The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections
Abstract
1. Introduction
2. The International Stock Centers
3. How Many Standard Strains Exist?
4. Linking Culture Center Isolates with Genetic Information
4.1. Small Subunit rRNA Genes
4.2. Connecting Isolate Information with the DNA Databases
4.3. Genome Projects for Isolates of the Culture Center in the DNA Databases
5. How Reliable Is the Sequence Information for Standard Strains of Acanthamoeba?
5.1. Assessing the Accuracy of Sequences in the DNA Database
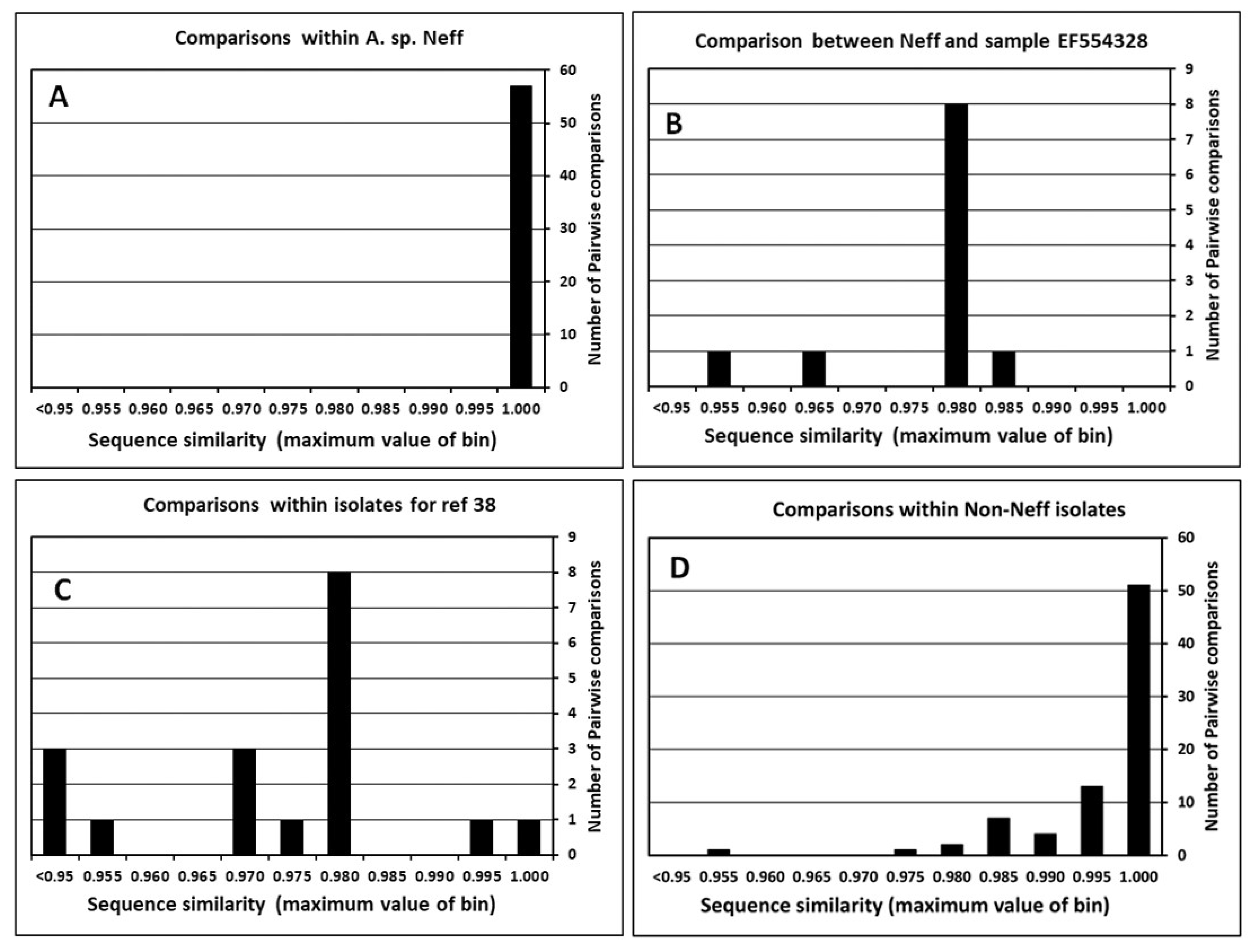
5.2. Accuracy of 18S Sequences in the DNA Database
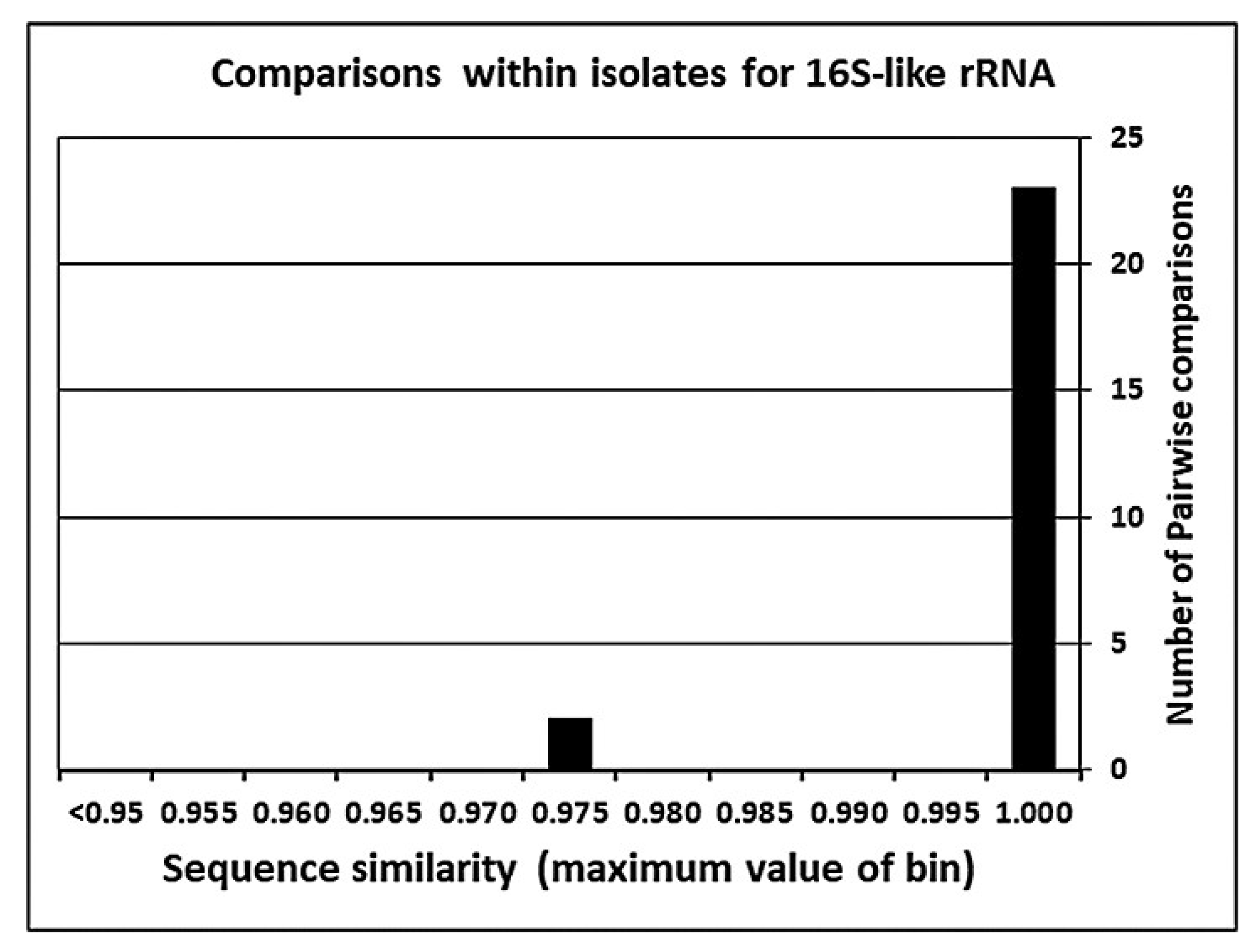
5.3. Reliability of Mitochondrial 16S-like Sequences in the DNA Database
5.4. rRNA Sequences for Other Acanthamoebidae
5.5. Phylogenetic Implications of rRNA Sequences from Culture Center Strains
6. Comparing Standard Strains to Isolates from Clinical or Environmental Studies?
6.1. What Proportion of the DNA Database Is Made up of Sequences Determined from the Standard Strains?
6.2. How Does the Classification of Standard Strains Mirror the Occurrence of Sequence Types in Samples Isolated in Clinical or Environmental Studies
7. The Relationship between Species Name, Species Type Sample, and Genotypic Information
7.1. Type Isolates Maintained in the Culture Centers—A. castellanii versus Neff
7.2. Type Isolates Maintained in the Culture Centers—Other Species
8. Discussion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, N.A.; Siddiqui, R. Predator vs aliens: Bacteria interactions with Acanthamoeba. Parasitology 2014, 141, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, T.K. Free-living pathogenic and nonpathogenic amoebae in Maryland soils. Appl. Environ. Microbiol. 1989, 55, 1074–1077. [Google Scholar] [CrossRef]
- Mergeryan, H. The prevalence of Acanthamoeba in the human environment. Rev. Infect. Dis. 1991, 13, S390–S391. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, R.; Khan, N.A. Biology and pathogenesis of Acanthamoeba. Parasites Vectors 2012, 5, 6. [Google Scholar] [CrossRef]
- Schuster, F.L.; Visvesvara, G.S. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 2004, 34, 1001–1027. [Google Scholar] [CrossRef] [PubMed]
- Castellani, A. An amoeba found in culture of yeast: Preliminary note. J. Trop Med. Hyg. 1930, 33, 160. [Google Scholar]
- Cursons, R.T.; Brown, T.J.; Keys, E.A.; Moriarty, K.M.; Till, D. Immunity to pathogenic free-living amoebae: Role of humoral antibody. Infect. Immun. 1980, 29, 401–407. [Google Scholar] [CrossRef]
- Chappell, C.L.; Wright, J.A.; Coletta, M.; Newsome, A.L. Standardized method of measuring Acanthamoeba antibodies in sera from healthy human subjects. Clin. Diagn. Lab. Immunol. 2001, 8, 724–730. [Google Scholar] [CrossRef]
- Schuster, F.L.; Honarmand, S.; Visvesvara, G.S.; Glaser, C.A. Detection of antibodies against free-living amoebae Balamuthia mandrillaris and Acanthamoeba species in a population of patients with encephalitis. Clin. Infect. Dis. 2006, 42, 1260–1265. [Google Scholar] [CrossRef]
- Brindley, N.; Matin, A.; Khan, N.A. Acanthamoeba castellanii: High antibody prevalence in racially and ethnically diverse populations. Exp. Parasitol. 2009, 121, 254–256. [Google Scholar] [CrossRef]
- Visvesvara, G.S. Classification of Acanthamoeba. Rev. Infect. Dis. 1991, 13, S369–S372. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, P.A.; Booton, G.C. Species, Sequence Types and alleles: Dissecting genetic variation in Acanthamoeba. Pathogens 2020, 9, 534. [Google Scholar] [CrossRef]
- Corsaro, D. Update on Acanthamoeba phylogeny. Parasitol. Res. 2020, 119, 3327–3338. [Google Scholar] [CrossRef] [PubMed]
- Putaporntip, C.; Kuamsab, N.; Nuprasert, W.; Rojrung, R.; Pattanawong, U.; Tia, T.; Yanmanee, S.; Jongwutiwes, S. Analysis of Acanthamoeba genotypes from public freshwater sources in Thailand reveals a new genotype, T23 Acanthamoeba bangkokensis sp. nov. Sci. Rep. 2021, 11, 17290. [Google Scholar] [CrossRef] [PubMed]
