Untargeted Metabolomic Profile for the Detection of Prostate Carcinoma—Preliminary Results from PARAFAC2 and PLS–DA Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Samples Collection
2.3. Sample Treatment and GC–MS Analysis
2.4. Statistical Analysis
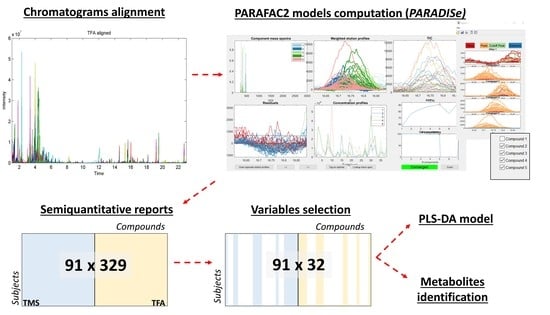
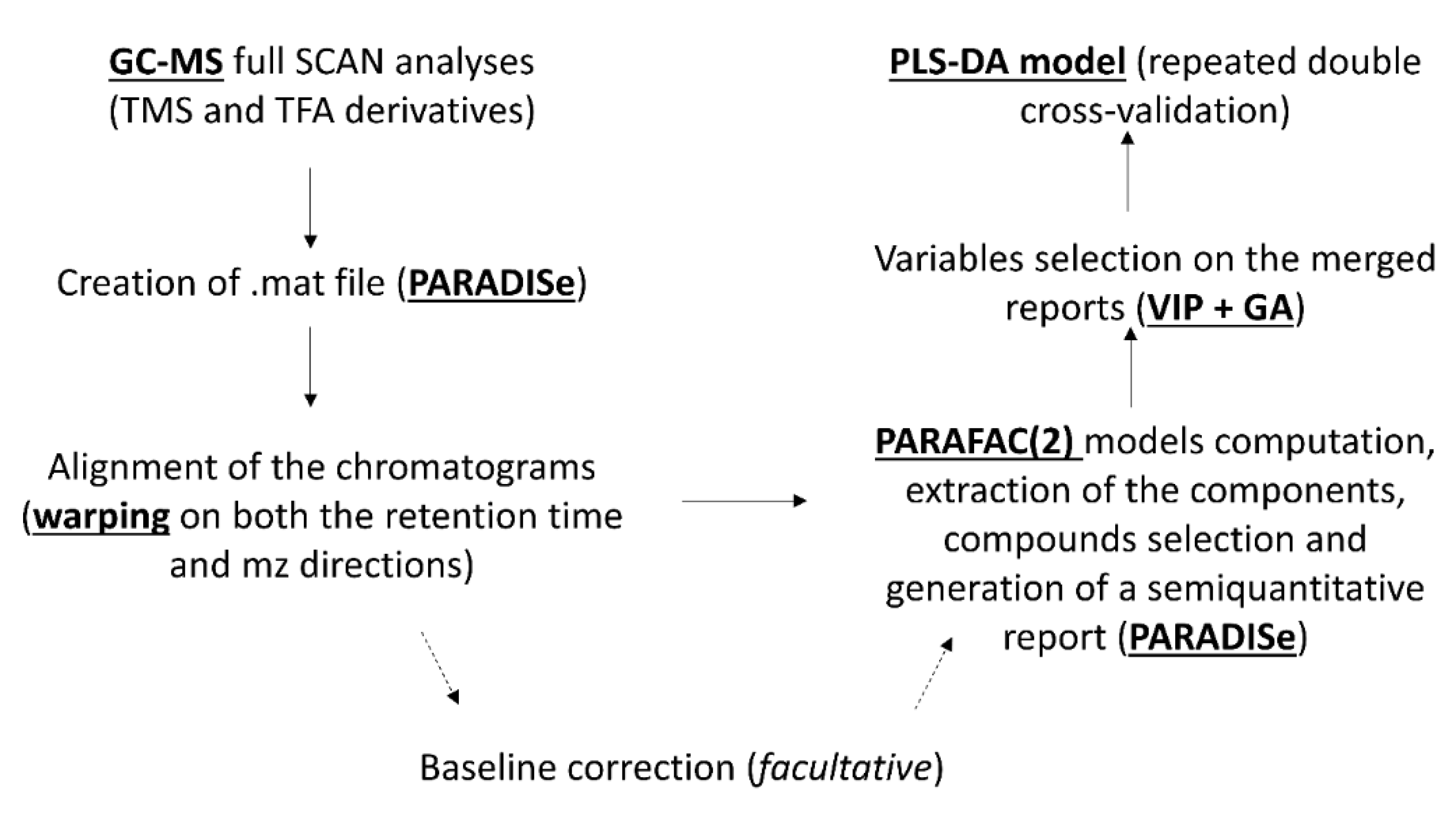
2.5. Pre-Treatment of the Raw Data
2.6. PARAFAC2 Models Computation and Molecular Identification
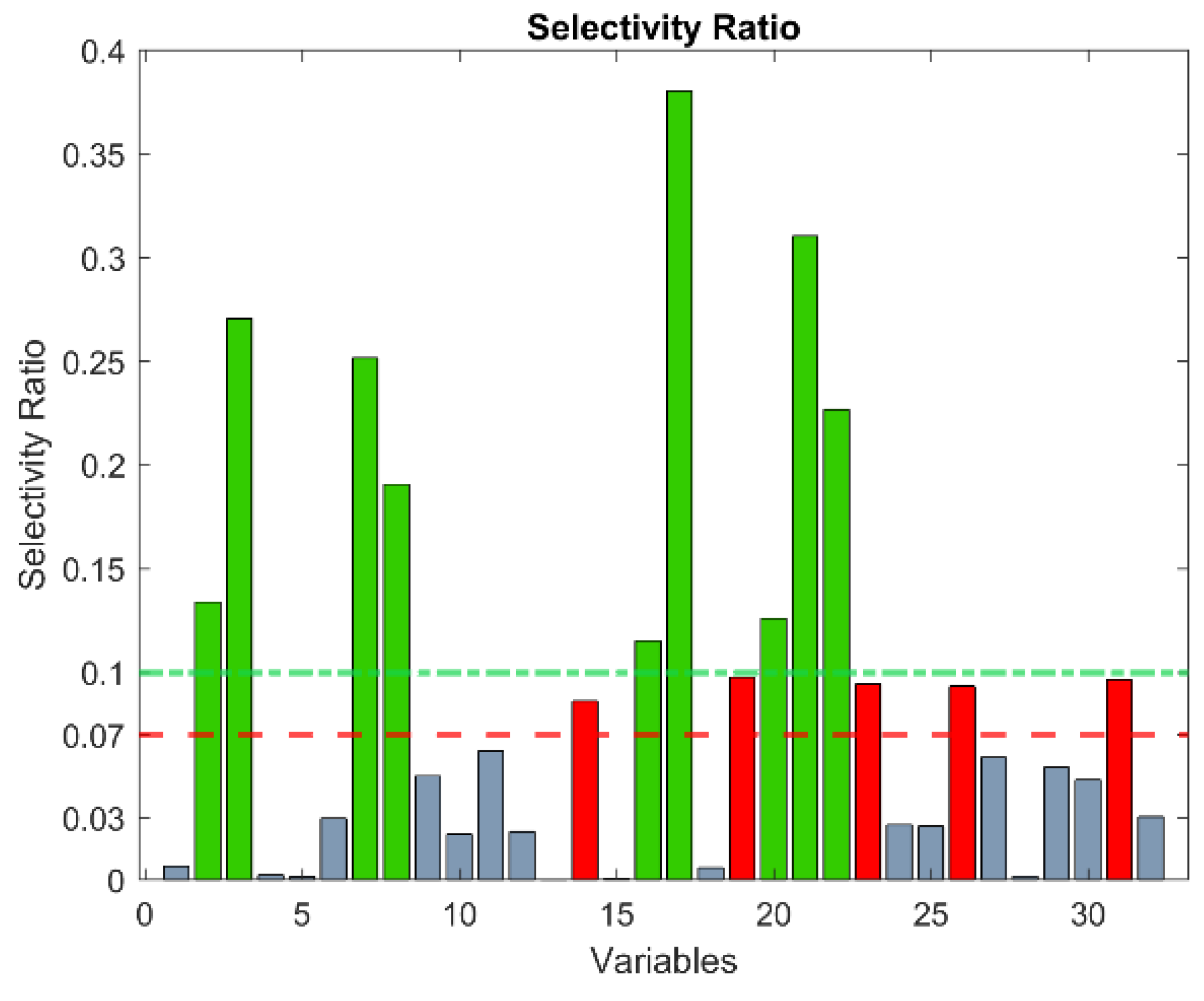
2.7. Classification Models
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Prenser, J.R.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Beyond PSA: The next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012, 4, 127rv3. [Google Scholar]
- Velonas, V.M.; Woo, H.H.; Dos Remedios, C.G.; Assinder, S.J. Current status of biomarkers for prostate cancer. Int. J. Mol. Sci. 2013, 14, 11034–11060. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; Van Oort, I.M.; Schalken, J.A. Blood-based and urinary prostate cancer biomarkers: A review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis. 2017, 20, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Etzioni, R.; Penson, D.F.; Legler, J.M.; Di Tommaso, D.; Boer, R.; Gann, P.H.; Feuer, E.J. Overdiagnosis due to prostate-specific antigen screening: Lessons from U.S. prostate cancer incidence trends. J. Natl. Cancer Inst. 2002, 94, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Sardana, G.; Dowell, B.; Diamandis, E.P. Emerging biomarkers for the diagnosis and prognosis of prostate cancer. Clin. Chem. 2008, 54, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Kramer, B.S.; Brown, M.L.; Prorok, P.C.; Potosky, A.L.; Gohagan, J.K. Prostate cancer screening: What we know and what we need to know. Ann. Intern. Med. 1993, 119, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Moyer, V.A. Screening for Prostate Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef]
- Gigerenzer, G.; Mata, J.; Frank, R. Public knowledge of benefits of breast and prostate cancer screening in Europe. J. Natl. Cancer Inst. 2009, 101, 1216–1220. [Google Scholar] [CrossRef]
- Tavoosidana, G.; Ronquist, G.; Darmanis, S.; Yan, J.; Carlsson, L.; Wu, D.; Conze, T.; Ek, P.; Semjonow, A.; Eltze, E.; et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Expert Rev. Anticancer Ther. 2011, 11, 1341–1343. [Google Scholar] [CrossRef]
