board. Therefore, no new ethics approval was required for this study.
Informed Consent Statement
Patient consent was waived because our analyses used publicly available GWAS summary statistics from FinnGen, PLCO, and Pan-UK Biobank. Informed consent has been obtained from the participants in all original studies. Therefore, no new informed consent was required for this study.
Data Availability Statement
Publicly available datasets were analyzed in this study. The GWAS summary statistics for LPH levels (the GWAS Catalog) are available at https://www.ebi.ac.uk/gwas/studies/GCST90248315 (accessed on 2 May 2023). The GWAS summary statistics for CRC are available at https://www.finngen.fi/en/access_results (accessed on 2 May 2023) for the FinnGen Study, https://exploregwas.cancer.gov/plco-atlas/#/ (accessed on 2 May 2023) for the PLCO Atlas, and https://pan.ukbb.broadinstitute.org/downloads/index.html (accessed on 2 May 2023) for the Pan-UK Biobank.
Acknowledgments
The authors express gratitude to all the participants from the GWAS cohorts whose contributions were invaluable to this study, as well as the investigators of the GWAS projects, including those from the Fenland Study, FinnGen Study, PLCO Atlas Project, and Pan-UK Biobank consortium, for their generosity in sharing the GWAS summary statistics.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| CRC: | Colorectal cancer |
| LPH: | Lactase-phlorizin hydrolase |
| LNP: | Lactase non-persistence |
| SNP: | Single nucleotide polymorphism |
| MR: | Mendelian Randomization |
| IV: | Instrumental variable |
| GWAS: | Genome-wide association study |
| PLCO: | the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial |
| STROBE-MR: | Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization |
| EBI: | European Bioinformatics Institute |
| NHGRI: | Human Genome Research Institute |
| SD: | Standard deviation |
| LD: | Linkage disequilibrium |
| MAF: | Minor allele frequency |
| EAF: | Effect allele frequency |
| IVW: | Inverse-variance weighted |
| InSIDE: | Instrument Strength Independent of Direct Effect |
| OR: | Odds ratio |
| CI: | Confidence interval |
| CaSR: | Calcium-sensing receptor |
| 15-PDGH: | 15-hydroxyprostaglandin dehydrogenase |
| VDR: | Vitamin D receptor |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Zhang, X.; Giovannucci, E. Calcium, vitamin D and colorectal cancer chemoprevention. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 485–494. [Google Scholar]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- de la Chapelle, A. Genetic predisposition to colorectal cancer. Nat. Rev. Cancer 2004, 4, 769–780. [Google Scholar] [CrossRef]
- Eklund, E.A.; Bode, L.; Freeze, H.H. 4.19-Diseases Associated with Carbohydrates/Glycoconjugates*. In Comprehensive Glycoscience; Kamerling, H., Ed.; Elsevier: Oxford, UK, 2007; pp. 339–371. [Google Scholar]
- Catanzaro, R.; Sciuto, M.; Marotta, F. Lactose intolerance: An update on its pathogenesis, diagnosis, and treatment. Nutr. Res. 2021, 89, 23–34. [Google Scholar] [CrossRef]
- Kuokkanen, M.; Enattah, N.S.; Oksanen, A.; Savilahti, E.; Orpana, A.; Järvelä, I. Transcriptional regulation of the lactase-phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut 2003, 52, 647–652. [Google Scholar] [CrossRef]
- Anguita-Ruiz, A.; Aguilera, C.M.; Gil, Á. Genetics of Lactose Intolerance: An Updated Review and Online Interactive World Maps of Phenotype and Genotype Frequencies. Nutrients 2020, 12, 2689. [Google Scholar] [CrossRef]
