Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS
Abstract
1. Introduction
2. Results and Discussion
2.1. Analytical Strategy
2.2. Mass Fragmentation Behavior of Eupatorin
2.3. Identification of Metabolites in Vivo and in Vitro
2.4. Metabolic Pathways of Eupatorin
2.5. Comparison of Metabolites in Vivo and in Vitro
2.6. Metabolite Activity of Eupatorin
3. Material and Methods
3.1. Chemicals and Materials
3.2. Instruments and Conditions
3.3. Metabolism in Vivo
3.3.1. Animals and Drug Administration
3.3.2. Bio-Sample Collection
3.3.3. Bio-Sample Pretreatment
3.4. Metabolism in Vitro by Rat Liver Microsomes
3.4.1. Phase I Metabolism
3.4.2. Phase II Metabolism
3.5. Metabolism in Vitro by Rat Intestinal Flora
3.5.1. Preparation of Anaerobic Culture Medium
3.5.2. Preparation of Intestinal Flora Culture Solution
3.5.3. Sample Preparation
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pariyani, R.; Ismail, I.S.; Azam, A.; Khatib, A.; Abas, F.; Shaari, K.; Hamza, H. Urinary metabolic profiling of cisplatin nephrotoxicity and nephroprotective effects of Orthosiphon stamineus leaves elucidated by 1H NMR spectroscopy. J. Pharm. Biomed. Anal. 2017, 135, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Rahman, S.M. Isolation and characterisation of flavonoids from the leaves of medicinal plant Orthosiphon stamineus. Arab. J. Chem. 2015, 8, 218–221. [Google Scholar] [CrossRef]
- Yuliana, N.D.; Khatib, A.; Link-Struensee, A.M.; Ijzerman, A.P.; Rungkat-Zakaria, F.; Choi, Y.H.; Verpoorte, R. Adenosine A1 receptor binding activity of methoxy flavonoids from Orthosiphon stamineus. Planta Med. 2009, 75, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-H.; Noryati, I.; Sulaiman, S.-F.; Rosma, A. In vitro antibacterial and antioxidant activities of Orthosiphon stamineus Benth. extracts against food-borne bacteria. Food Chem. 2010, 122, 1168–1172. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, Q.; Qian, K.; Feng, Y.; Luo, Y.; Tan, T. Metabolic profile of rosmarinic acid from Java tea (Orthosiphon stamineus) by ultra-high-performance liquid chromatography coupled to quadrupole-time-of-flight tandem mass spectrometry with a three-step data mining strategy. Biomed. Chromatogr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liang, X.; Xie, Y. Qualitative and quantitative analysis on the chemical constituents in Orthosiphon stamineus Benth. using ultra high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 2019, 164, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Saidan, N.H.; Aisha, A.F.A.; Hamil, M.S.R.; Majid, A.M.S.A.; Ismail, Z. A novel reverse phase high-performance liquid chromatography method for standardization of Orthosiphon stamineus leaf extracts. Pharmacogn. Res. 2015, 7, 23–31. [Google Scholar]
- Yam, M.F.; Mohamed, E.A.H.; Ang, L.F.; Pei, L.; Darwis, Y.; Mahmud, R.; Asmawi, M.Z.; Basir, R.; Ahmad, M. A simple isocratic HPLC method for the simultaneous determination of sinensetin, eupatorin, and 3′-hydroxy-5,6,7,4′-tetramethoxyflavone in Orthosiphon stamineus extracts. J. Acupunct. Meridian Stud. 2012, 5, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Akowuah, G.; Zhari, I.; Norhayati, I.; Sadikun, A.; Khamsah, S. Sinensetin, eupatorin, 3′-hydroxy-5,6,7,4′-tetramethoxyflavone and rosmarinic acid contents and antioxidative effect of Orthosiphon stamineus from Malaysia. Food Chem. 2004, 87, 559–566. [Google Scholar] [CrossRef]