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Morales, J.; Martín-Navarro, C.M.; López-Arencibia, A.; Arnalich-Montiel, F.; Piñero, J.E.; Valladares, B. Acanthamoeba keratitis: An emerging disease gathering importance worldwide? Trends Parasitol. 2013, 29, 181–187. [Google Scholar] [CrossRef]
- Lorenzo-Morales, J.; Khan, N.A.; Walochnik, J. An update on Acanthamoeba keratitis: Diagnosis, pathogenesis and treatment. Parasite 2015, 22, 10. [Google Scholar] [CrossRef]
- Rowbotham, T.J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 1980, 33, 1179–1183. [Google Scholar] [CrossRef]
- Michel, R.; Hauröder-Philippczyk, B.; Müller, K.; Weishaar, I. Acanthamoeba from human nasal mucosa infected with an obligate intracellular parasite. Eur. J. Protistol. 1994, 30, 104–110. [Google Scholar] [CrossRef]
- Scheid, P. Relevance of free-living amoebae as hosts for phylogenetically diverse microorganisms. Parasitol. Res. 2014, 113, 2407–2414. [Google Scholar] [CrossRef]
- Balczun, C.; Scheid, P.L. Free-Living Amoebae as Hosts for and Vectors of Intracellular Microorganisms with Public Health Significance. Viruses 2017, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Rayamajhee, B.; Willcox, M.D.; Henriquez, F.L.; Petsoglou, C.; Subedi, D.; Carnt, N. Acanthamoeba, an environmental phagocyte enhancing survival and transmission of human pathogens. Trends Parasitol. 2022, 38, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Pussard, M. Le genre Acanthamoeba Volkonsky 1931 (Hartmannellidae-Amoebida). Protistologica 1966, 2, 71–93. [Google Scholar]
- Page, F.C. Re-Definition of Genus Acanthamoeba with Descriptions of 3 Species. J. Protozool. 1967, 14, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Daggett, P.M.; Lipscomb, D.; Sawyer, T.K.; Nerad, T.A. A Molecular Approach to the Phylogeny of Acanthamoeba. BioSystems 1985, 18, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Stothard, D.R.; Schroeder-Diedrich, J.M.; Awwad, M.H.; Gast, R.J.; Ledee, D.R.; Rodriguez-Zaragoza, S.; Dean, C.L.; Fuerst, P.A.; Byers, T.J. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene Sequence Types. J. Eukaryot. Microbiol. 1998, 45, 45–54. [Google Scholar] [CrossRef]
- Ledee, D.R.; Booton, G.C.; Awwad, M.H.; Sharma, S.; Aggarwal, R.K.; Niszl, I.A.; Markus, M.B.; Fuerst, P.A.; Byers, T.J. Advantages of using mitochondrial 16S rDNA sequences to classify clinical isolates of Acanthamoeba. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1142–1149. [Google Scholar] [CrossRef]
- Gast, R.J.; Ledee, D.R.; Fuerst, P.A.; Byers, T.J. Subgenus systematics of Acanthamoeba: Four nuclear 18S rDNA Sequence Types. J. Eukaryot. Microbiol. 1996, 43, 498–504. [Google Scholar] [CrossRef]
- Fuerst, P.A. Insights from the DNA databases: Approaches to the phylogenetic structure of Acanthamoeba. Exp. Parasitol. 2014, 145, S39–S45. [Google Scholar] [CrossRef] [PubMed]
- Bogler, S.A.; Zarley, C.D.; Burianek, L.L.; Fuerst, P.A.; Byers, T.J. Interstrain mitochondrial DNA polymorphism detected in Acanthamoeba by restriction endonuclease analysis. Mol. Biochem. Parasitol. 1983, 8, 145–163. [Google Scholar] [CrossRef]
- Kong, H.; Chung, D. PCR and RFLP variation of conserved region of small subunit ribosomal DNA among Acanthamoeba isolates assigned to either A. castellanii or A. polyphaga. Korean J. Parasitol. 1996, 34, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Yagita, K.; Endo, T.; De Jonckheere, J.F. Clustering of Acanthamoeba isolates from human eye infections by means of mitochondrial DNA digestion patterns. Parasitol. Res. 1999, 85, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hwang, M.; Kim, T.; Yun, H.; Kim, T.; Kong, H.; Chung, D. Phylogenetic relationships among Acanthamoeba spp. based on PCR-RFLP analyses of mitochondrial small subunit rRNA gene. Korean J. Parasitol. 1999, 37, 181. [Google Scholar] [CrossRef][Green Version]
