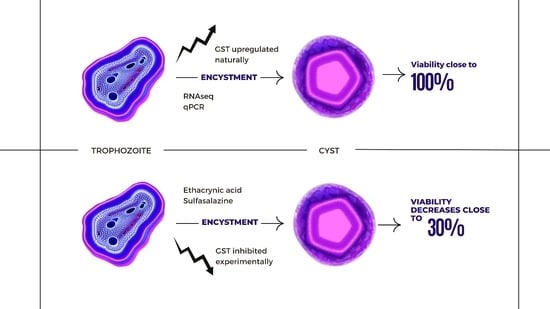
mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation
Abstract
1. Introduction
2. Materials and Methods
2.1. Acanthamoeba Cell Culture
2.2. Encystation and RNA Extraction
2.3. mRNA Sequencing
2.4. Differential Expression Analysis
2.5. qPCR Expression Analysis
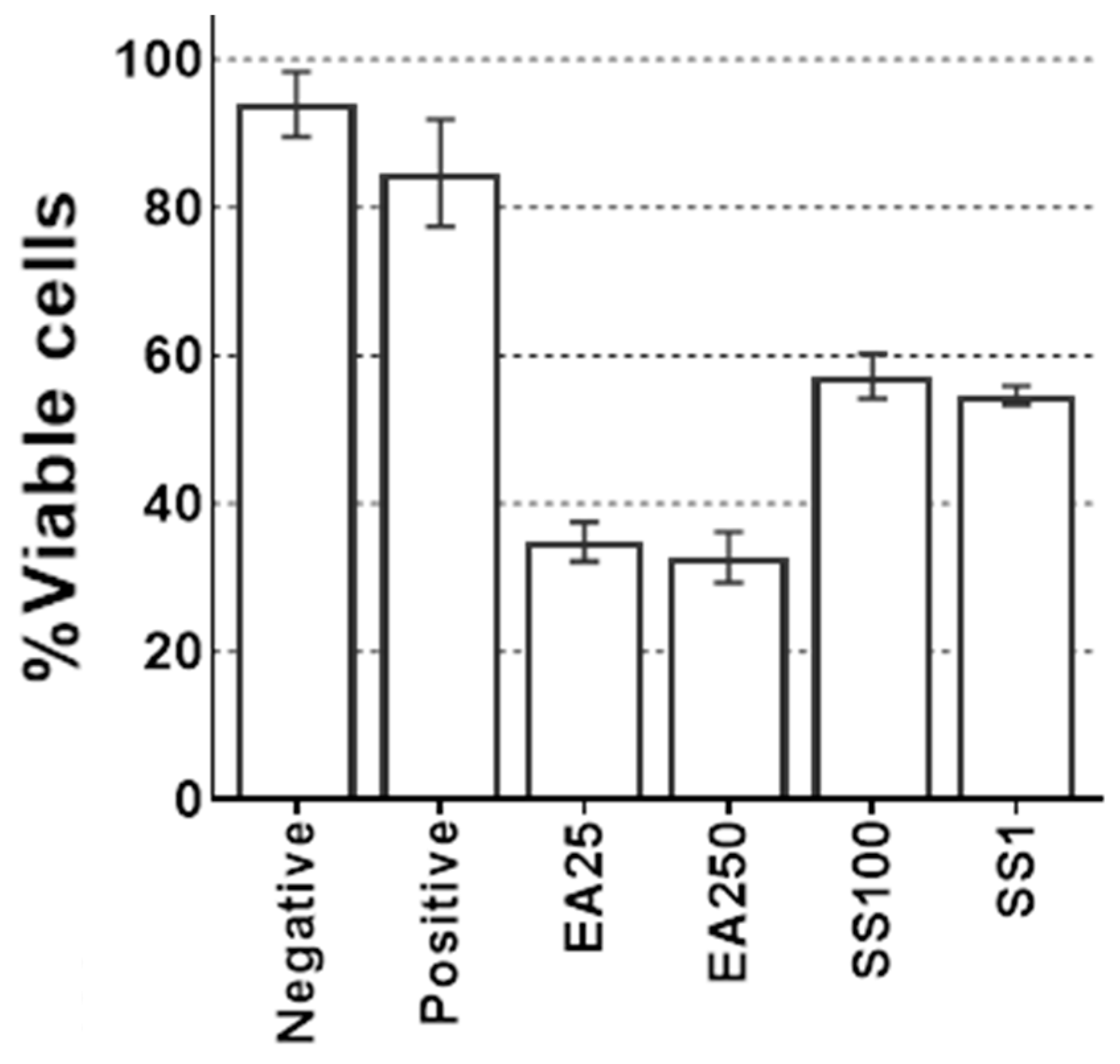
2.6. Glutathione S-Transferase (GST) Inhibitors
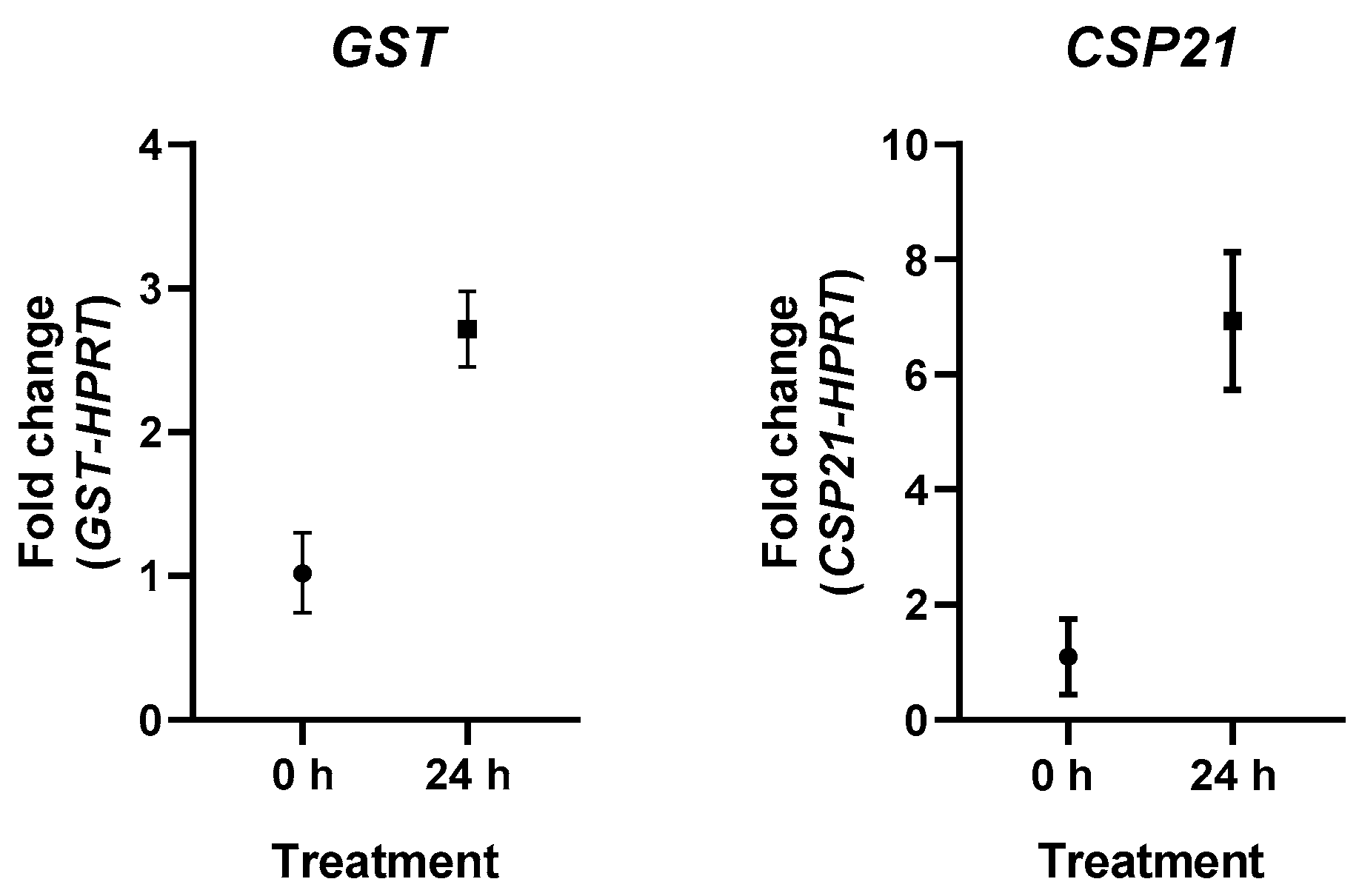
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martinez, A.J. Is Acanthamoeba encephalitis an opportunistic infection? Neurology 1980, 30, 567. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara, G.S. Amebic meningoencephalitides and keratitis: Challenges in diagnosis and treatment. Curr. Opin. Infect. Dis. 2010, 23, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Morales, J.; Khan, N.A.; Walochnik, J. An update on Acanthamoeba keratitis: Diagnosis, pathogenesis and treatment. Parasite 2015, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Morales, J.; Martín-Navarro, C.M.; López-Arencibia, A.; Arnalich-Montiel, F.; Piñero, J.E.; Valladares, B. Acanthamoeba keratitis: An emerging disease gathering importance worldwide? Trends Parasitol. 2013, 29, 181–187. [Google Scholar] [CrossRef]
- Leitsch, D.; Köhsler, M.; Marchetti-Deschmann, M.; Deutsch, A.; Allmaier, G.; Duchêne, M.; Walochnik, J. Major role for cysteine proteases during the early phase of Acanthamoeba castellanii encystment. Eukaryot. Cell 2010, 9, 611–618. [Google Scholar] [CrossRef]
- Moon, E.K.; Chung, D.I.; Hong, Y.C.; Ahn, T.I.; Kong, H.H. Acanthamoeba castellanii: Gene profile of encystation by ESTs analysis and KOG assignment. Exp. Parasitol. 2008, 119, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Moon, E.K.; Chung, D.I.; Hong, Y.C.; Kong, H.H. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Mol. Biochem. Parasitol. 2009, 168, 43–48. [Google Scholar] [CrossRef]
- Song, S.M.; Han, B.I.; Moon, E.K.; Lee, Y.R.; Yu, H.S.; Jha, B.K.; Danne, D.B.S.; Kong, H.H.; Chung, D.I.; Hong, Y. Autophagy protein 16-mediated autophagy is required for the encystation of Acanthamoeba castellanii. Mol. Biochem. Parasitol. 2012, 183, 158–165. [Google Scholar] [CrossRef]