- Carroccio, A.; Montalto, G.; Cavera, G.; Notarbatolo, A. Lactose intolerance and self-reported milk intolerance: Relationship with lactose maldigestion and nutrient intake. Lactase Deficiency Study Group. J. Am. Coll. Nutr. 1998, 17, 631–636. [Google Scholar] [CrossRef]
- Dewiasty, E.; Setiati, S.; Agustina, R.; Roosheroe, A.G.; Abdullah, M.; Istanti, R.; de Groot, L.C. Prevalence of lactose intolerance and nutrients intake in an older population regarded as lactase non-persistent. Clin. Nutr. ESPEN 2021, 43, 317–321. [Google Scholar] [CrossRef]
- Kato, K.; Ishida, S.; Tanaka, M.; Mitsuyama, E.; Xiao, J.-Z.; Odamaki, T. Association between functional lactase variants and a high abundance of Bifidobacterium in the gut of healthy Japanese people. PLoS ONE 2018, 13, e0206189. [Google Scholar]
- Heine-Bröring, R.C.; Winkels, R.M.; Renkema, J.M.; Kragt, L.; van Orten-Luiten, A.C.; Tigchelaar, E.F.; Chan, D.S.; Norat, T.; Kampman, E. Dietary supplement use and colorectal cancer risk: A systematic review and meta-analyses of prospective cohort studies. Int. J. Cancer 2015, 136, 2388–2401. [Google Scholar] [CrossRef]
- Chau, R.; Dashti, S.G.; Ait Ouakrim, D.; Buchanan, D.D.; Clendenning, M.; Rosty, C.; Winship, I.M.; Young, J.P.; Giles, G.G.; Macrae, F.A.; et al. Multivitamin, calcium and folic acid supplements and the risk of colorectal cancer in Lynch syndrome. Int. J. Epidemiol. 2016, 45, 940–953. [Google Scholar] [CrossRef]
- Cruz-Pierard, S.M.; Nestares, T.; Amaro-Gahete, F.J. Vitamin D and Calcium as Key Potential Factors Related to Colorectal Cancer Prevention and Treatment: A Systematic Review. Nutrients 2022, 14, 4934. [Google Scholar] [CrossRef]
- Arayici, M.E.; Basbinar, Y.; Ellidokuz, H. Vitamin D Intake, Serum 25-Hydroxyvitamin-D (25(OH)D) Levels, and Cancer Risk: A Comprehensive Meta-Meta-Analysis Including Meta-Analyses of Randomized Controlled Trials and Observational Epidemiological Studies. Nutrients 2023, 15, 2722. [Google Scholar] [CrossRef]
- Fratila, T.D.; Ismaiel, A.; Dumitrascu, D.L. Microbiome modulation in the prevention and management of colorectal cancer: A systematic review of clinical interventions. Med. Pharm. Rep. 2023, 96, 131–145. [Google Scholar] [CrossRef]
- Tsilidis, K.K.; Branchini, C.; Guallar, E.; Helzlsouer, K.J.; Erlinger, T.P.; Platz, E.A. C-reactive protein and colorectal cancer risk: A systematic review of prospective studies. Int. J. Cancer 2008, 123, 1133–1140. [Google Scholar]
- Murphy, N.; Carreras-Torres, R.; Song, M.; Chan, A.T.; Martin, R.M.; Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Bradbury, K.E. Circulating levels of insulin-like growth factor 1 and insulin-like growth factor binding protein 3 associate with risk of colorectal cancer based on serologic and mendelian randomization analyses. Gastroenterology 2020, 158, 1300–1312.e1320. [Google Scholar]
- Mehta, R.S.; Song, M.; Bezawada, N.; Wu, K.; Garcia-Albeniz, X.; Morikawa, T.; Fuchs, C.S.; Ogino, S.; Giovannucci, E.L.; Chan, A.T. A prospective study of macrophage inhibitory cytokine-1 (MIC-1/GDF15) and risk of colorectal cancer. J. Natl. Cancer Inst. 2014, 106, dju016. [Google Scholar]
- Kakourou, A.; Koutsioumpa, C.; Lopez, D.S.; Hoffman-Bolton, J.; Bradwin, G.; Rifai, N.; Helzlsouer, K.J.; Platz, E.A.; Tsilidis, K.K. Interleukin-6 and risk of colorectal cancer: Results from the CLUE II cohort and a meta-analysis of prospective studies. Cancer Causes Control. 2015, 26, 1449–1460. [Google Scholar]