- Amaral Zettler, L.A.; Anderson, O.R.; Nerad, T.A.; Sogin, M.L. The Phylogenetic Position of Comandonia operculata and its Implications for the Taxonomy of the Genus Acanthamoeba. In Proceedings of the IXth International Meeting on the Biology and Pathogenicity of Free-Living Amoebae Proceedings, Paris, France, 8–14 July 2000; pp. 235–242. [Google Scholar]
- Pernin, P.; Pussard, M. Etude en microscopie photonique et electronique d’une amibe voisine du genre Acanthamoeba: Comandonia operculata n. gen., n. sp. (Amoebida, Acanthamoebidae). Protistologica 1979, 15, 87–102. [Google Scholar]
- Kudryavtsev, A.; Wylezich, C.; Schlegel, M.; Walochnik, J.; Michel, R. Ultrastructure, SSU rRNA gene sequences and phylogenetic relationships of Flamella Schaeffer, 1926 (Amoebozoa), with description of three new species. Protist 2009, 160, 21–40. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2022, 50, D161–D164. [Google Scholar] [CrossRef]
- Arita, M.; Karsch-Mizrachi, I. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021, 49, D121–D124. [Google Scholar] [CrossRef] [PubMed]
- Köhsler, M.; Leitsch, D.; Fürnkranz, U.; Duchêne, M.; Aspöck, H.; Walochnik, J. Acanthamoeba strains lose their abilities to encyst synchronously upon prolonged axenic culture. Parasitol. Res. 2008, 102, 1069–1072. [Google Scholar] [CrossRef]
- Schroeder, J.M.; Booton, G.C.; Hay, J.; Niszl, I.A.; Seal, D.V.; Markus, M.B.; Fuerst, P.A.; Byers, T.J. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of Acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 2001, 39, 1903–1911. [Google Scholar] [CrossRef]
- Khan, N.A.; Jarroll, E.L.; Paget, T.A. Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Curr. Microbiol. 2002, 45, 197–202. [Google Scholar] [CrossRef]
- Johnson, A.M.; Fielke, R.; Christy, P.E.; Robinson, B.; Baverstock, P.R. Small subunit ribosomal RNA evolution in the genus Acanthamoeba. J. Gen. Microbiol. 1990, 136, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Booton, G.C.; Lares-Villa, F.; Kelly, D.J.; Fuerst, P.A.; Byers, T.J. Multiple alleles of Acanthamoeba nuclear small subunit ribosomal RNA genes: Further evidence of possible genetic exchange between closely related strains. In Proceedings of the Xth International Meeting on the Biology and Pathogenicity of Free-Living Amoebae Proceedings, ITSON-DIEP, Obregon, Mexico, 5–10 October 2003; pp. 83–91. [Google Scholar]
- Booton, G.C.; Kelly, D.J.; Chu, Y.; Seal, D.V.; Houang, E.; Lam, D.; Byers, T.J.; Fuerst, P.A. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J. Clin. Microbiol. 2002, 40, 1621–1625. [Google Scholar] [CrossRef] [PubMed]
- Matthey-Doret, C.; Colp, M.J.; Escoll, P.; Thierry, A.; Curtis, B.; Sarrasin, M.; Gray, M.W.; Lang, B.F.; Archibald, J.M.; Buchrieser, C. Chromosome-scale assemblies of Acanthamoeba castellanii genomes provide insights into Legionella pneumophila infection-related chromatin re-organization. bioRxiv 2021. [Google Scholar] [CrossRef]
- Crary, M.L.; Lares-Villa, F.; Booton, G.; Joslin, C.; Tu, E.; Fuerst, P.A. Multilocus sequence typing of Acanthamoeba keratitis-associated clinical isolates from a Chicago outbreak. In Proceedings of the XIVth International Meeting on the Biology and Pathogenicity of Free-living Amoebae, Montego Bay, Jamaica, 10 October 2011; p. 11. [Google Scholar]
- Köhsler, M.; Corsaro, D.; Kniha, E.; Hafeli, I.; Walochnik, J. Barcoding Acanthamoeba spp.: Evaluation of COI and ND5 for Identification and Differentiation of Acanthamoeba genotypes. 2020; unpublished. [Google Scholar]
- Crary, M.J.; Booton, G.C.; Fuerst, P.A. Cytochrome C Oxidase Subunit I (COI) Taxon Barcoding of the Opportunistically Pathogenic Protists Acanthamoeba and Balamuthia. 2019; unpublished. [Google Scholar]
- Malavin, S.; Shmakova, L. Isolates from ancient permafrost help to elucidate species boundaries in Acanthamoeba castellanii complex (Amoebozoa: Discosea). Eur. J. Protistol. 2020, 73, UNSP 125671. [Google Scholar] [CrossRef]