- Bachrach, U. Polyamines and cancer: Minireview article. Amino Acids 2004, 26, 307–309. [Google Scholar] [CrossRef]
- Schipper, R.G.; Romijn, J.C.; Cuijpers, V.M.; Verhofstad, A.A. Polyamines and prostatic cancer. Biochem. Soc. Trans. 2003, 31, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, E.; Huang, S.P.; Audet-Walsh, E.; Lacombe, L.; Bao, B.Y.; Fradet, Y.; Laverdière, I.; Rouleau, M.; Huang, C.Y.; Yu, C.C.; et al. Molecular markers in key steroidogenic pathways, circulating steroid levels, and prostate cancer progression. Clin. Cancer Res. 2013, 19, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Gnanapragasam, V.J.; Robson, C.N.; Leung, H.Y.; E Neal, D. Androgen receptor signalling in the prostate. BJU Int. 2000, 86, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Kelloff, G.J.; Lieberman, R.; Steele, V.E.; Boone, C.W.; Lubet, R.A.; Kopelovich, L.; Malone, W.A.; Crowell, J.A.; Higley, H.R.; Sigman, C.C. Agents, biomarkers, and cohorts for chemopreventive agent development in prostate cancer. Urology 2001, 57, 46–51. [Google Scholar] [CrossRef]
- De Luca, S.; Fiori, C.; Manfredi, M.; Amante, E. Preliminary results of prospective evaluation of urinary endogenous steroid profile and prostatic carcinoma-induced deviation. J. Urol. 2019, 201, e263–e264. [Google Scholar] [CrossRef]
- Amante, E.; Alladio, E.; Salomone, A.; Vincenti, M.; Marini, F.; Alleva, G.; De Luca, S.; Porpiglia, F. Correlation between chronological and physiological age of males from their multivariate urinary endogenous steroid profile and prostatic carcinoma-induced deviation. Steroids 2018, 139, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, G.; Berg, F.V.D.; Andersson, C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemom. 2004, 18, 231–241. [Google Scholar] [CrossRef]
- Johnsen, L.G.; Skou, P.B.; Khakimov, B.; Bro, R. Gas chromatography—Mass spectrometry data processing made easy. J. Chromatogr. A 2017, 1503, 57–64. [Google Scholar] [CrossRef]
- Amigo, J.M.; Skov, T.; Bro, R.; Coello, J.; Maspoch, S. Solving GC-MS problems with PARAFAC2. TrAC Trends Anal. Chem. 2008, 27, 714–725. [Google Scholar] [CrossRef]