- Piepoli, A.; Schirru, E.; Mastrorilli, A.; Gentile, A.; Cotugno, R.; Quitadamo, M.; Merla, A.; Congia, M.; Usai Satta, P.; Perri, F. Genotyping of the lactase-phlorizin hydrolase c/t-13910 polymorphism by means of a new rapid denaturing high-performance liquid chromatography-based assay in healthy subjects and colorectal cancer patients. J. Biomol. Screen 2007, 12, 733–739. [Google Scholar] [CrossRef]
- Rasinperä, H.; Forsblom, C.; Enattah, N.S.; Halonen, P.; Salo, K.; Victorzon, M.; Mecklin, J.P.; Järvinen, H.; Enholm, S.; Sellick, G.; et al. The C/C-13910 genotype of adult-type hypolactasia is associated with an increased risk of colorectal cancer in the Finnish population. Gut 2005, 54, 643–647. [Google Scholar] [CrossRef]
- Gençdal, G.; Salman, E.; Özütemiz, Ö.; Akarca, U.S. Association of LCT-13910 C/T Polymorphism and Colorectal Cancer. Ann. Coloproctol. 2017, 33, 169–172. [Google Scholar] [CrossRef]
- Bácsi, K.; Hitre, E.; Kósa, J.P.; Horváth, H.; Lazáry, A.; Lakatos, P.L.; Balla, B.; Budai, B.; Lakatos, P.; Speer, G. Effects of the lactase 13910 C/T and calcium-sensor receptor A986S G/T gene polymorphisms on the incidence and recurrence of colorectal cancer in Hungarian population. BMC Cancer 2008, 8, 317. [Google Scholar] [CrossRef]
- Tarabra, E.; Pazienza, P.; Borghesio, E.; Actis, G.C.; Tappero, G.; Framarin, L.; Ayoubi, M.; Castellino, F.; Leone, N.; Sansoè, G.; et al. LCT-13910C>T polymorphism-associated lactose malabsorption and risk for colorectal cancer in Italy. Dig. Liver Dis. 2010, 42, 741–743. [Google Scholar] [CrossRef]
- Larsson, S.C.; Mason, A.M.; Kar, S.; Vithayathil, M.; Carter, P.; Baron, J.A.; Michaëlsson, K.; Burgess, S. Genetically proxied milk consumption and risk of colorectal, bladder, breast, and prostate cancer: A two-sample Mendelian randomization study. BMC Med. 2020, 18, 370. [Google Scholar] [CrossRef]
- Lumsden, A.L.; Mulugeta, A.; Hyppönen, E. Milk consumption and risk of twelve cancers: A large-scale observational and Mendelian randomisation study. Clin. Nutr. 2023, 42, 1–8. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Lomer, M.C.; Gibson, P.R. Short-chain carbohydrates and functional gastrointestinal disorders. Am. J. Gastroenterol. 2013, 108, 707–717. [Google Scholar] [CrossRef]
- Misselwitz, B.; Pohl, D.; Frühauf, H.; Fried, M.; Vavricka, S.R.; Fox, M. Lactose malabsorption and intolerance: Pathogenesis, diagnosis and treatment. United Eur. Gastroenterol. J. 2013, 1, 151–159. [Google Scholar] [CrossRef]
- Holma, R.; Laatikainen, R.; Orell, H.; Joensuu, H.; Peuhkuri, K.; Poussa, T.; Korpela, R.; Österlund, P. Consumption of Lactose, Other FODMAPs and Diarrhoea during Adjuvant 5-Fluorouracil Chemotherapy for Colorectal Cancer. Nutrients 2020, 12, 407. [Google Scholar]
- Österlund, P.; Ruotsalainen, T.; Peuhkuri, K.; Korpela, R.; Ollus, A.; Ikonen, M.; Joensuu, H.; Elomaa, I. Lactose intolerance associated with adjuvant 5-fluorouracil-based chemotherapy for colorectal cancer. Clin. Gastroenterol. Hepatol. 2004, 2, 696–703. [Google Scholar] [CrossRef]
- Wright, E.M.; Martín, M.N.G.; Turk, E. Intestinal absorption in health and disease—Sugars. Best Pract. Res. Clin. Gastroenterol. 2003, 17, 943–956. [Google Scholar]
- Grover, S.; Del Greco, M.F.; Stein, C.M.; Ziegler, A. Mendelian Randomization. Methods Mol. Biol. 2017, 1666, 581–628. [Google Scholar] [CrossRef]
- Smith, G.D.; Timpson, N.; Ebrahim, S. Strengthening causal inference in cardiovascular epidemiology through Mendelian randomization. Ann. Med. 2008, 40, 524–541. [Google Scholar] [CrossRef]
- Hingorani, A.; Humphries, S. Nature’s randomised trials. Lancet 2005, 366, 1906–1908. [Google Scholar] [CrossRef]