- Crary, M.J.; Narayanan, S.; Visvesvara, G.; Joslin, C.; Tu, E.; Panjwani, N.; Hopkins, A.; Booton, G.; Fuerst, P. Genotyping Acanthamoeba Keratitis (AK) Infections: Cytochrome Oxidase I (COI) Barcoding and Phylogenetic Analysis. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5832. [Google Scholar]
- Corsaro, D. Exploring LSU and ITS rDNA sequences for Acanthamoeba identification and phylogeny. Microorganisms 2022, 10, 1776. [Google Scholar] [CrossRef]
- Corsaro, D. Acanthamoeba Mannose and Laminin Binding Proteins Variation across Species and Genotypes. Microorganisms 2022, 10, 2162. [Google Scholar] [CrossRef]
- Crary, M.J. Genetic variability and its relationship to Acanthamoeba pathogenesis; Chapter 5. Multilocus Sequence Typing of Acanthamoeba Keratitis-Associated Clinical Isolates from a Chicago Outbreak. Ph.D. Thesis, The Ohio State University, Columbus, OH, USA, 2012. [Google Scholar]
- Qvarnstrom, Y.; Nerad, T.A.; Visvesvara, G.S. Characterization of a new pathogenic Acanthamoeba species, A. byersi n. sp., isolated from a human with fatal amoebic encephalitis. J. Eukaryot. Microbiol. 2013, 60, 626–633. [Google Scholar] [CrossRef]
- Corsaro, D.; Walochnik, J.; Köhsler, M.; Rott, M.B. Acanthamoeba misidentification and multiple labels: Redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli sp nov (genotype T19). Parasitol. Res. 2015, 114, 2481–2490. [Google Scholar] [CrossRef] [PubMed]
- Tice, A.K.; Shadwick, L.L.; Fiore-Donno, A.M.; Geisen, S.; Kang, S.; Schuler, G.A.; Spiegel, F.W.; Wilkinson, K.A.; Bonkowski, M.; Dumack, K.; et al. Expansion of the molecular and morphological diversity of Acanthamoebidae (Centramoebida, Amoebozoa) and identification of a novel life cycle type within the group. Biol. Direct. 2016, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.; Fritsche, T.R.; Gautom, R.K.; Schleifer, K.; Wagner, M. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1999, 1, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Gast, R.J. Development of an Acanthamoeba-specific reverse dot-blot and the discovery of a new ribotype. J. Eukaryot. Microbiol. 2001, 48, 609–615. [Google Scholar] [PubMed]
- Hewett, M.K.; Robinson, B.S.; Monis, P.T.; Saint, C.P. Identification of a new Acanthamoeba 18S rRNA gene Sequence Type, corresponding to the species Acanthamoeba jacobsi Sawyer, Nerad and Visvesvara, 1992 (Lobosea: Acanthamoebidae). Acta Protozool. 2003, 42, 325–329. [Google Scholar]
- Corsaro, D.; Venditti, D. Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida). Parasitol. Res. 2010, 107, 233–238. [Google Scholar] [CrossRef]
- Nuprasert, W.; Putaporntip, C.; Pariyakanok, L.; Jongwutiwes, S. Identification of a novel T17 genotype of Acanthamoeba from environmental isolates and T10 genotype causing keratitis in Thailand. J. Clin. Microbiol. 2010, 48, 4636–4640. [Google Scholar] [CrossRef]
- De Jonckheere, J.F.; Michel, R. Species identification and virulence of Acanthamoeba strains from human nasal mucosa. Parasitol. Res. 1988, 74, 314–316. [Google Scholar] [CrossRef]
- Costas, M.; Griffiths, A.J. Enzyme composition and the taxonomy of Acanthamoeba. J. Protozool. 1985, 32, 604–607. [Google Scholar] [CrossRef]
- Badenoch, P.R.; Adams, M.; Coster, D.J. Corneal virulence, cytopathic effect on human keratocytes and genetic characterization of Acanthamoeba. Int. J. Parasitol. 1995, 25, 229–239. [Google Scholar] [CrossRef]
- Fuerst, P.A.; Booton, G.C.; Crary, M. Phylogenetic Analysis and the Evolution of the 18S rRNA Gene Typing System of Acanthamoeba. J. Eukaryot. Microbiol. 2015, 62, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Volkonsky, M. Hartmannella castellani Douglas et classification des Hartmannelles. Arc. Zoolog. Exp. Gen. 1931, 72, 317–339. [Google Scholar]
- Pussard, M.; Pons, R. Morphologie de la Paroi Kystique et Taxonomie du Genre Acanthamoeba (Protozoa, Amoebida). Protistologica 1977, 13, 557–598. [Google Scholar]