- Razak, N.A.; Abu, N.; Ho, W.Y.; Zamberi, N.R.; Tan, S.W.; Alitheen, N.B.; Long, K.; Yeap, S.K. Cytotoxicity of eupatorin in MCF-7 and MDA-MB-231 human breast cancer cells via cell cycle arrest, anti-angiogenesis and induction of apoptosis. Sci. Rep. 2019, 9, 1514. [Google Scholar] [CrossRef]
- Lee, K.; Hyun Lee, D.; Jung, Y.J.; Shin, S.Y.; Lee, Y.H. The natural flavone eupatorin induces cell cycle arrest at the G2/M phase and apoptosis in HeLa cells. Appl. Biol. Chem. 2016, 59, 193–199. [Google Scholar] [CrossRef]
- Estevez, S.; Marrero, M.T.; Quintana, J.; Estevez, F. Eupatorin-induced cell death in human leukemia cells is dependent on caspases and activates the mitogen-activated protein kinase pathway. PLoS ONE 2014, 9, e112536. [Google Scholar] [CrossRef] [PubMed]
- Androutsopoulos, V.; Arroo, R.R.J.; Hall, J.F.; Surichan, S.; Potter, G.A. Antiproliferative and cytostatic effects of the natural product eupatorin on MDA-MB-468 human breast cancer cells due to CYP1-mediated metabolism. Breast Cancer Res. 2008, 10. [Google Scholar] [CrossRef] [PubMed]
- Doleckova, I.; Rarova, L.; Gruz, J.; Vondrusova, M.; Strnad, M.; Krystof, V. Antiproliferative and antiangiogenic effects of flavone eupatorin, an active constituent of chloroform extract of Orthosiphon stamineus leaves. Fitoterapia 2012, 83, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Laavola, M.; Nieminen, R.; Yam, M.F.; Sadikun, A.; Asmawi, M.Z.; Basir, R.; Welling, J.; Vapaatalo, H.; Korhonen, R.; Moilanen, E. Flavonoids eupatorin and sinensetin present in Orthosiphon stamineus leaves inhibit inflammatory gene expression and STAT1 activation. Planta Med. 2012, 78, 779–786. [Google Scholar] [CrossRef]
- Yam, M.F.; Lim, V.; Salman, I.M.; Ameer, O.Z.; Ang, L.F.; Rosidah, N.; Abdulkarim, M.F.; Abdullah, G.Z.; Basir, R.; Sadikun, A.; et al. HPLC and anti-inflammatory studies of the flavonoid rich chloroform extract fraction of Orthosiphon stamineus leaves. Molecules 2010, 15, 4452–4466. [Google Scholar] [CrossRef]
- Yam, M.F.; Tan, C.S.; Ahmad, M.; Shibao, R. Mechanism of vasorelaxation induced by eupatorin in the rats aortic ring. Eur. J. Pharmacol. 2016, 789, 27–36. [Google Scholar] [CrossRef]
- Liao, M.; Cheng, X.; Diao, X.; Sun, Y.; Zhang, L. Metabolites identificaion of two bioactive constituents in Trollius ledebourii in rats using ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. J. Chromatogr. B 2017, 1068, 297–312. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Jia, W. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin. Drug Metab. Toxicol. 2016, 12, 31–40. [Google Scholar] [CrossRef]