- Gill, D.; Georgakis, M.K.; Laffan, M.; Sabater-Lleal, M.; Malik, R.; Tzoulaki, I.; Veltkamp, R.; Dehghan, A. Genetically determined FXI (factor XI) levels and risk of stroke. Stroke 2018, 49, 2761–2763. [Google Scholar]
- Gill, D.; Burgess, S. Use of a genetic variant related to circulating FXa (Activated Factor X) levels to proxy the effect of FXa inhibition on cardiovascular outcomes. Circ. Genom. Precis. Med. 2020, 13, 551–553. [Google Scholar]
- Gill, D.; Georgakis, M.K.; Walker, V.M.; Schmidt, A.F.; Gkatzionis, A.; Freitag, D.F.; Finan, C.; Hingorani, A.D.; Howson, J.M.; Burgess, S. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021, 6, 16. [Google Scholar]
- Schmidt, A.F.; Finan, C.; Gordillo-Marañón, M.; Asselbergs, F.W.; Freitag, D.F.; Patel, R.S.; Tyl, B.; Chopade, S.; Faraway, R.; Zwierzyna, M. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 2020, 11, 3255. [Google Scholar]
- Gkatzionis, A.; Burgess, S.; Newcombe, P.J. Statistical methods for cis-Mendelian randomization with two-sample summary-level data. Genet. Epidemiol. 2023, 47, 3–25. [Google Scholar]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian randomization. JAMA 2017, 318, 1925–1926. [Google Scholar]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Güneş, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ 2021, 375, n2233. [Google Scholar] [CrossRef]
- Pietzner, M.; Wheeler, E.; Carrasco-Zanini, J.; Cortes, A.; Koprulu, M.; Wörheide, M.A.; Oerton, E.; Cook, J.; Stewart, I.D.; Kerrison, N.D.; et al. Mapping the proteo-genomic convergence of human diseases. Science 2021, 374, eabj1541. [Google Scholar] [CrossRef]
- Pietzner, M.; Wheeler, E.; Carrasco-Zanini, J.; Raffler, J.; Kerrison, N.D.; Oerton, E.; Auyeung, V.P.W.; Luan, J.; Finan, C.; Casas, J.P.; et al. Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nat. Commun. 2020, 11, 6397. [Google Scholar] [CrossRef]
- Chen, Z.; Boehnke, M.; Wen, X.; Mukherjee, B. Revisiting the genome-wide significance threshold for common variant GWAS. G3 2021, 11, jkaa056. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Luo, J.; Thomassen, J.Q.; Bellenguez, C.; Grenier-Boley, B.; De Rojas, I.; Castillo, A.; Parveen, K.; Küçükali, F.; Nicolas, A.; Peters, O. Genetic Associations Between Modifiable Risk Factors and Alzheimer Disease. JAMA Netw. Open 2023, 6, e2313734. [Google Scholar]
- Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Martin, R.M.; Lewis, S.J.; Kazmi, N.; Robinson, T.M.; Albanes, D.; Aleksandrova, K.; et al. Physical activity and risks of breast and colorectal cancer: A Mendelian randomisation analysis. Nat. Commun. 2020, 11, 597. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Use of allele scores as instrumental variables for Mendelian randomization. Int. J. Epidemiol. 2013, 42, 1134–1144. [Google Scholar] [CrossRef]
- Machiela, M.J.; Huang, W.-Y.; Wong, W.; Berndt, S.I.; Sampson, J.; De Almeida, J.; Abubakar, M.; Hislop, J.; Chen, K.-L.; Dagnall, C.; et al. GWAS Explorer: An open-source tool to explore, visualize, and access GWAS summary statistics in the PLCO Atlas. Sci. Data 2023, 10, 25. [Google Scholar] [CrossRef]
- Kurki, M.I.; Karjalainen, J.; Palta, P.; Sipilä, T.P.; Kristiansson, K.; Donner, K.M.; Reeve, M.P.; Laivuori, H.; Aavikko, M.; Kaunisto, M.A.; et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023, 613, 508–518. [Google Scholar] [CrossRef]
- Pan-UKB Team. Available online: https://pan.ukbb.broadinstitute.org (accessed on 2 May 2023).