- Simitzis-Le Flohic, A.M.; Le Goff, F.; Quiviger, J. Essai de taxonomie des Amibes libres du genre Acanthamoeba (Protozoa, Amoebida) par l’analyse des caractères kystiques en microscopie électronique à balayage. Ann. Parasitol. Hum. Comparée 1981, 56, 563–573. [Google Scholar] [CrossRef][Green Version]
- Costas, M.; Griffiths, A.J. The esterases and acid-phosphatases of Acanthamoeba (Amoebida, Acanthamoebidae). Protistologica 1984, 20, 33–41. [Google Scholar]
- Booton, G.C.; Visvesvara, G.S.; Byers, T.J.; Kelly, D.J.; Fuerst, P.A. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J. Clin. Microbiol. 2005, 43, 1689–1693. [Google Scholar] [CrossRef]
- Shi, Y.; Queller, D.C.; Tian, Y.; Zhang, S.; Yan, Q.; He, Z.; He, Z.; Wu, C.; Wang, C.; Shu, L. The ecology and evolution of amoeba-bacterium interactions. Appl. Environ. Microbiol. 2021, 87, 1866. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Toenshoff, E.R.; Haider, S.; Heinz, E.; Hoenninger, V.M.; Wagner, M.; Horn, M. Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl. Environ. Microbiol. 2008, 74, 5822–5831. [Google Scholar] [CrossRef]
- Thomas, V.; Herrera-Rimann, K.; Blanc, D.S.; Greub, G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 2006, 72, 2428–2438. [Google Scholar] [CrossRef]
- Rayamajhee, B.; Subedi, D.; Peguda, H.K.; Willcox, M.D.; Henriquez, F.L.; Carnt, N. A systematic review of intracellular microorganisms within Acanthamoeba to understand potential impact for infection. Pathogens 2021, 10, 225. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Gomes, K.X.; Cortines, J.R.; Peralta, J.M.; Peralta, R.H.S. Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol. Res. 2016, 193, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Anacarso, I.; de Niederhäusern, S.; Messi, P.; Guerrieri, E.; Iseppi, R.; Sabia, C.; Bondi, M. Acanthamoeba polyphaga, a potential environmental vector for the transmission of food-borne and opportunistic pathogens. J. Basic Microbiol. 2012, 52, 261–268. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Chao, E.E.; Lewis, R. 187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol. Phylogenet. Evol. 2016, 99, 275–296. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Chao, E.E.; Lewis, R. Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: Contrasting cell organisation of sister phyla Cercozoa and Retaria. Protoplasma 2018, 255, 1517–1574. [Google Scholar] [CrossRef] [PubMed]
- Tekle, Y.; Wang, F.; Wood, F.; Anderson, O.; Smirnov, A. New Insights on the Evolutionary Relationships Between the Major Lineages of Amoebozoa. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Tekle, Y.I.; Wood, F.C. Longamoebia is not monophyletic: Phylogenomic and cytoskeleton analyses provide novel and well-resolved relationships of amoebozoan subclades. Mol. Phylogenet. Evol. 2017, 114, 249–260. [Google Scholar] [CrossRef]
- Page, F.C. An Illustrated Key to Freshwater and Soil Amoebae: With Notes on Cultivation and Ecology; Freshwater Biological Association: Ambleside, UK, 1976; 155p. [Google Scholar]
- Page, F.C. A New Key to Freshwater and Soil Gymnamoebae: With Instructions for Culture; Freshwater Biological Association: Ambleside, UK, 1988; 122p. [Google Scholar]
- Gunderson, J.H.; Sogin, M.L. Length variation in eukaryotic rRNAs: Small subunit rRNAs from the protists Acanthamoeba castellanii and Euglena gracilis. Gene 1986, 44, 63–70. [Google Scholar] [CrossRef]
- Glücksman, E.; Snell, E.A.; Berney, C.; Chao, E.E.; Bass, D.; Cavalier-Smith, T. The Novel Marine Gliding Zooflagellate Genus Mantamonas (Mantamonadida ord. n.: Apusozoa). Protist 2011, 162, 207–221. [Google Scholar] [CrossRef]
- Clarke, M.; Lohan, A.J.; Liu, B.; Lagkouvardos, I.; Roy, S.; Zafar, N.; Bertelli, C.; Schilde, C.; Kianianmomeni, A.; Bürglin, T.R.; et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013, 14, 1–15. [Google Scholar] [CrossRef]
- Liu, Y. Acanthamoeba castellanii strain: Neff genome Project, Baylor College of Medicine. 2011. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA20303 (accessed on 20 December 2022).