- Noh, K.; Kang, Y.R.; Nepal, M.R.; Shakya, R.; Kang, M.J.; Kang, W.; Lee, S.; Jeong, H.G.; Jeong, T.C. Impact of gut microbiota on drug metabolism: An update for safe and effective use of drugs. Arch. Pharm. Res. 2017, 40, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Wang, Z.J.; Li, Y.; Liu, Y.; Cai, W.; Li, C.; Lu, J.-Q.; Qiao, Y.-J. A strategy for comprehensive identification of sequential constituents using ultra-high-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometer, application study on chlorogenic acids in Flos Lonicerae Japonicae. Talanta 2016, 147, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Wang, Y.; Huo, C.; Wu, Y.; Zhang, M.; Li, L.; Shi, Q.; Kiyota, H.; Shi, X. Study on the metabolites of isoalantolactone in vivo and in vitro by ultra performance liquid chromatography combined with Triple TOF mass spectrometry. Food Chem. 2017, 214, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, X.; Li, L.; Zhang, X.; Song, K.; Diao, X.; Sun, Y.; Zhang, L. UHPLC-Q-TOF-MS/MS method based on four-step strategy for metabolites of hinokiflavone in vivo and in vitro. J. Pharm. Biomed. Anal. 2019, 169, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liang, C.; Diao, X.; Cheng, X.; Liao, M.; Zhang, L. Metabolism studies on hydroxygenkwanin and genkwanin in human liver microsomes by UHPLC-Q-TOF-MS. Xenobiotica 2018, 48, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yin, J.; Liang, C.; Sun, Y.; Zhang, L. A simple and sensitive UHPLC-Q-TOF-MS/MS method for sophoricoside metabolism study in vitro and in vivo. J. Chromatogr. B 2017, 1061, 193–208. [Google Scholar] [CrossRef]
- Ma, Y.; Xie, W.; Tian, T.; Jin, Y.; Xu, H.; Zhang, K.; Du, Y. Identification and comparative oridonin metabolism in different species liver microsomes by using UPLC-Triple-TOF-MS/MS and PCA. Anal. Biochem. 2016, 511, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, X.; Diao, X.; Liao, M.; Sun, Y.; Zhang, L. Metabolism profiling of nevadensin in vitro and in vivo by UHPLC-Q-TOF-MS/MS. J. Chromatogr. B 2018, 1084, 69–79. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, J.; Liang, C.; Sun, Y.; Zhang, L. UHPLC-Q-TOF-MS/MS Method Based on Four-Step Strategy for Metabolism Study of Fisetin in vitro and in vivo. J. Agric. Food Chem. 2017, 65, 10959–10972. [Google Scholar] [CrossRef]
- Chen, X.; Han, R.; Hao, P.; Wang, L.; Liu, M.; Jin, M.; Kong, D.; Li, X. Nepetin inhibits IL-1beta induced inflammation via NF-kappaB and MAPKs signaling pathways in ARPE-19 cells. Biomed. Pharmacother. 2018, 101, 87–93. [Google Scholar] [CrossRef]
- Clavin, M.; Gorzalczany, S.; Macho, A.; Muñoz, E.; Ferraro, G.; Acevedo, C.; Martino, V. Anti-inflammatory activity of flavonoids from Eupatorium arnottianum. J. Ethnopharmacol. 2007, 112, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Yang, W.K.; Yee, S.M.; Kim, S.M.; Park, Y.C.; Shin, H.J.; Han, C.K.; Lee, Y.C.; Kang, H.S.; Kim, S.H. KGC3P attenuates ovalbumin-induced airway inflammation through downregulation of p-PTEN in asthmatic mice. Phytomedicine 2019, 62, 152942. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Diao, X.; Cheng, X.; Sun, Y.; Zhang, L. Nontargeted SWATH acquisition mode for metabolites identification of osthole in rats using ultra-high-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. RSC Adv. 2018, 8, 14925–14935. [Google Scholar] [CrossRef]