- Bro, R.; Andersson, C.A.; Kiers, H.A.L. PARAFAC2—Part II. Modeling chromatographic data with retention time shifts. J. Chemom. 1999, 13, 295–309. [Google Scholar] [CrossRef]
- Amigo, J.M.; Popielarz, M.J.; Callejón, R.M.; Morales, M.L.; Troncoso, A.M.; Petersen, M.A.; Toldam-Andersen, T.B.; Morales, M.L. Comprehensive analysis of chromatographic data by using PARAFAC2 and principal components analysis. J. Chromatogr. A 2010, 1217, 4422–4429. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Wold, S.; Johansson, E.; Cocchi, M. PLS: Partial Least Squares Projections to Latent Structures. In 3D QSAR in Drug Design: Theory, Methods and Applications; KLUWER ESCOM Science Publisher: Heidelberg, Germany, 1993; pp. 523–550. [Google Scholar]
- Zou, W.; Tolstikov, V.V. Probing genetic algorithms for feature selection in comprehensive metabolic profiling approach. Rapid Commun. Mass Spectrom. 2008, 22, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Filzmoser, P.; Liebmann, B.; Varmuza, K. Repeated double cross validation. J. Chemom. 2009, 23, 160–171. [Google Scholar] [CrossRef]
- Wise, B.; Gallagher, N.; Bro, R. PLS_Toolbox 8.5; Eigenvector Research, Inc.: Manson, WA, USA, 2017; Available online: http://eigenvector.com/software/pls-toolbox/ (accessed on 22 August 2019).
- Choi, M.H.; Moon, J.-Y.; Cho, S.-H.; Chung, B.C.; Lee, E.J. Metabolic alteration of urinary steroids in pre- and post-menopausal women, and men with papillary thyroid carcinoma. BMC Cancer 2011, 11, 342. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.K.M. The Biological Actions of Dehydroepiandrosterone. Drug Metab. Rev. 2006, 38, 89–116. [Google Scholar]
- Arlt, W.; Biehl, M.; Taylor, A.E.; Hahner, S.; Libé, R.; Hughes, B.A.; Schneider, P.; Smith, D.J.; Stiekema, H.; Krone, N.; et al. Urine steroid metabolomics as a biomarker tool for detecting malignancy in adrenal tumors. J. Clin. Endocrinol. Metab. 2011, 96, 3775–3784. [Google Scholar] [CrossRef] [PubMed]
- Arlt, W.; Stewart, P.M. Adrenal corticosteroid biosynthesis, metabolism, and action. Endocrinol. Metab. Clin. N. Am. 2005, 34, 293–313. [Google Scholar] [CrossRef] [PubMed]
- Lionetto, L.; Lostia, A.M.; Stigliano, A.; Cardelli, P.; Simmaco, M. HPLC-mass spectrometry method for quantitative detection of neuroendocrine tumor markers: Vanillylmandelic acid, homovanillic acid and 5-hydroxyindoleacetic acid. Clin. Chim. Acta 2008, 398, 53–56. [Google Scholar] [CrossRef]