- Mbugua, M.W. Characterization of Unusual Gymnamoebae Isolated from the Marine Environment. Master’s Thesis, Marshall University, Huntington, WV, USA, 2008. Available online: https://mds.marshall.edu/cgi/viewcontent.cgi?article=1723&context=etd (accessed on 20 December 2022).
- De Jonckheere, J.F. Genotyping of Acanthamoeba reference strains. 2013, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Kilvington, S. Acanthamoeba reference sequences. 2008, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Burger, G.; Plante, I.; Longeran, K.M.; Gray, M.W. The Mitochondrial-DNA of the Ameboid Protozoan, Acanthamoeba castellanii - Complete Sequence, Gene Content and Genome Organization. J. Mol. Biol 1995, 245, 522–537. [Google Scholar] [CrossRef] [PubMed]
- Mafra, C.S.P.; Carrijo-Carvalho, L.C.; Chudzinski-Tavassi, A.M.; Taguchi, F.M.d.C.; Foronda, A.S.; Carvalho, F.R.d.S.; de Freitas, D. Antimicrobial action of biguanides on the viability of Acanthamoeba cysts and assessment of cell toxicity. Invest. Ophthalmol Vis. Sci 2013, 54, 6363–6372. [Google Scholar] [CrossRef] [PubMed]
- Chelkha, N.; Jardot, P.; Moussaoui, I.; Levasseur, A.; La Scola, B.; Colson, P. Core gene-based molecular detection and identification of Acanthamoeba species. Scientific reports 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Hertz-Fowler, C.; Noyes, H.; Brenchley, R.; Goodhead, I.; Hall, N.; D’Amore, R. Phylogenomics of Acanthamoeba species: The Acanthamoeba whole genome shotgun (WGS) project. 2015. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJEB7687 (accessed on 20 December 2022).
- Xuan, Y.; Chung, B.; Hong, Y.; Kong, H.; Hahn, T.; Chung, D. Keratitis by Acanthamoeba triangularis: Report of cases and characterization of isolates. Korean J. Parasitol. 2008, 46, 157–164. [Google Scholar] [CrossRef]
- Scheid, P.L.; Gruen, A.-L.; Michel, R. Genotyping of Acanthamoeba isolates from clinical and environmentalsamples in Germany. 2014; unpublished. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KJ476526.1 (accessed on 20 December 2022).
- Ives, J.M.P. Caracterização e filogenia moleculares de Acanthamoeba. Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brazil, 2001. Available online: https://www.teses.usp.br/teses/disponiveis/42/42135/tde-01112001-123050/publico/tde.pdf (accessed on 20 December 2022).
- Byers, T.J.; Gast, R.J.; Ledee, D.R.; Stothard, D.R.; Fuerst, P.A. Genotyping of Acanthamoeba reference strains. 2010, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Alcon Research, L. Acanthamoeba ATCC 30461 transcriptome kinetics on contact lens materials. 2022. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA905484 (accessed on 20 December 2022).
- Crary, M.; Booton, G.B.; Fuerst, P.A. The 18S rRNA sequence from Acanthamoeba jacobsi ATCC 30732. 2015, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Schroeder-Diedrich, J.M.; Fuerst, P.A.; Byers, T.J. Group-I introns with unusual sequences occur at three sites in nuclear 18S rRNA genes of Acanthamoeba lenticulata. Curr. Genet. 1998, 34, 71–78. [Google Scholar] [CrossRef]
- Hewett, M.K. Characterisation of bacterial symbionts in amoebae. Ph.D. Thesis, University of South Australia and CRC for Water Quality and Treatment Centre, Boliver, SA, Australia, 2006. [Google Scholar]
- Patterson, D.J.; Sogin, M.L. Eukaryote origins and protistan diversity. In The origin and evolution of prokaryotic and eukaryotic cells; World Scientific: Hackensack, NJ, USA, 1993; pp. 13–46. [Google Scholar]
- Liu, H.; Moon, E.; Yu, H.; Jeong, H.; Hong, Y.; Kong, H.; Chung, D. Evaluation of taxonomic validity of four species of Acanthamoeba: A. divionensis, A. paradivionensis, A. mauritaniensis, and A. rhysodes, inferred from molecular analyses. Korean J. Parasitol 2005, 43, 7–13. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Ha, Y.; Lee, S.; Hong, Y.; Kong, H.; Chung, D. Genetic diversity of Acanthamoeba isolates from ocean sediments. Korean J. Parasitol 2006, 44, 117–125. [Google Scholar] [CrossRef][Green Version]
- Coronado-Velazquez, D.; Silva-Olivares, A.; Castro-Munozledo, F.; Lares-Jimenez, L.F.; Rodriguez-Anaya, L.Z.; Shibayama, M.; Serrano-Luna, J. Acanthamoeba mauritaniensis genotype T4D: An environmental isolate displays pathogenic behavior. Parasitol Int 2020, 74, 102002. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, G.; Sharma, S.; Garg, P.; Aggarwal, R.K. Use of 18S rRNA gene-based PCR assay for diagnosis of Acanthamoeba keratitis in non-contact lens wearers in India. J. Clin. Microbiol. 2003, 41, 3206–3211. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.; Molestina, R. Acanthamoeba 18S rRNA gene sequences obtained by ATCC staff. 2014, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Crary, M.J.; Booton, G.C.; Lares-Villa, F.; Shoff, M.; Joslin, C.; Tu, E.; Fuerst, P.A. Legionella and Pseudomonas Associated with the Protist Acanthamoeba during a Keratitis Outbreak. 2014; unpublished. Available online: https://www.ncbi.nlm.nih.gov/popset?DbFrom=nuccore&Cmd=Link&LinkName=nuccore_popset&IdsFromResult=1707530989 (accessed on 20 December 2022).
- Sriram, R.; Shoff, M.; Booton, G.; Fuerst, P.; Visvesvara, G.S. Survival of Acanthamoeba cysts after desiccation for more than 20 years. J. Clin. Microbiol. 2008, 46, 4045–4048. [Google Scholar] [CrossRef]
- Gast, R.J.; Byers, T.J.; Fuerst, P.A. 18S rRNA gene sequences from isolates of Acanthamoeba associated with keratitis. 2017, unpublished. Available online: https://u.osu.edu/acanthamoeba/acanthamoeba-isolates-at-atcc-or-ccap/ (accessed on 20 December 2022).