- Potter, J.L.; Weisman, R.A. Correlation of cellulose synthesis in vivo and in vitro during the encystment of Acanthamoeba. Dev. Biol. 1972, 28, 472–479. [Google Scholar] [CrossRef]
- Hirukawa, Y.; Nakato, H.; Izumi, S.; Tsuruhara, T.; Tomino, S. Structure and expression of a cyst specific protein of Acanthamoeba castellanii. Biochim. Biophys. Acta 1998, 1398, 47–56. [Google Scholar] [CrossRef]
- Rubin, R.W.; Hill, M.C.; Hepworth, P.; Boehmer, J. Isolation and electrophoretic analysis of nucleoli, phenol-soluble nuclear proteins and outer cyst walls from Acanthamoeba castellanii during encystation initiation. J. Cell Biol. 1976, 68, 740–751. [Google Scholar] [CrossRef]
- Moon, E.-K.; Xuan, Y.-H.; Chung, D.-I.; Hong, Y.; Kong, H.-H. Microarray analysis of differentially expressed genes between cysts and trophozoites of Acanthamoeba castellanii. Korean J. Parasitol. 2011, 49, 341–347. [Google Scholar] [CrossRef]
- De Cádiz, A.E.; Jeelani, G.; Nakada-Tsukui, K.; Caler, E.; Nozaki, T. Transcriptome analysis of encystation in Entamoeba invadens. PLoS ONE 2013, 8, e74840. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkaufer, G.M.; Haque, R.; Hackney, J.A.; Eichinger, D.J.; Singh, U. Identification of developmentally regulated genes in Entamoeba histolytica: Insights into mechanisms of stage conversion in a protozoan parasite. Cell. Microbiol. 2007, 9, 1426–1444. [Google Scholar] [CrossRef] [PubMed]
- Moon, E.K.; Chung, D.I.; Hong, Y.C.; Kong, H.H. Differentially expressed genes of Acanthamoeba castellanii during encystation. Korean J. Parasitol. 2007, 45, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Makioka, A.; Kumagai, M.; Ohtomo, H.; Kobayashi, S.; Takeuchi, T. Entamoeba invadens: Protein kinase C inhibitors block the growth and encystation. Exp. Parasitol. 2000, 95, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Makioka, A.; Kumagai, M.; Ohtomo, H.; Kobayashi, S.; Takeuchi, T. Effect of calcium antagonists, calcium channel blockers and calmodulin inhibitors on the growth and encystation of Entamoeba histolytica and E. invadens. Parasitol. Res. 2001, 87, 833–837. [Google Scholar] [CrossRef]