- Burgess, S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int. J. Epidemiol. 2014, 43, 922–929. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar]
- Burgess, S.; Bowden, J.; Dudbridge, F.; Thompson, S.G. Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv 2016, arXiv:1606.03729. [Google Scholar]
- Slob, E.A.W.; Burgess, S. A comparison of robust Mendelian randomization methods using summary data. Genet. Epidemiol. 2020, 44, 313–329. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar]
- Hartwig, F.P.; Davey Smith, G.; Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 2017, 46, 1985–1998. [Google Scholar] [CrossRef]
- Rees, J.M.B.; Wood, A.M.; Dudbridge, F.; Burgess, S. Robust methods in Mendelian randomization via penalization of heterogeneous causal estimates. PLoS ONE 2019, 14, e0222362. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 6 January 2023).
- Yavorska, O.O.; Burgess, S. MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017, 46, 1734–1739. [Google Scholar]
- Balduzzi, S.; Rücker, G.; Schwarzer, G. How to perform a meta-analysis with R: A practical tutorial. BMJ Ment. Health 2019, 22, 153–160. [Google Scholar]
- Vissers, L.E.T.; Sluijs, I.; van der Schouw, Y.T.; Forouhi, N.G.; Imamura, F.; Burgess, S.; Barricarte, A.; Boeing, H.; Bonet, C.; Chirlaque, M.D.; et al. Dairy Product Intake and Risk of Type 2 Diabetes in EPIC-InterAct: A Mendelian Randomization Study. Diabetes Care 2019, 42, 568–575. [Google Scholar] [CrossRef]
- Bergholdt, H.K.M.; Larsen, M.K.; Varbo, A.; Nordestgaard, B.G.; Ellervik, C. Lactase persistence, milk intake, hip fracture and bone mineral density: A study of 97 811 Danish individuals and a meta-analysis. J. Intern. Med. 2018, 284, 254–269. [Google Scholar] [CrossRef]
- Yuan, S.; Sun, J.; Lu, Y.; Xu, F.; Li, D.; Jiang, F.; Wan, Z.; Li, X.; Qin, L.Q.; Larsson, S.C. Health effects of milk consumption: Phenome-wide Mendelian randomization study. BMC Med. 2022, 20, 455. [Google Scholar] [CrossRef]
- Cho, E.; Smith-Warner, S.A.; Spiegelman, D.; Beeson, W.L.; van den Brandt, P.A.; Colditz, G.A.; Folsom, A.R.; Fraser, G.E.; Freudenheim, J.L.; Giovannucci, E.; et al. Dairy foods, calcium, and colorectal cancer: A pooled analysis of 10 cohort studies. J. Natl. Cancer Inst. 2004, 96, 1015–1022. [Google Scholar] [CrossRef]
- Aune, D.; Lau, R.; Chan, D.S.M.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dairy products and colorectal cancer risk: A systematic review and meta-analysis of cohort studies. Ann. Oncol. 2012, 23, 37–45. [Google Scholar] [CrossRef]
- Thorning, T.K.; Raben, A.; Tholstrup, T.; Soedamah-Muthu, S.S.; Givens, I.; Astrup, A. Milk and dairy products: Good or bad for human health? An assessment of the totality of scientific evidence. Food Nutr. Res. 2016, 60, 32527. [Google Scholar] [CrossRef]
- Vieira, A.R.; Abar, L.; Chan, D.S.M.; Vingeliene, S.; Polemiti, E.; Stevens, C.; Greenwood, D.; Norat, T. Foods and beverages and colorectal cancer risk: A systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR Continuous Update Project. Ann. Oncol. 2017, 28, 1788–1802. [Google Scholar] [CrossRef]