- Ledee, D.R.; Hay, J.; Byers, T.J.; Seal, D.V.; Kirkness, C.M. Acanthamoeba griffini. Molecular characterization of a new corneal pathogen. Investig. Ophthalmol Vis. Sci. 1996, 37, 544–550. [Google Scholar]
- Tegel, L.; De Llano, E.; Schoenthaler, S.; Walochnik, J.; Michel, R.; Barisic, I. Draft Whole Genome Sequence of Acanthamoeba lenticulata Strain 72/2. 2017. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA360052 (accessed on 20 December 2022).
- Horn, M.; Fritsche, T.R.; Linner, T.; Gautom, R.K.; Harzenetter, M.D.; Wagner, M. Obligate bacterial endosymbionts of Acanthamoeba spp. related to the beta-Proteobacteria: Proposal of ‘Candidatus Procabacter acanthamoebae’gen. nov., sp. nov. Int J. Syst Evol Microbiol 2002, 52, 599–605. [Google Scholar] [CrossRef]
- Walochnik, J.; Obwaller, A.; Aspöck, H. Correlations between morphological, molecular biological, and physiological characteristics in clinical and nonclinical isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 2000, 66, 4408–4413. [Google Scholar] [CrossRef]
- Walochnik, J.; Hassl, A.; Simon, K.; Benyr, G.; AspoÈck, H. Isolation and identification by partial sequencing of the 18S ribosomal gene of free-living amoebae from necrotic tissue of Basiliscus plumifrons (Sauria: Iguanidae). Parasitol Res. 1999, 85, 601–603. [Google Scholar] [CrossRef]
- Walochnik, J.; Haller-Schober, E.; Kölli, H.; Picher, O.; Obwaller, A.; Aspöck, H. Discrimination between Clinically Relevant and Nonrelevant Acanthamoeba Strains Isolated from Contact Lens-Wearing Keratitis Patients in Austria. J. Clin. Microbiol. 2000, 38, 3932–3936. [Google Scholar] [CrossRef] [PubMed]
- Heinz, E.; Kolarov, I.; Kästner, C.; Toenshoff, E.R.; Wagner, M.; Horn, M. An Acanthamoeba sp. containing two phylogenetically different bacterial endosymbionts. Environ. Microbiol. 2007, 9, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Michel, R.; Steinert, M.; Zoeller, L.; Hauroeder, B.; Henning, K. Free-living amoebae may serve as hosts for the Chlamydia-like bacterium Waddlia chondrophila isolated from an aborted bovine foetus. Acta Protozool. 2004, 43, 37–42. [Google Scholar]
- Karlyshev, A.V. Remarkable features of mitochondrial DNA of Acanthamoeba polyphaga Linc Ap-1, revealed by whole-genome sequencing. Microbiol. Resour. Announc. 2019, 8, e00430-19. [Google Scholar] [CrossRef] [PubMed]
- McKellar, M.S.; Mehta, L.R.; Greenlee, J.E.; Hale, D.C.; Booton, G.C.; Kelly, D.J.; Fuerst, P.A.; Sriram, R.; Visvesvara, G.S. Fatal granulomatous Acanthamoeba encephalitis mimicking a stroke, diagnosed by correlation of results of sequential magnetic resonance imaging, biopsy, in vitro culture, immunofluorescence analysis, and molecular analysis. J. Clin. Microbiol. 2006, 44, 4265–4269. [Google Scholar] [CrossRef] [PubMed]
- Ledee, D.R.; Seal, D.V.; Byers, T.J. Confirmatory evidence from 18S rRNA gene analysis for in vivo development of propamidine resistance in a temporal series of Acanthamoeba ocular isolates from a patient. Antimicrob. Agents Chemother. 1998, 42, 2144–2145. [Google Scholar] [CrossRef]
| Types of Molecular Data Collected for an Isolate | Cultures Available from ATCC | Available in BEI (Not in ATCC) | Available in CCAP (Not in ATCC) | Discontinued or Unavailable from ATCC or BEI or CCAP | Totals |
|---|---|---|---|---|---|
| Both 18S and 16S-like rRNA | 48 | 0 | 3 | 1 | 52 |
| only 18S rRNA | 79 | 9 | 4 | 5 | 97 |
| only 16S-like rRNA | 2 | 0 | 0 | 0 | 2 |
| only allozymes and/or RFLP | 14 | 0 | 0 | 0 | 14 |
| no molecular studies | 20 | 0 | 1 | 4 | 25 |
| total | 163 | 9 | 8 | 10 | 190 |
| ATCC/CCAP ID | Strain/Species Name | Whole Genome Shotgun Project (WGS) |
|---|---|---|
| ATCC 30010 | AEYA | |
| A. castellanii Neff | AHJI | |
| JAJGAP | ||
| ATCC 30137 | A. astronyxis | CDFH 1 |
| ATCC 30171 | A. culbertsoni strain A1 | CDFF |
| ATCC 30461 | A. polyphaga | SRX18334599 |
| ATCC 30841 | A. lenticulata isolate PD2S | CDFG |
| ATCC 30870 | A. palestinensis Reich | CDFA 2 |
| ATCC 30872 | A. sp. strain Page 45 | CDFK 3 |
| ATCC 30973 | A. rhysodes isolate Singh | CDFC |
| ATCC 50240 | A. lugdenensis isolate L3a | CDFB |
| ATCC 50241 | A. quina isolate Vil3 | CDFN |
| ATCC 50253 | A. mauritaniensis isolate 1652 | CDFE |
| ATCC 50254 | A. triangularis isolate SH621 | CDFD 4 CACVKS |