- Makioka, A.; Kumagai, M.; Ohtomo, H.; Kobayashi, S.; Takeuchi, T. Effect of proteasome inhibitors on the growth, encystation, and excystation of Entamoeba histolytica and Entamoeba invadens. Parasitol. Res. 2002, 88, 454–459. [Google Scholar] [CrossRef]
- Dierickx, P.J.; Almar, M.M.; De Jonckheere, J.F. Glutathione transferase activity in some flagellates and amoebae, and purification of the soluble glutathione transferases from Acanthamoeba. Biochem. Int. 1990, 22, 593–600. [Google Scholar]
- Aon, M.A.; Roussel, M.R.; Cortassa, S.; O’Rourke, B.; Murray, D.B.; Beckmann, M.; Lloyd, D. The scale-free dynamics of eukaryotic cells. PLoS ONE 2008, 3, e3624. [Google Scholar] [CrossRef]
- Chen, Z.; Odstrcil, E.A.; Tu, B.P.; McKnight, S.L. Restriction of DNA replication to the cycle protects genome integrity. Science 2007, 316, 1916–1919. [Google Scholar] [CrossRef]
- Ondarza, R. Drug targets from human pathogenic amoebas: Entamoeba histolytica, Acanthamoeba polyphaga and Naegleria fowleri. Infect. Disord. Drug Targets 2007, 7, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.F.; Alberts, D.W.; Rush, G.F. Role of glutathione reductase during menadione-induced nadph oxidation in isolated rat hepatocytes. Biochem. Pharmacol. 1987, 36, 3879–3884. [Google Scholar] [CrossRef]
- Tu, C.D.; Akgül, B. Drosophila glutathione S-transferases. In Gluthione Transferases and Gamma-Glutamyl Transpeptidases; Sies, H., Packer, L.B.T.-M.E., Eds.; Academic Press: Cambridge, MA, USA, 2005; Volume 401, pp. 204–226. ISBN 00766879. [Google Scholar]
- Harwaldt, P.; Rahlfs, S.; Becker, K. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: Characterization of a potential drug target. Biol. Chem. 2002, 383, 821–830. [Google Scholar] [CrossRef]
- Neff, R.J.; Ray, S.A.; Benton, W.F.; Wilborn, M. Chapter 4 Induction of synchronous encystment (differentiation) in Acanthamoeba sp. In Methods in Cell Biology; Academic Press: Cambridge, MA, USA, 1964; pp. 55–83. [Google Scholar]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2017, 46, D802–D808. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Köhsler, M.; Leitsch, D.; Müller, N.; Walochnik, J. Validation of reference genes for the normalization of RT-qPCR gene expression in Acanthamoeba spp. Sci. Rep. 2020, 10, 10362. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 1997, 21, A.3B.1–A.3B.2. [Google Scholar] [CrossRef]