- Diao, X.; Liao, M.; Cheng, X.; Liang, C.; Sun, Y.; Zhang, X.; Zhang, L. Identification of metabolites of Helicid in vivo using ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Biomed. Chromatogr. 2018, 32, e4263. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Jin, Y.; Hou, L.; Ma, Y.; Xu, H.; Zhang, K.; Zhang, L.; Du, Y. A practical strategy for the characterization of ponicidin metabolites in vivo and in vitro by UHPLC-Q-TOF-MS based on nontargeted SWATH data acquisition. J. Pharm. Biomed. Anal. 2017, 145, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Jin, Y.; Ma, Y.; Xie, W.; Xu, H.; Zhang, K.; Zhang, L.; Du, Y. Identification of metabolites of oridonin in rats with a single run on UPLC-Triple-TOF-MS/MS system based on multiple mass defect filter data acquisition and multiple data processing techniques. J. Chromatogr. B 2015, 1006, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, C.; Yin, J.; Sun, Y.; Zhang, L. Identification of metabolites of liquiritin in rats by UHPLC-Q-TOF-MS/MS: Metabolic profiling and pathway comparison in vitro and in vivo. RSC Adv. 2018, 8, 11813–11827. [Google Scholar] [CrossRef]
- Jia, P.; Zhang, Y.; Zhang, Q.; Sun, Y.; Yang, H.; Shi, H.; Zhang, X.; Zhang, L. Metabolism studies on prim-O-glucosylcimifugin and cimifugin in human liver microsomes by ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry. Biomed. Chromatogr. 2016, 30, 1498–1505. [Google Scholar] [CrossRef]
- Yisimayili, Z.; Guo, X.; Liu, H.; Xu, Z.; Abdulla, R.; Aisa, H.A.; Huang, C. Metabolic profiling analysis of corilagin in vivo and in vitro using high-performance liquid chromatography quadrupole time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2019, 165, 251–260. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |

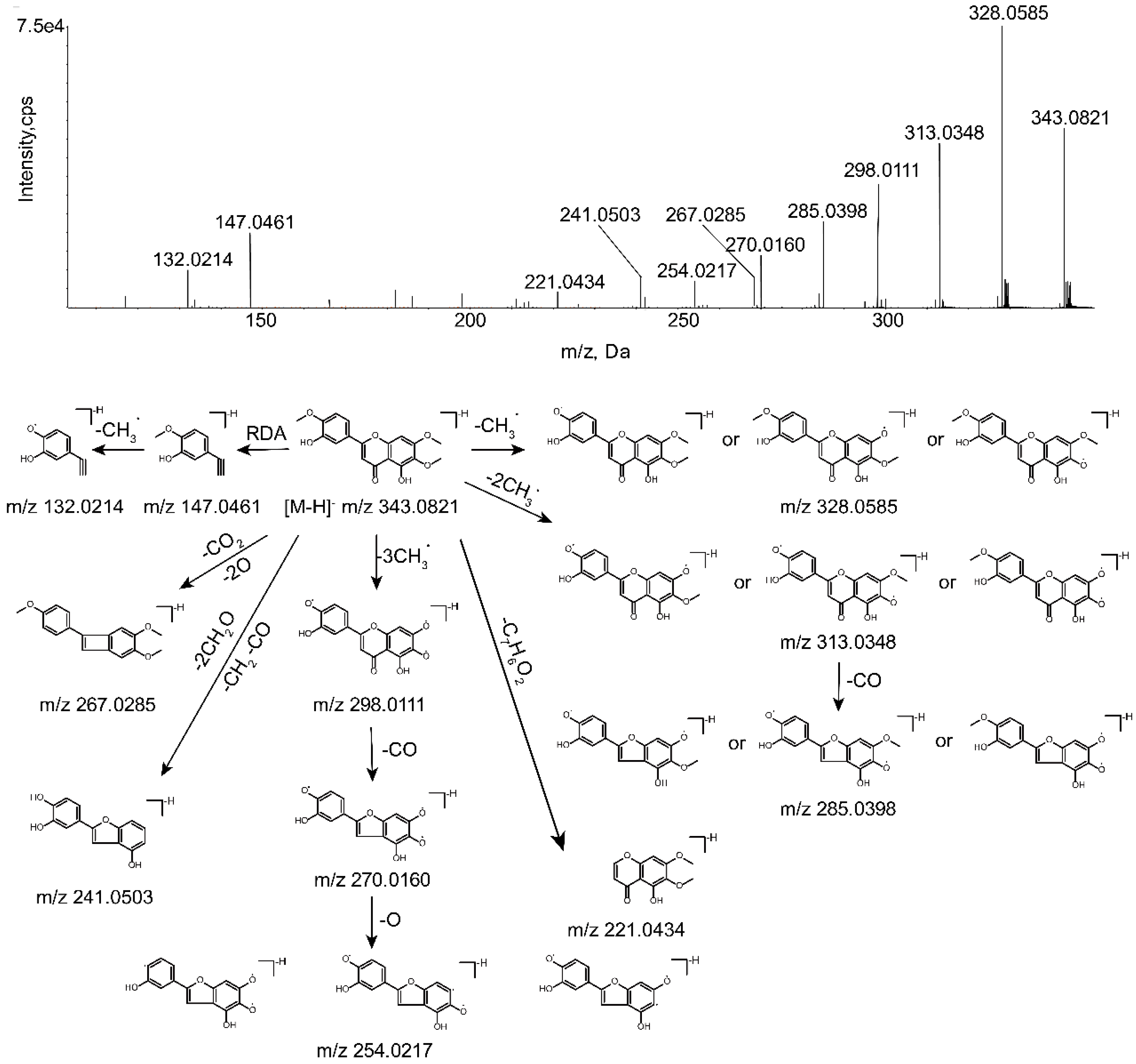
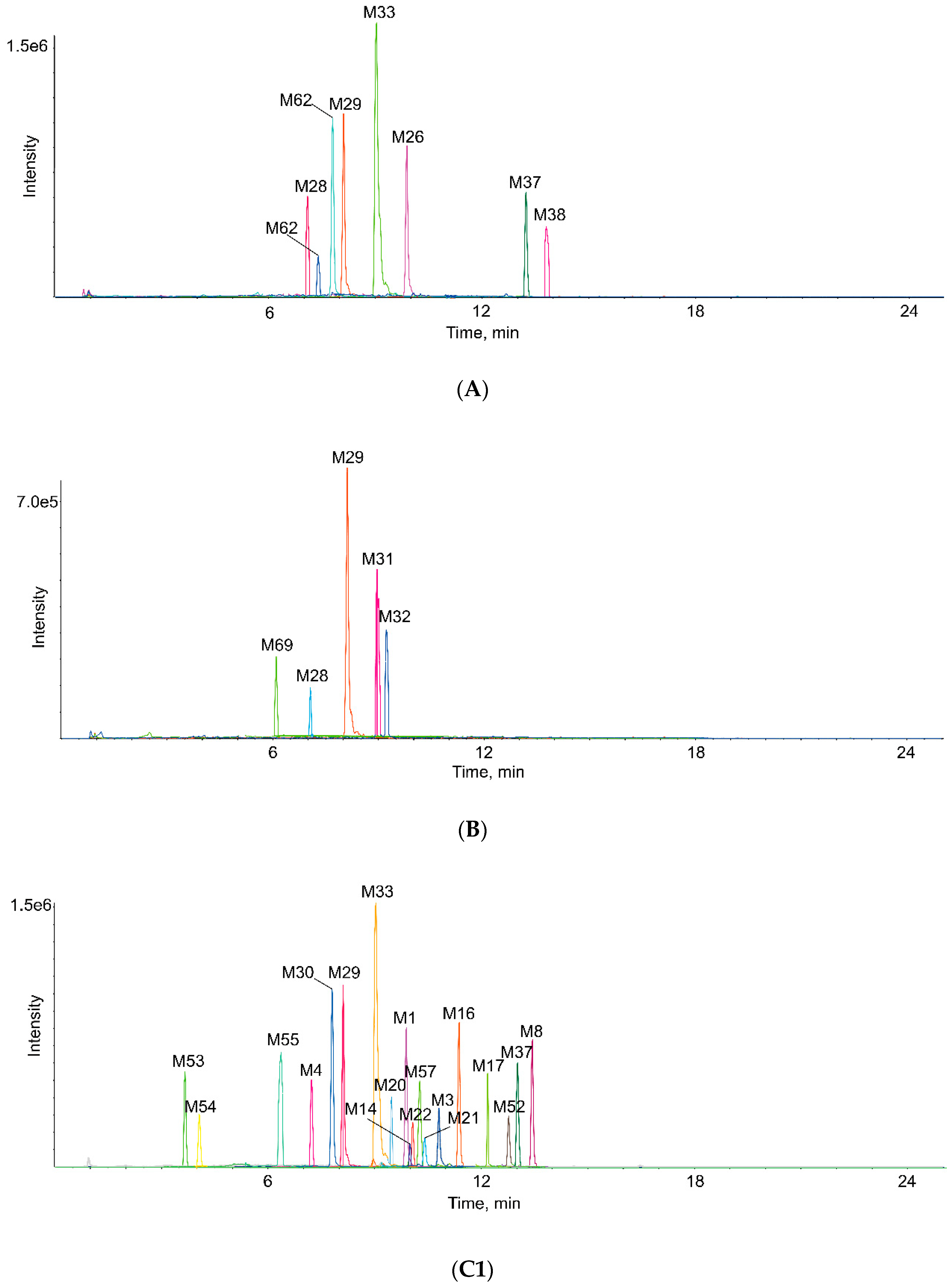
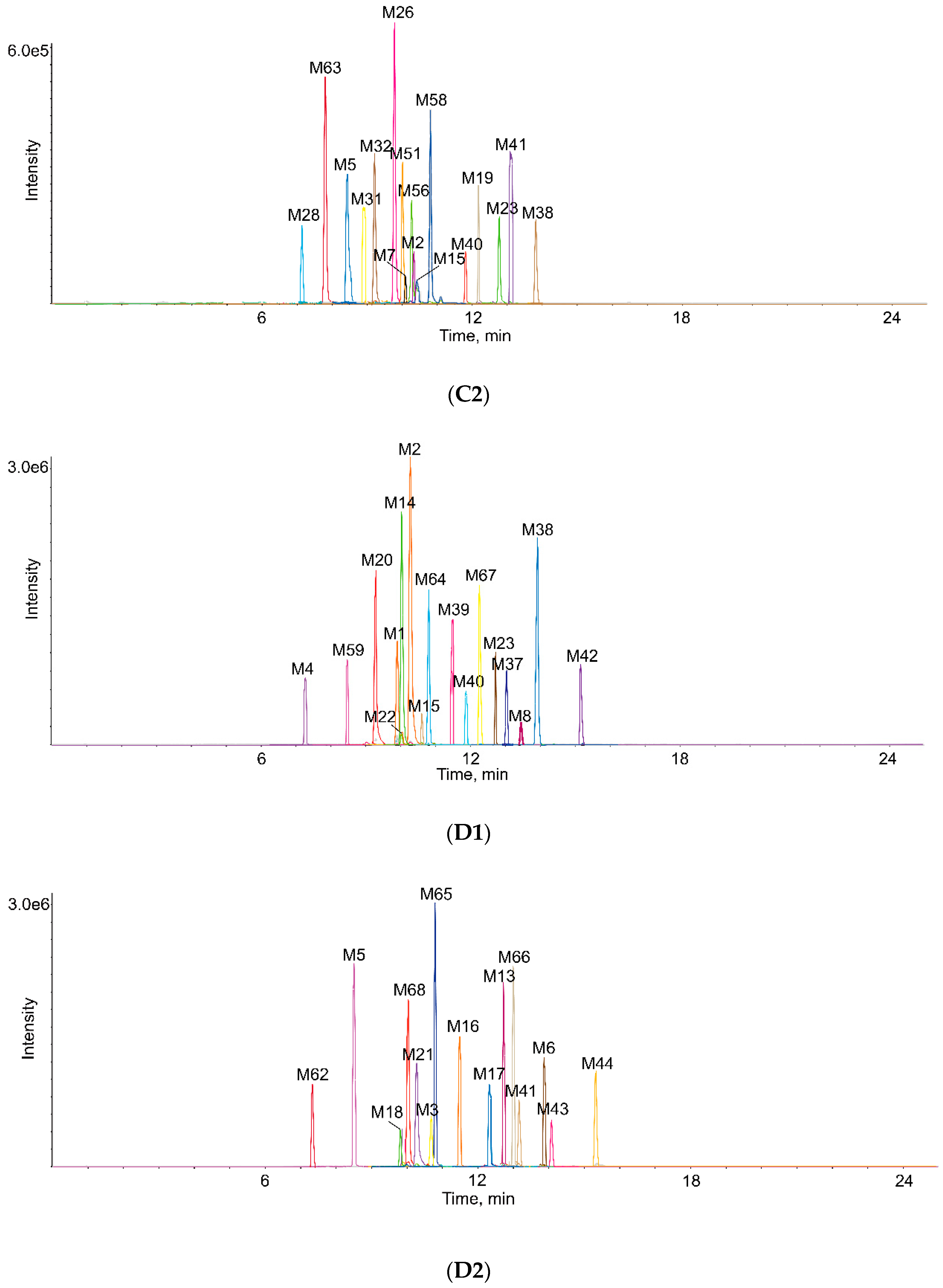
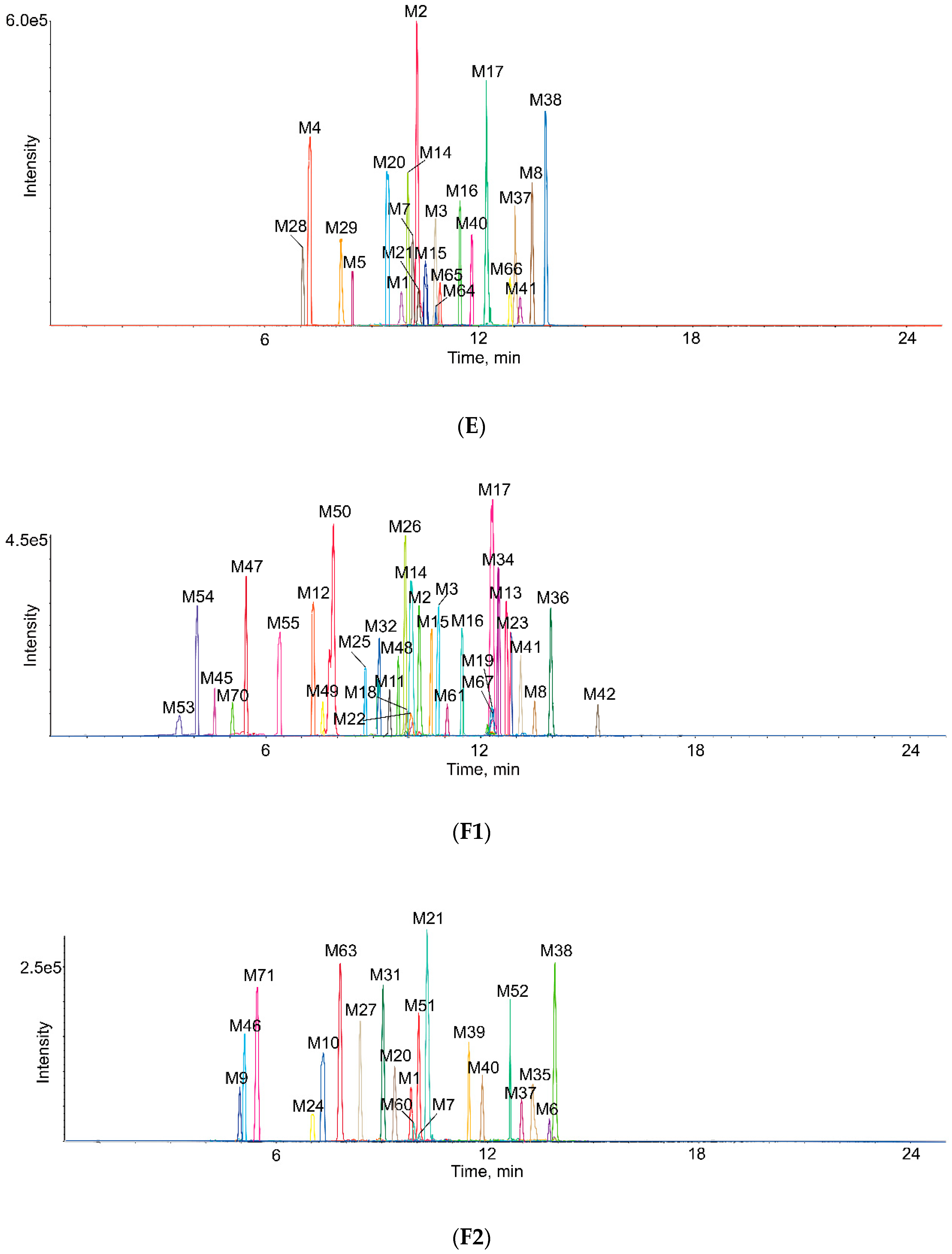
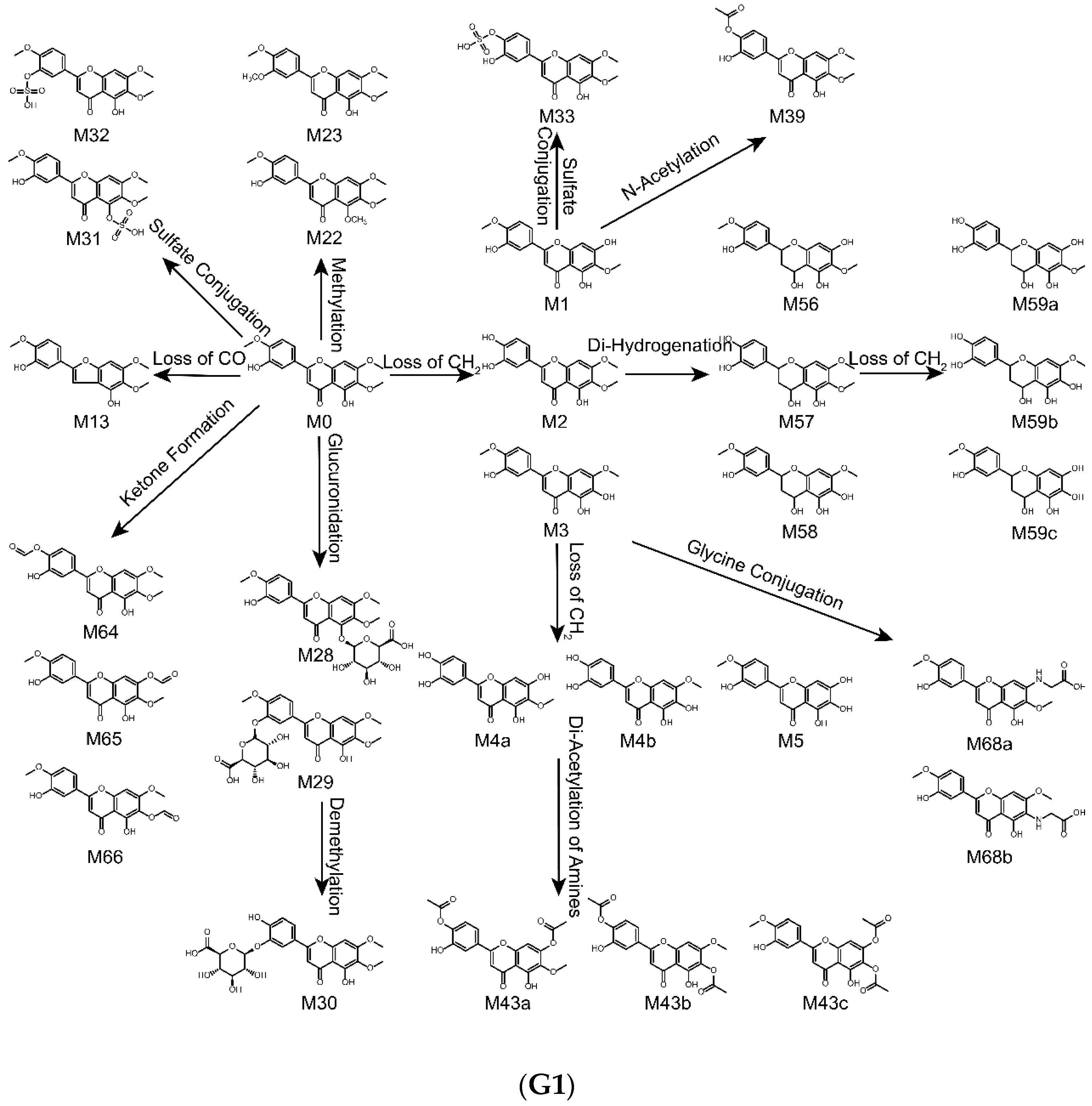
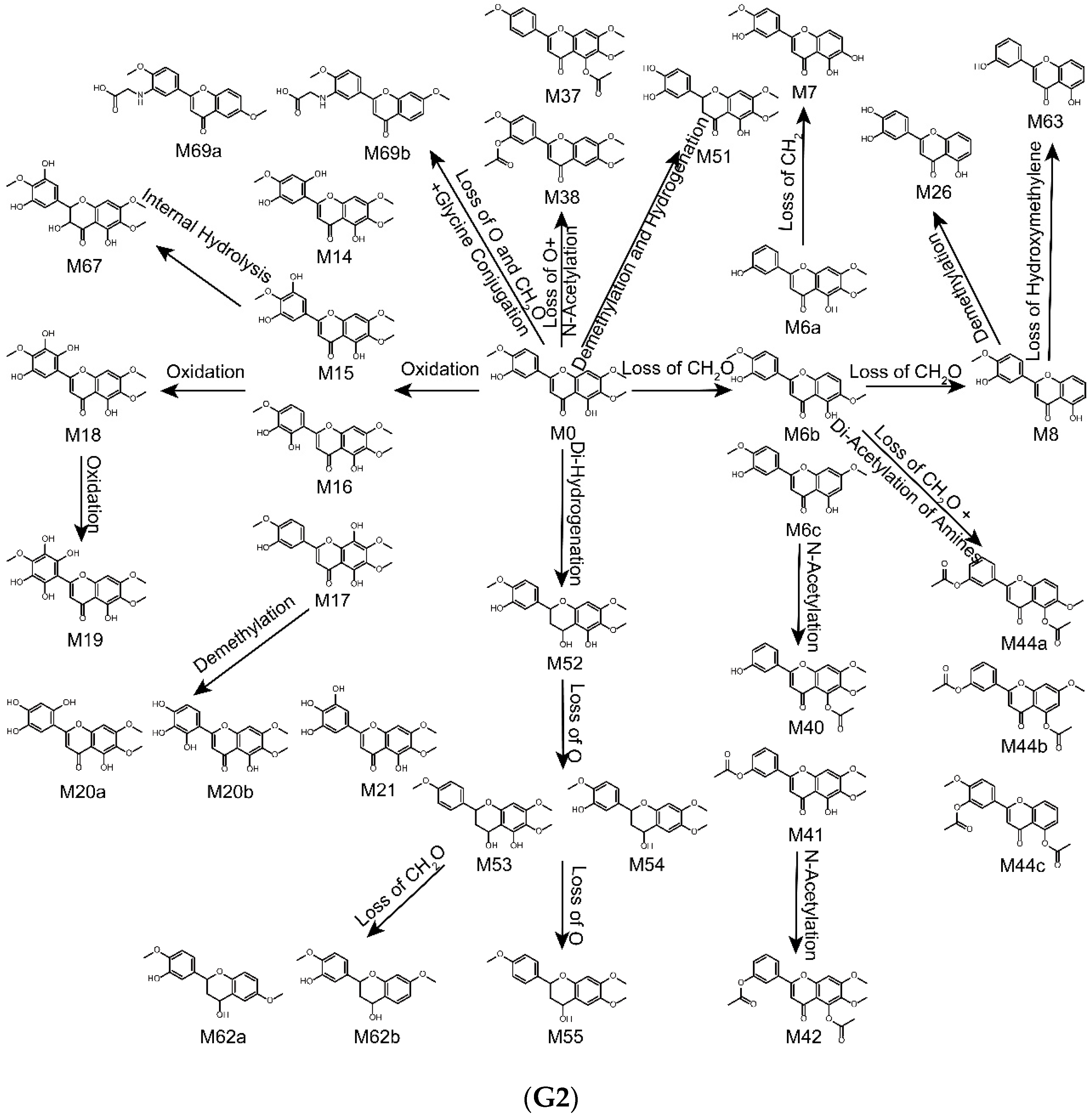


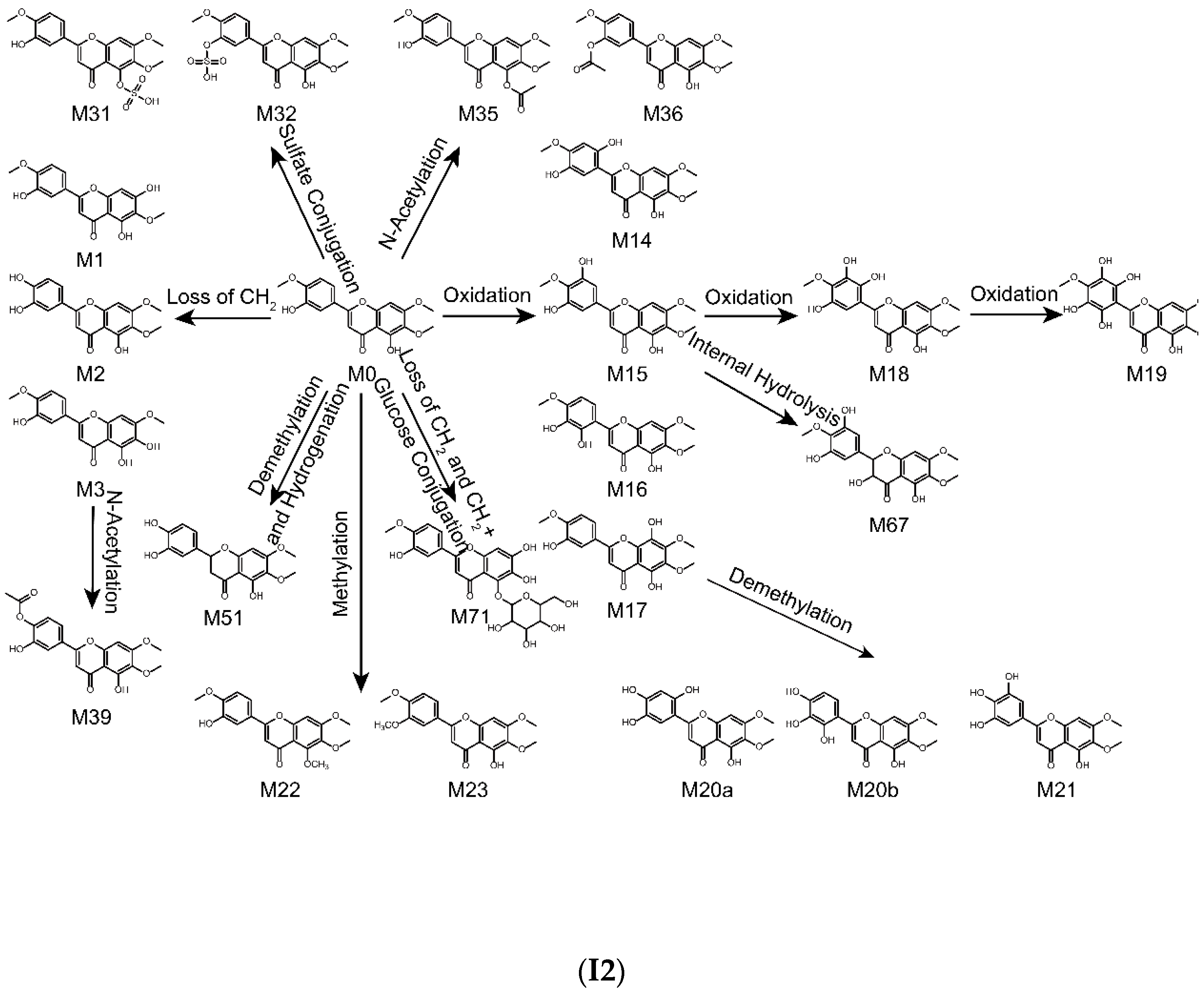
| Metabolites ID | Composition | Formula | m/z | Error (ppm) | R.T. (min) | MS/MS Fragments | Clog P | Score (%) | Plasma | Bile | Urine | Feces | RLMs | RIF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | Loss of CH2 [M-H]− | C17H14O7 | 329.0660 | −2.0 | 9.93 | 314.0427, 313.0384, 299.0188, 285.0371, 207.7129 | 2.26422 | 83.3 | - | - | +a,b | + a,b | +I,II | + a,b |
| M2 | Loss of CH2 [M-H]− | C17H14O7 | 329.0668 | 0.5 | 10.27 | 314.0423, 313.0344, 299.0189, 285.0393, 133.0287 | 2.26434 | 91.8 | - | - | + a,b | + a,b | + I,II | + a,b |
| M3 | Loss of CH2 [M-H]− | C17H14O7 | 329.0662 | −1.4 | 10.79 | 314.0436,313.0357, 299.0204, 285.0396, 207.7166 | 2.51422 | 96.3 | - | - | + a,b | + a,b | + I,II | + a,b |
| M4a | Loss of CH2 and CH2 [M-H]− | C16H12O7 | 315.0500 | −3.3 | 7.26 | 300.0279, 297.1740, 269.1760, 251.1269, 133.0270 | 1.82034 | 85.4 | - | - | + a,b | + a,b | +I | - |
| M4b | 2.07034 | |||||||||||||
| M5 | Loss of CH2 and CH2 [M-H]− | C16H12O7 | 315.0504 | −2.0 | 8.50 | 300.0275, 297.0411, 269.1755, 251.1658, 147.0821 | 2.18513 | 93.3 | - | - | + a,b | + a,b | + I | - |
| M6a | Loss of CH2O [M-H]− | C17H14O6 | 313.0713 | −1.4 | 13.86 | 298.0483, 283.0250, 221.0632, 147.0078, 117.0364 | 2.86048 | 90.9 | - | - | - | + a,b | - | + a,b |