| ATCC 50370 | A. castellanii (isolate Ma) | CDFL |
| ATCC 50496 | A. sp. (strain Galka) | CDFJ 5 |
| ATCC 50704 | A. lenticulata strain 72/2 | MSTW |
| ATCC 50739 | A. castellanii strain C3 | JAJGAO |
| ATCC PRA-287 | A. comandoni strain Pb30/40 | MRZZ |
| CCAP 1501/18 | A. polyphaga strain Linc Ap-1 | LQHA |
| CCAP 1501/19 | A. pyriformis CR15 | SRA SRX2163158 |
| Sequence Type | # | % Standard Strains | % Survey Strains |
|---|---|---|---|
| T1 | 2 | 1.32% | 0.43% |
| T2 | 3 | 1.98% | 2.20% |
| T2/6a | 2 | 1.32% | 0.65% |
| T2/6b | 0 | 0.00% | 0.43% |
| T2/6c | 1 | 0.66% | 1.72% |
| T3 | 5 | 3.31% | 5.60% |
| T4 (total) | 98 | 64.90% | 70.30% |
| T4A | 42 | 27.81% | 25.52% |
| T4B | 18 | 11.92% | 12.59% |
| T4C | 5 | 3.31% | 7.11% |
| T4D | 12 | 8.6% | 10.60% |
| T4E | 13 | 9.62% | 6.08% |
| T4F | 2 | 1.32% | 2.23% |
| T4-neff | 6 | 7.05% | 4.98% |
| T5 | 18 | 11.92% | 6.98% |
| T6 | 4 | 2.65% | 1.75% |
| T7 | 1 | 0.66% | 0.18% |
| T8 | 1 | 0.66% | 0.03% |
| T9 | 2 | 1.32% | 0.44% |
| T10 | 1 | 0.66% | 0.32% |
| T11 | 3 | 1.92% | 1.91% |
| T12 | 2 | 0.64% | 0.43% |
| T13 | 2 | 0.64% | 1.06% |
| T14 | 0 | 0.00% | 0.08% |
| T15 | 1 | 0.64% | 2.60% |
| T16 | 0 | 0.00% | 0.35% |
| T17 | 0 | 0.00% | 0.31% |
| T18 | 1 | 0.64% | 0.50% |
| T19 | 1 | 0.64% | 0.10% |
| T20 | 0 | 0.00% | 0.55% |
| T21 | 1 | 0.64% | 0.02% |
| T22 | 0 | 0.00% | 0.02% |
| T23 | 0 | 0.00% | 0.03% |
| Morphological Group | Species Name | ATCC Strain # | CCAP Strain # | Strain Name | Nuclear 18S rRNA Gene | Mito. 16S-like Gene | Genotype Group |
|---|---|---|---|---|---|---|---|
| Group 1 | A. astronyxis | 30137 | 1534/1 | Y | Y | T7 | |
| A. byersi | PRA-411 | Y | N | T18 | |||
| A. comandoni | 30135 | 1501/5 | Y | Y | T9 | ||
| A. echinulata(1) | 50239 | 278 | Y (1) | N | T4D? (1) | ||
| A. tubiashi | 30867 | OC-15C | Y | Y | T8 | ||
| Group 2 | A. castellanii | 30011 | 1501/2a | Ac30 | Y | Y | T4A |
| A. divionensis | 50238 | AA2 | Y | Y | T4D | ||
| A. griffini | 30731 | 1501/4 | S-7 | Y | Y | T3 | |
| A. hatchetti | 30730 | BH-2 | Y | N | T11 | ||
| A. lugdunensis | 50240 | L3a | Y | N | T4A | ||
| A. mauritaniensis | 50253 | 1652 | Y | Y | T4D | ||
| A. paradivionensis | 50251 | AA1 | Y | Y | T4D | ||
| A. pearcei | 50435 | 205-1 | Y | N | T3 | ||
| A. polyphaga | 30871 | 1501/3a | Page-23 | Y | N | T4E | |
| A. quina | 50241 | Vil3 | Y | N | T4A | ||
| A. rhysodes(2) | 30973 | 1534/3 | Singh | Y | ? | T4D | |
| A. stevensoni | 50388 | RB-F-1 | Y | N | T11 | ||
| A. terricola | 30134 | N? | Y | T4-neff | |||
| A. triangularis | 50254 | SH 621 | Y | N | T4F | ||
| C. operculata | 50243 | Y | N | T6 | |||
| A. sawyeri(3) | 50656 | NR46460 | CDC:0484:V017 | Y | Y | T4B | |
| Group 3 | A. culbertsoni | 30171 | 1501/6 | Lilly A1 | Y | Y | T10 |
| A. healyi | 30866 | OC-3A | Y | Y | T12 | ||
| A. jacobsi | 30732 | Y | N | T15 | |||
| A. lenticulata | 30841 | PD2 | Y | Y | T5 | ||
| A. palestinensis | 30870 | 1547/1 | Reich | Y | Y | T2 | |
| A. pustulosa | 50252 | GE 3a | Y | N | T2 | ||
| A. royreba | 30884 | OR (Oak Ridge) | Y | Y | T4D | ||
| unknown | A. giganteum(4) | 50670 | 25-349-MX | Y | N | T4A |
| Species | # Sequence Types | Sequence Types |
|---|---|---|
| A. astronyxis | 2 | T7 T9 |
| A. castellanii | 6 | T1 T4A T4B T4C T4D T4-neff |
| A. culbertsoni | 3 | T4A T4B T10 |
| A. hatchetti | 4 | T4B T4E T6 T11 |
| A. mauritaniensis | 2 | T4D T4E |
| A. palestinensis | 3 | T2 T6 T2/6C |
| A. polyphaga | 6 | T3 T4A T4B T4D T4E T2/6A |
| A. rhysodes | 2 | T4A T4D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuerst, P.A. The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections. Microorganisms 2023, 11, 295. https://doi.org/10.3390/microorganisms11020295
Fuerst PA. The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections. Microorganisms. 2023; 11(2):295. https://doi.org/10.3390/microorganisms11020295
Chicago/Turabian StyleFuerst, Paul A. 2023. "The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections" Microorganisms 11, no. 2: 295. https://doi.org/10.3390/microorganisms11020295
APA StyleFuerst, P. A. (2023). The Status of Molecular Analyses of Isolates of Acanthamoeba Maintained by International Culture Collections. Microorganisms, 11(2), 295. https://doi.org/10.3390/microorganisms11020295