- Flanagan, J.U.; Smythe, M.L. Sigma-class glutathione transferases. Drug Metab. Rev. 2011, 43, 194–214. [Google Scholar] [CrossRef]
- Garcerá, A.; Barreto, L.; Piedrafita, L.; Tamarit, J.; Herrero, E. Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem. J. 2006, 398, 187–196. [Google Scholar] [CrossRef]
- Oakley, A.J. Glutathione transferases: New functions. Curr. Opin. Struct. Biol. 2005, 15, 716–723. [Google Scholar] [CrossRef]
- Oakley, A.J.; Lo Bello, M.; Nuccetelli, M.; Mazzetti, A.P.; Parker, M.W. The ligandin (non-substrate) binding site of human pi class glutathione transferase is located in the electrophile binding site (H-site). J. Mol. Biol. 1999, 291, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Veal, E.A.; Toone, W.M.; Jones, N.; Morgan, B.A. Distinct roles for glutathione S-transferases in the oxidative stress response in Schizosaccharomyces pombe. J. Biol. Chem. 2002, 277, 35523–35531. [Google Scholar] [CrossRef]
- Smith, G.A.; Lin, T.H.; Sheehan, A.E.; Van der Goes van Naters, W.; Neukomm, L.J.; Graves, H.K.; Bis-Brewer, D.M.; Züchner, S.; Freeman, M.R. Glutathione S-transferase regulates mitochondrial populations in axons through increased glutathione oxidation. Neuron 2019, 103, 52–65.e6. [Google Scholar] [CrossRef]
- Müller, M.; Lu, K.; Reichert, A.S. Mitophagy and mitochondrial dynamics in Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Klionsky, D.J. Mitochondria removal by autophagy. Autophagy 2011, 7, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Kanki, T. How autophagy eats large mitochondria: Autophagosome formation coupled with mitochondrial fragmentation. Autophagy 2017, 13, 980–981. [Google Scholar] [CrossRef]
- Kim, S.-H.; Moon, E.-K.; Hong, Y.; Chung, D.-I.; Kong, H.-H. Autophagy protein 12 plays an essential role in Acanthamoeba encystation. Exp. Parasitol. 2015, 159, 46–52. [Google Scholar] [CrossRef]
- Lloyd, D. Encystment in Acanthamoeba castellanii: A review. Exp. Parasitol. 2014, 145, S20–S27. [Google Scholar] [CrossRef]