- Barrubés, L.; Babio, N.; Becerra-Tomás, N.; Rosique-Esteban, N.; Salas-Salvadó, J. Association Between Dairy Product Consumption and Colorectal Cancer Risk in Adults: A Systematic Review and Meta-Analysis of Epidemiologic Studies. Adv. Nutr. 2019, 10, S190–S211. [Google Scholar] [CrossRef]
- Kim, J.W. Lactose Intolerance and Colorectal Cancer. Ann. Coloproctol. 2017, 33, 157–158. [Google Scholar] [CrossRef]
- Campbell, B.J.; Finnie, I.A.; Hounsell, E.F.; Rhodes, J.M. Direct demonstration of increased expression of Thomsen-Friedenreich (TF) antigen in colonic adenocarcinoma and ulcerative colitis mucin and its concealment in normal mucin. J. Clin. Investig. 1995, 95, 571–576. [Google Scholar] [CrossRef]
- Evans, R.C.; Fear, S.; Ashby, D.; Hackett, A.; Williams, E.; Van Der Vliet, M.; Dunstan, F.D.; Rhodes, J.M. Diet and colorectal cancer: An investigation of the lectin/galactose hypothesis. Gastroenterology 2002, 122, 1784–1792. [Google Scholar]
- Yang, W.; Liu, L.; Masugi, Y.; Qian, Z.R.; Nishihara, R.; Keum, N.; Wu, K.; Smith-Warner, S.; Ma, Y.; Nowak, J.A.; et al. Calcium intake and risk of colorectal cancer according to expression status of calcium-sensing receptor (CASR). Gut 2018, 67, 1475–1483. [Google Scholar] [CrossRef]
- Fleet, J.C.; Desmet, M.; Johnson, R.; Li, Y. Vitamin D and cancer: A review of molecular mechanisms. Biochem. J. 2012, 441, 61–76. [Google Scholar]
- Norat, T.; Riboli, E. Dairy products and colorectal cancer. A review of possible mechanisms and epidemiological evidence. Eur. J. Clin. Nutr. 2003, 57, 1–17. [Google Scholar] [CrossRef]
- Kozu, T.; Iinuma, G.; Ohashi, Y.; Saito, Y.; Akasu, T.; Saito, D.; Alexander, D.B.; Iigo, M.; Kakizoe, T.; Tsuda, H. Effect of Orally Administered Bovine Lactoferrin on the Growth of Adenomatous Colorectal Polyps in a Randomized, Placebo-Controlled Clinical TrialEffect of bLF on Colorectal Polyps. Cancer Prev. Res. 2009, 2, 975–983. [Google Scholar]
- Pufulete, M. Intake of dairy products and risk of colorectal neoplasia. Nutr. Res. Rev. 2008, 21, 56–67. [Google Scholar]
- Abdelali, H.; Cassand, P.; Soussotte, V.; Daubeze, M.; Bouley, C.; Narbonne, J.F. Effect of dairy products on initiation of precursor lesions of colon cancer in rats. Nutr. Cancer 1995, 24, 121–132. [Google Scholar]
- Parodi, P. Conjugated linoleic acid and other anticarcinogenic agents of bovine milk fat. J. Dairy Sci. 1999, 82, 1339–1349. [Google Scholar]
- Velázquez, O.C.; Zhou, D.; Seto, R.W.; Jabbar, A.; Choi, J.; Lederer, H.M.; Rombeau, J.L. In vivo crypt surface hyperproliferation is decreased by butyrate and increased by deoxycholate in normal rat colon: Associated in vivo effects on c-fos and c-jun expression. J. Parenter. Enter. Nutr. 1996, 20, 243–250. [Google Scholar]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Man Lei, Y.; Jabri, B.; Alegre, M.-L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016, 40, 597–608. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).