| M6b | 3.08571 | |||||||||||||
| M6c | 3.33571 | |||||||||||||
| M7 | Loss of CH2O and CH2 [M-H]− | C16H12O6 | 299.0562 | 0.3 | 10.10 | 284.0326, 281.1787, 251.1281, 146.9687 | 2.74964 | 83.8 | - | - | + a | - | + I | + b |
| M8 | Loss of CH2O and CH2O [M-H]− | C16H12O5 | 283.0614 | 0.8 | 13.60 | 268.0379, 240.0428, 267.0306, 161.0025, 146.9655 | 3.30833 | 93.5 | - | - | + a,b | + b | + I | + b |
| M9 | Loss of O [M-H]− | C18H16O6 | 327.0882 | 2.4 | 4.98 | 308.9931, 299.1274, 281.2489, 205.0025, 146.9380 | 2.45814 | 75.3 | - | - | - | - | - | + b |
| M10 | Loss of O [M-H]− | C18H16O6 | 327.0872 | −0.8 | 7.47 | 309.0800, 299.0957, 281.2493, 221.0452, 130.9716 | 3.44497 | 83.5 | - | - | - | - | - | + b |
| M11 | Loss of O and O [M-H]− | C18H16O5 | 311.0930 | 1.5 | 9.55 | 250.9816, 204.9868, 174.9556, 130.9658 | 3.19475 | 75.7 | - | - | - | - | - | + b |
| M12a | Loss of O and CH2O [M-H]− | C17H14O5 | 297.0768 | −0.2 | 7.33 | 267.1016, 253.0865, 175.0394, 147.0452, 145.0305 | 2.78433 | 82.1 | - | - | - | - | - | + b |
| M12b | ||||||||||||||
| M13 | Loss of CO [M-H]− | C17H16O6 | 315.0862 | −3.8 | 12.74 | 300.0633, 285.0401, 270.0144, 193.0503, 147.0445 | 2.84747 | 87.9 | - | - | - | + a,b | - | + a,b |
| M14 | Oxidation [M-H]− | C18H16O8 | 359.0772 | −0.2 | 10.01 | 344.0542, 329.0304, 314.0064, 220.9817, 163.0384 | 1.79518 | 82.9 | - | - | + a,b | + a,b | + I | + a,b |
| M15 | Oxidation [M-H]− | C18H16O8 | 359.0768 | −1.3 | 10.50 | 344.0529, 329.0306, 314.0061, 221.0098, 163.0368 | 1.84518 | 82.8 | - | - | + a,b | + a,b | + I | + a,b |
| M16 | Oxidation [M-H]− | C18H16O8 | 359.0767 | −1.6 | 11.47 | 344.0536, 329.0296, 314.0066, 221.0762, 163.0019 | 1.86518 | 81.9 | - | - | + a,b | + a,b | + I | + a,b |
| M17 | Oxidation [M-H]− | C18H16O8 | 359.0767 | −1.4 | 12.23 | 344.0542, 329.0315, 314.0085, 237.0375, 147.0130 | 1.87123 | 85.5 | - | - | + a,b | + a,b | + I | + a,b |
| M18 | Di-Oxidation [M-H]− | C18H16O9 | 375.0709 | −3.3 | 9.90 | 329.0669, 314.0434, 299.0191, 221.1216, 178.9947 | 0.9644 | 91.2 | - | - | - | + a,b | - | + a |
| M19 | Tri-Oxidation [M-H]− | C18H16O10 | 391.0673 | 0.5 | 12.26 | 345.0869, 330.0636, 315.0393, 221.0399, 195.0289 | 0.25226 | 77.1 | - | - | + b | - | - | + a,b |
| M20a | Demethylation and Oxidation [M-H]− | C17H14O8 | 345.0605 | −3.0 | 9.43 | 330.0379, 301.1825, 221.1270, 149.0245, 125.0237 | 1.29734 | 84.9 | - | - | + a,b | + b | + I | + b |
| M20b | ||||||||||||||
| M21 | Demethylation and Oxidation [M-H]− | C17H14O8 | 345.0606 | −2.8 | 10.29 | 330.0384, 301.0719, 221.0028, 149.0234, 125.0311 | 1.59734 | 87.1 | - | - | + a,b | + b | + I | + b |
| M22 | Methylation [M-H]− | C19H18O7 | 357.0972 | −2.1 | 10.02 | 342.0740, 327.0503, 312.0266, 235.0434, 147.0433 | 2.06632 | 80.6 | - | - | + a,b | + a,b | - | + a,b |
| M23 | Methylation [M-H]− | C19H18O7 | 357.0969 | −3.1 | 12.86 | 342.0737, 327.0508, 312.0266, 221.0766, 161.0269 | 3.18323 | 78.6 | - | - | + a,b | + a,b | - | + a,b |
| M24 | Loss of O+Methylation [M-H]− | C19H18O6 | 341.1025 | −1.5 | 7.15 | 326.1073, 311.0918, 235.0607, 130.9906, 107.0440 | 2.80306 | 82.8 | - | - | - | - | - | + b |