- Moon, E.K.; Hong, Y.; Chung, D.I.; Kong, H.H. Cysteine protease involving in autophagosomal degradation of mitochondria during encystation of Acanthamoeba. Mol. Biochem. Parasitol. 2012, 185, 121–126. [Google Scholar] [CrossRef]
- Gorsich, S.W.; Shaw, J.M. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 2004, 15, 4369–4381. [Google Scholar] [CrossRef]
- Scheckhuber, C.Q.; Erjavec, N.; Tinazli, A.; Hamann, A.; Nyström, T.; Osiewacz, H.D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007, 9, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Vasiliou, V. Analysis of the glutathione S-transferase (GST) gene family. Hum. Genom. 2004, 1, 460. [Google Scholar] [CrossRef] [PubMed]
- Joachim, A.; Lautscham, E.; Christoffers, J.; Ruttkowski, B. Oesophagostomum dentatum: Effect of glutathione S-transferase (GST) inhibitors on GST activity and larval development. Exp. Parasitol. 2011, 127, 762–767. [Google Scholar] [CrossRef]
- Borst, P.; Ouellette, M. New mechanisms of drug resistance in parasitic protozoa. Annu. Rev. Microbiol. 1995, 49, 427–460. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.-L.; Epstein, D.L.; de Kater, A.W.; Shahsafaei, A.; Erickson-Lamy, K.A. Ethacrynic Acid Increases Facility of Outflow in the Human Eye In Vitro. Arch. Ophthalmol. 1992, 110, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Kotas-Neumann, R.; Barak, A.; Epstein, D.L. The Effect of Intracamerally Injected Ethacrynic Acid on Intraocular Pressure in Patients with Glaucoma. Am. J. Ophthalmol. 1992, 113, 508–512. [Google Scholar] [CrossRef]
- Lin, C.-W.; Gonzalez, P.; Yuan, F. Cellular pharmacokinetic and pharmacodynamic analyses of ethacrynic acid: Implications in topical drug delivery in the eye. Mol. Vis. 2011, 17, 2507–2515. [Google Scholar]
- Cynkowska, G.; Cynkowski, T.; Al-Ghananeem, A.A.; Guo, H.; Ashton, P.; Crooks, P.A. Novel antiglaucoma prodrugs and codrugs of ethacrynic acid. Bioorg. Med. Chem. Lett. 2005, 15, 3524–3527. [Google Scholar] [CrossRef]
- Christopher Kent Glaucoma Drugs: The Search for New Options. Available online: https://www.reviewofophthalmology.com/article/glaucoma-drugs-the-search-for-new-options (accessed on 3 April 2023).