- Stephens, F.O.; Unit, O. Phytoestrogens and prostate cancer: Possible preventive role. Med. J. Aust. 1997, 167, 138–140. [Google Scholar] [CrossRef]
- Hedelin, M.; Klint, Å.; Chang, E.T.; Bellocco, R.; Johansson, J.E.; Andersson, S.O.; Heinonen, S.M.; Adlercreutz, H.; Adami, H.O.; Grönberg, H.; et al. Dietary phytoestrogen, serum enterolactone and risk of prostate cancer: The Cancer Prostate Sweden Study (Sweden). Cancer Causes Control 2006, 17, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Rajalahti, T.; Arneberg, R.; Berven, F.S.; Myhr, K.-M.; Ulvik, R.J.; Kvalheim, O.M. Biomarker discovery in mass spectral profiles by means of selectivity ratio plot. Chemom. Intell. Lab. Syst. 2009, 95, 35–48. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |


| Compound | Derivatization | Retention Time (min) | Match with NIST | HMDB ID | KEGG ID | Expression in PCa Patients | ||
|---|---|---|---|---|---|---|---|---|
| TMS derivatives | 1 | 5-Hydroxyindoleacetic acid | TMS | 5.26 | 893 | HMDB0000763 | C05635 | overexpression |
| 2 | Unknown 1 | TMS | 5.86 | - | - | - | overexpression | |
| 3 | Unknown 2 | TMS | 7.44 | - | - | - | underexpression | |
| 4 | Androsterone | TMS | 8.14 | 912 | HMDB0000031 | C00523 | overexpression | |
| 5 | 16-Hydroxydehydroisoandrosterone | TMS | 9.23 | 888 | HMDB0000352 | C05139 | overexpression | |
| 6 | Unknown 3 | TMS | 9.84 | - | - | - | comparable | |
| 7 | Unknown 4 | TMS | 10.31 | - | - | - | underexpression | |
| 8 | Unknown 5 | TMS | 10.61 | - | - | - | underexpression | |
| 9 | Unknown 6 | TMS | 11.29 | - | - | - | underexpression | |
| 10 | Unknown 7 | TMS | 11.32 | - | - | - | comparable | |
| 11 | Enterodiol | TMS | 12.19 | 826 | HMDB0005056 | C18166 | underexpression | |
| 12 | 5β-pregnanediol | TMS | 12.53 | 853 | HMDB0005943 | Not available | underexpression | |
| 13 | Unknown 8 | TMS | 13.6 | - | - | - | overexpression | |
| 14 | Unknown 9 | TMS | 13.67 | - | - | - | comparable | |
| 15 | Pregnanetriol | TMS | 13.73 | 904 | HMDB0006070 | Not available | underexpression | |
| 16 | Unknown 10 | TMS | 14.03 | - | - | - | underexpression | |