| M25 | Loss of O+Methylation [M-H]− | C19H18O6 | 341.1027 | −1.2 | 8.79 | 326.0798, 311.0451, 204.9196, 161.0595, 137.0553 | 2.9313 | 82.1 | - | - | - | - | - | + b |
| M26 | Loss of CH2O and CH2O+Demethylation [M-H]− | C15H10O5 | 269.0459 | 1.3 | 9.89 | 253.0124, 241.0500, 225.0555, 133.0298, 117.0349 | 2.88784 | 95.6 | + a,b | - | + a,b | - | - | + a,b |
| M27 | Loss of CH2O and CH2+Demethylation [M-H]− | C15H10O6 | 285.0402 | −0.9 | 8.45 | 267.0130, 241.0462, 221.0063, 177.0189, 133.0307 | 2.31115 | 90.2 | - | - | - | - | - | + b |
| M28 | Glucuronidation [M-H]− | C24H24O13 | 519.1140 | −0.8 | 7.10 | 397.0442, 343.0822, 328.0587, 313.0346, 146.9662 | −0.495 | 81.6 | + a | + b | + a,b | - | + II | - |
| M29 | Glucuronidation [M-H]− | C24H24O13 | 519.1151 | 1.3 | 8.14 | 343.0824, 328.0588, 323.0173, 313.0354, 221.0262 | 0.62193 | 83.1 | + a | + b | + a,b | - | + II | - |
| M30 | Demethylation and Glucuronide Conjugation [M-H]− | C23H22O13 | 505.0979 | −1.8 | 7.88 | 329.0669, 309.0687, 299.0165, 285.0735 | 0.14693 | 81.3 | - | - | + a,b | - | - | - |
| M31 | Sulfate Conjugation [M-H]− | C18H16O10S | 423.0391 | 0.1 | 8.93 | 343.0830, 328.0593, 313.0355, 285.0413, 147.0037 | 0.27032 | 86.1 | - | + a,b | + a,b | - | - | + a,b |
| M32 | Sulfate Conjugation [M-H]− | C18H16O10S | 423.0387 | −0.9 | 9.20 | 343.0836, 328.0606, 313.0371, 285.0457, 227.0084 | 1.38723 | 89.6 | - | + a,b | + a,b | - | - | + a,b |
| M33 | Loss of CH2+Sulfate Conjugation [M-H]− | C17H14O10S | 409.0233 | −0.4 | 9.01 | 329.0670, 314.0432, 299.0198, 212.0456, 132.0208 | 0.81223 | 91.5 | + a | - | + a,b | - | - | - |
| M34 | Loss of O+Sulfate Conjugation [M-H]− | C18H16O9S | 407.0434 | −2.1 | 12.65 | 327.0826, 301.0034, 220.9818, 131.0573 | 1.00706 | 63.3 | - | - | - | - | - | + b |
| M35 | N-Acetylation [M-H]− | C20H18O8 | 385.0917 | −3.1 | 13.36 | 370.0729, 355.0427, 343.0846, 263.0551, 147.0513 | 1.49632 | 74.4 | - | - | - | - | - | + b |
| M36 | N-Acetylation [M-H]− | C20H18O8 | 385.0925 | −0.9 | 13.90 | 370.0735, 355.0492, 343.0874, 221.0781, 189.0551 | 2.61323 | 77.9 | - | - | - | - | - | + b |
| M37 | Loss of O+N-Acetylation [M-H]− | C20H18O7 | 369.0987 | 2.1 | 13.02 | 327.2227, 279.0680, 263.1681, 237.1098, 130.9934 | 2.23306 | 75.3 | + a | - | + a,b | + a,b | + I | + a,b |
| M38 | Loss of O+N-Acetylation [M-H]− | C20H18O7 | 369.0975 | −1.3 | 13.90 | 354.0755, 339.0542, 311.0594, 174.9586, 164.9289 | 2.3613 | 76.7 | + a | - | + a,b | + a,b | + I | + a,b |
| M39 | Loss of CH2+N-Acetylation [M-H]− | C19H16O8 | 371.0761 | −3.2 | 11.45 | 329.0680, 314.0439, 299.0196, 220.9869, 175.0389 | 2.13823 | 76.8 | - | - | - | + a,b | - | + b |
| M40 | Loss of CH2O+N-Acetylation [M-H]− | C19H16O7 | 355.0813 | −2.9 | 11.86 | 340.0587, 325.0335, 313.1107, 263.0361, 117.0329 | 1.64857 | 78.2 | - | - | + a,b | + a,b | + I | + a,b |
| M41 | Loss of CH2O+N-Acetylation [M-H]− | C19H16O7 | 355.0814 | −2.6 | 13.15 | 340.0589, 325.0356, 313.0337, 221.0027, 159.1093 | 2.87497 | 78.1 | - | - | + a,b | + a,b | + I | + a,b |
| M42 | Loss of CH2O+Di-Acetylation of Amines [M-H]− | C21H18O8 | 397.0918 | −2.6 | 15.21 | 382.0691, 367.0426, 313.2553, 263.0559, 159.0462 | 1.66306 | 74.8 | - | - | - | + b | - | + b |