- Benitez-Del-Castillo, J.M.; Garcia-Sanchez, J.; Iradier, T.; Bañares, A. Sulfasalazine in the prevention of anterior uveitis associated with ankylosing spondylitis. Eye 2000, 14, 340–343. [Google Scholar] [CrossRef]
- Muñoz-Fernández, S.; Hidalgo, V.; Fernández-Melón, J.; Schlincker, A.; Bonilla, G.; Ruiz-Sancho, D.; Fonseca, A.; Gijón-Baños, J.; Martín-Mola, E. Sulfasalazine reduces the number of flares of acute anterior uveitis over a one-year period. J. Rheumatol. 2003, 30, 1277–1279. [Google Scholar]
- Doan, S.; Lerouic, J.-F.; Robin, H.; Prost, C.; Savoldelli, M.; Hoang-Xuan, T. Treatment of ocular cicatricial pemphigoid with sulfasalazine. Ophthalmology 2001, 108, 1565–1568. [Google Scholar] [CrossRef]
- Joo, C.-K.; Choi, J.-S. The Effect of Sulfasalazine—Hyaluronic Acid Complex on Posterior Capsule Opacification. Invest. Ophthalmol. Vis. Sci. 2013, 54, 2955. [Google Scholar]
- Galperin, M.Y.; Koonin, E.V. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 1999, 10, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Bakker, B.M.; Assmus, H.E.; Bruggeman, F.; Haanstra, J.R.; Klipp, E.; Westerhoff, H. Network-based selectivity of antiparasitic inhibitors. Mol. Biol. Rep. 2002, 29, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cassidy-Stone, A.; Chipuk, J.E.; Ingerman, E.; Song, C.; Yoo, C.; Kuwana, T.; Kurth, M.J.; Shaw, J.T.; Hinshaw, J.E.; Green, D.R.; et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell 2008, 14, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.; Kristensen, B.; Degn, H. Oxidative detoxification of hydrogen sulphide detected by mass spectrometry in the soil amoeba Acanthamoeba castellanii. J. Gen. Microbiol. 1981, 126, 167–170. [Google Scholar] [CrossRef]
| Gene | Sequence (5′-3′) | Tm | Amplicon Lenght | Source or Accesion Number |
|---|---|---|---|---|
| GST | F: CAAGTGCTACCCCAAGGAC | 57.75 | 162 bp | NW_004457554 |
| R: CCCTTCTCGTCCGGGTAG | 58.48 | |||
| CSP21 | F: ACTTTGGCGACAAGGTGTG | 58.6 | 80 bp | XM_004337011 |
| R: CGACACGTCGTCCCTCT | 58.31 | |||
| HPRT | F: GGAGCGGATCGTTCTCTG | 58.4 | 201 bp | [35] |
| R: ATCTTGGCGTCGACGTGC | 58.4 |
| Gene_ID | Description | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| ACA1_116240 | GST C-terminal domain containing protein | 4.7154 | 0.4877 | 0.4623 |
| ACA1_075240 | Cyst-specific protein 21 | 6.5435 | 3.5723 | 1.7860 |
| ACA1_022350 | Hypothetical protein | 8.4458 | 3.9417 | 1.4994 |
| ACA1_096640 | Hypothetical protein | 7.0754 | 4.2638 | 1.8371 |
| ACA1_188370 | Hypothetical protein | 10.0624 | 6.1620 | 3.4728 |
| ACA1_247090 | Hypothetical protein | 7.5066 | 2.7215 | 0.8404 |
| ACA1_374130 | Hypothetical protein | 6.8663 | 2.5504 | 0.2569 |
| Gene_ID | Predicted Protein | Predicted RNA | Genomic |
|---|---|---|---|
| ACA1_188370 | 74% | 73% | 75% |
| ACA1_022350 | NA | 75% | 75% |
| ACA1_247090 | 54% | 72% | 68% |
| ACA1_096640 | 54% | 74% | 73% |
| ACA1_374130 | 79% | 81% | 79% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Obeso Fernández del Valle, A.; Scheckhuber, C.Q.; Chavaro-Pérez, D.A.; Ortega-Barragán, E.; Maciver, S.K. mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation. Microorganisms 2023, 11, 992. https://doi.org/10.3390/microorganisms11040992
de Obeso Fernández del Valle A, Scheckhuber CQ, Chavaro-Pérez DA, Ortega-Barragán E, Maciver SK. mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation. Microorganisms. 2023; 11(4):992. https://doi.org/10.3390/microorganisms11040992
Chicago/Turabian Stylede Obeso Fernández del Valle, Alvaro, Christian Quintus Scheckhuber, David Armando Chavaro-Pérez, Erandi Ortega-Barragán, and Sutherland K. Maciver. 2023. "mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation" Microorganisms 11, no. 4: 992. https://doi.org/10.3390/microorganisms11040992
APA Stylede Obeso Fernández del Valle, A., Scheckhuber, C. Q., Chavaro-Pérez, D. A., Ortega-Barragán, E., & Maciver, S. K. (2023). mRNA Sequencing Reveals Upregulation of Glutathione S-Transferase Genes during Acanthamoeba Encystation. Microorganisms, 11(4), 992. https://doi.org/10.3390/microorganisms11040992