| 17 | Unknown 11 | TMS | 14.50 | - | - | - | underexpression | |
| 18 | Unknown 12 | TMS | 14.53 | - | - | - | underexpression | |
| 19 | Unknown 13 | TMS | 14.6 | - | - | - | overexpression | |
| 20 | Unknown 14 | TMS | 14.66 | - | - | - | underexpression | |
| 21 | Unknown 15 | TMS | 15.04 | - | - | - | underexpression | |
| TFA derivatives | 22 | Unknown 16 | TFA | 1.63 | - | - | - | underexpression |
| 23 | Unknown 17 | TFA | 1.71 | - | - | - | comparable | |
| 24 | Vanillyl alcohol | TFA | 3.37 | 860 | HMDB0032012 | C06317 | overexpression | |
| 25 | Unknown 18 | TFA | 4.97 | - | - | - | comparable | |
| 26 | Unknown 19 | TFA | 5.71 | - | - | - | underexpression | |
| 27 | Unknown 20 | TFA | 3.32 | - | - | - | underexpression | |
| 28 | Epiandrosterone | TFA | 15.61 | 925 | HMDB0000365 | C07635 | comparable | |
| 29 | Unknown 21 | TFA | 16.32 | - | - | - | underexpression | |
| 30 | Unknown 22 | TFA | 17.87 | - | - | - | underexpression | |
| 31 | Unknown 23 | TFA | 18.11 | - | - | - | overexpression | |
| 32 | Unknown 24 | TFA | 18.24 | - | - | - | underexpression | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amante, E.; Salomone, A.; Alladio, E.; Vincenti, M.; Porpiglia, F.; Bro, R. Untargeted Metabolomic Profile for the Detection of Prostate Carcinoma—Preliminary Results from PARAFAC2 and PLS–DA Models. Molecules 2019, 24, 3063. https://doi.org/10.3390/molecules24173063
Amante E, Salomone A, Alladio E, Vincenti M, Porpiglia F, Bro R. Untargeted Metabolomic Profile for the Detection of Prostate Carcinoma—Preliminary Results from PARAFAC2 and PLS–DA Models. Molecules. 2019; 24(17):3063. https://doi.org/10.3390/molecules24173063
Chicago/Turabian StyleAmante, Eleonora, Alberto Salomone, Eugenio Alladio, Marco Vincenti, Francesco Porpiglia, and Rasmus Bro. 2019. "Untargeted Metabolomic Profile for the Detection of Prostate Carcinoma—Preliminary Results from PARAFAC2 and PLS–DA Models" Molecules 24, no. 17: 3063. https://doi.org/10.3390/molecules24173063
APA StyleAmante, E., Salomone, A., Alladio, E., Vincenti, M., Porpiglia, F., & Bro, R. (2019). Untargeted Metabolomic Profile for the Detection of Prostate Carcinoma—Preliminary Results from PARAFAC2 and PLS–DA Models. Molecules, 24(17), 3063. https://doi.org/10.3390/molecules24173063