| M43a | Loss of CH2 and CH2+Di-Acetylation of Amines [M-H]− | C20H16O9 | 399.0704 | −4.3 | 14.14 | 384.0497, 357.0641, 315.0523, 175.0034, 147.0316 | 1.56823 | 71.8 | - | - | - | + b | - | - |
| M43b | ||||||||||||||
| M43c | ||||||||||||||
| M44a | Loss of CH2O and CH2O+Di-Acetylation of Amines [M-H]− | C20H16O7 | 367.0806 | −4.8 | 15.24 | 283.0237, 233.1253, 202.9904, 189.0633, 159.0348 | 2.05475 | 71.9 | - | - | - | + a,b | - | - |
| M44b | 2.11133 | |||||||||||||
| M44c | 2.40475 | |||||||||||||
| M45 | Loss of O+Hydrogenation [M-H]− | C18H18O6 | 329.1028 | −0.8 | 4.57 | 314.0861, 299.1136, 283.2624, 223.0900, 130.9874 | 1.71027 | 75.9 | - | - | - | - | - | + a,b |
| M46 | Loss of O+Hydrogenation [M-H]− | C18H18O6 | 329.1029 | −0.5 | 5.14 | 314.0905, 299.1189, 283.1288, 207.0777, 147.0535 | 1.7953 | 76.7 | - | - | - | - | - | + a,b |
| M47 | Loss of O+Hydrogenation [M-H]− | C18H18O6 | 329.1029 | −0.3 | 5.43 | 314.0384, 299.0986, 283.2612, 207.0618, 149.0538 | 2.28982 | 78.7 | - | - | - | - | - | + a,b |
| M48 | Loss of O+Hydrogenation [M-H]− | C18H18O6 | 329.1029 | −0.6 | 9.69 | 314.0226, 299.0298, 283.0982, 223.0742, 133.0731 | 2.89116 | 75.8 | - | - | - | - | - | + a,b |
| M49 | Loss of O and O+Hydrogenation [M-H]− | C18H18O5 | 313.1080 | −0.3 | 7.61 | 298.0759, 269.1191, 239.1066, 206.9936, 130.9677 | 2.5321 | 88.5 | - | - | - | - | - | + a,b |
| M50 | Loss of O and O+Hydrogenation [M-H]− | C18H18O5 | 313.1086 | 1.6 | 7.88 | 298.0767, 269.1189, 239.1061, 207.0818, 133.0654 | 3.02662 | 90.7 | - | - | - | - | - | + a,b |
| M51 | Demethylation and Hydrogenation [M-H]− | C17H16O7 | 331.0824 | 0.3 | 10.05 | 316.0595, 313.1396, 301.0343, 223.1657, 135.0450 | 1.70816 | 90.5 | - | - | + a,b | - | - | + a |
| M52 | Di-Hydrogenation [M-H]− | C18H20O7 | 347.1140 | 1.0 | 12.78 | 332.0913, 317.0676, 225.0074, 149.0591, 123.0079 | 0.81747 | 60.2 | - | - | + a,b | - | - | + b |
| M53 | Loss of O+Di-Hydrogenation [M-H]− | C18H20O6 | 331.1186 | −0.2 | 3.60 | 301.1097, 299.1257, 285.1728, 225.0501, 133.0325 | 1.55427 | 53.1 | - | - | + b | - | - | + b |
| M54 | Loss of O+Di-Hydrogenation [M-H]− | C18H20O6 | 331.1185 | −0.7 | 4.07 | 301.1088, 299.1304, 285.0945, 209.0813, 149.0680 | 1.6393 | 52.8 | - | - | + b | - | - | + b |
| M55 | Loss of O and O+Di-Hydrogenation [M-H]− | C18H20O5 | 315.1214 | −1.6 | 6.36 | 285.1155, 271.1557, 269.1318, 241.1035, 133.0693 | 2.3761 | 83.6 | - | - | + a | - | - | + a,b |
| M56 | Loss of CH2+Di-Hydrogenation [M-H]− | C17H18O7 | 333.0972 | −2.3 | 10.18 | 318.0813, 317.1125, 303.0579, 211.0624, 149.0642 | 0.32475 | 94.4 | - | - | + a,b | - | - | - |
| M57 | Loss of CH2+Di-Hydrogenation [M-H]− | C17H18O7 | 333.0982 | 0.6 | 10.24 | 315.0034, 225.2199, 179.1064, 135.1164, 109.0679 | 0.37127 | 63.2 | - | - | + a,b | - | - | - |
| M58 | Loss of CH2+Di-Hydrogenation [M-H]− | C17H18O7 | 333.0979 | −0.3 | 10.80 | 315.0884, 300.0638, 285.0374, 211.0614, 149.0693 | 0.64475 | 90.6 | - | - | + a,b | - | - | - |
| M59a | Loss of CH2 and CH2+Di-Hydrogenation [M-H]− | C16H16O7 | 319.0815 | −2.6 | 8.47 | 301.0701, 211.0353, 197.0452, 149.0269, 135.0443 | 0.19855 | 89.0 | - | - | - | + a,b | - | - |
| M59b | −0.1214 | |||||||||||||
| M59c | 0.22768 | |||||||||||||
| M60 | Loss of CH2O and CH2O+Di-Hydrogenation [M-H]− | C16H16O5 | 287.0923 | −0.8 | 9.97 | 272.0689, 241.0875, 195.0653, 119.0500, 93.0325 | 1.35527 | 80.7 | - | - | - | - | - | + b |
| M61 | Loss of CH2O and CH2O+Di-Hydrogenation [M-H]− | C16H16O5 | 287.0927 | 0.5 | 11.07 | 272.0695, 241.2138, 165.0166, 149.0683, 123.0117 | 1.4214 | 85.8 | - | - | - | - | - | + b |
| M62a | Loss of O and CH2O+Di-Hydrogenation [M-H]− | C17H18O5 | 301.1078 | −1.2 | 7.37 | 271.1359, 255.0541, 241.0814, 179.0711, 149.0605 | 1.9972 | 87.9 | + a | - | - | + a,b | - | - |
| M62b | ||||||||||||||
| M63 | Loss of CH2O and CH2O+Loss of Hydroxymethylene [M-H]− | C15H10O4 | 253.0512 | 2.1 | 7.81 | 225.0558, 209.0606, 161.0249, 117.0351, 101.0246 | 3.4753 | 70.9 | + b | - | + a,b | - | - | + b |
| M64 | Ketone Formation [M-H]− | C18H14O8 | 357.0607 | −2.5 | 10.79 | 342.0374, 327.0132, 313.0304, 221.0224, 161.0174 | 2.17223 | 79.1 | - | - | - | + b | + I | - |
| M65 | Ketone Formation [M-H]− | C18H14O8 | 357.0609 | −1.8 | 10.81 | 342.0379, 327.0146, 313.0349, 235.0241, 147.0012 | 2.27223 | 76.1 | - | - | - | + b | + I | - |
| M66 | Ketone Formation [M-H]− | C18H14O8 | 357.0610 | −1.6 | 12.99 | 342.0385, 327.0198, 313.0421, 235.0267, 147.0503 | 2.52223 | 77.4 | - | - | - | + b | + I | - |
| M67 | Oxidation and Internal Hydrolysis [M-H]− | C18H18O9 | 377.0875 | −0.8 | 12.29 | 362.0508, 349.0873, 239.0435, 181.0136, 139.0054 | 0.21452 | 83.4 | - | - | - | + a,b | - | + a,b |
| M68a | Loss of CH2+Glycine Conjugation [M-H]− | C19H17NO8 | 386.0865 | −4.2 | 10.00 | 329.0662, 314.0421, 299.0189, 264.1192, 147.0973 | 2.15391 | 80.8 | - | - | - | + b | - | - |
| M68b | 2.40391 | |||||||||||||
| M69a | Loss of O and CH2O+Glycine Conjugation [M-H]− | C19H17NO6 | 354.0992 | 2.5 | 6.09 | 324.2008, 250.9077, 221.0751, 204.0387, 174.9553 | 2.66894 | 73.9 | - | + a | - | - | - | - |
| M69b | ||||||||||||||
| M70 | Loss of CH2O+Glutamine Conjugation [M-H]− | C22H22N2O8 | 441.1302 | −0.2 | 5.07 | 312.8496, 245.1021, 221.5883, 117.2304 | 0.99254 | 50.9 | - | - | - | - | - | + b |
| M71 | Loss of CH2 and CH2+Glucose Conjugation [M-H]− | C22H22O12 | 477.1036 | −0.6 | 5.49 | 355.0661, 315.6277, 192.9548, 146.9654, 123.0795 | −0.3982 | 52.9 | - | - | - | - | - | + b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Chen, Y.; Feng, X.; Yin, J.; Li, S.; Sun, Y.; Zhang, L. Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS. Molecules 2019, 24, 2658. https://doi.org/10.3390/molecules24142658
Li L, Chen Y, Feng X, Yin J, Li S, Sun Y, Zhang L. Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS. Molecules. 2019; 24(14):2658. https://doi.org/10.3390/molecules24142658
Chicago/Turabian StyleLi, Luya, Yuting Chen, Xue Feng, Jintuo Yin, Shenghao Li, Yupeng Sun, and Lantong Zhang. 2019. "Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS" Molecules 24, no. 14: 2658. https://doi.org/10.3390/molecules24142658
APA StyleLi, L., Chen, Y., Feng, X., Yin, J., Li, S., Sun, Y., & Zhang, L. (2019). Identification of Metabolites of Eupatorin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS. Molecules, 24(14), 2658. https://doi.org/10.3390/molecules24142658





