Cell-Free Demineralized Bone Matrix for Mesenchymal Stem Cells Survival and Colonization
Abstract
1. Introduction
2. Results
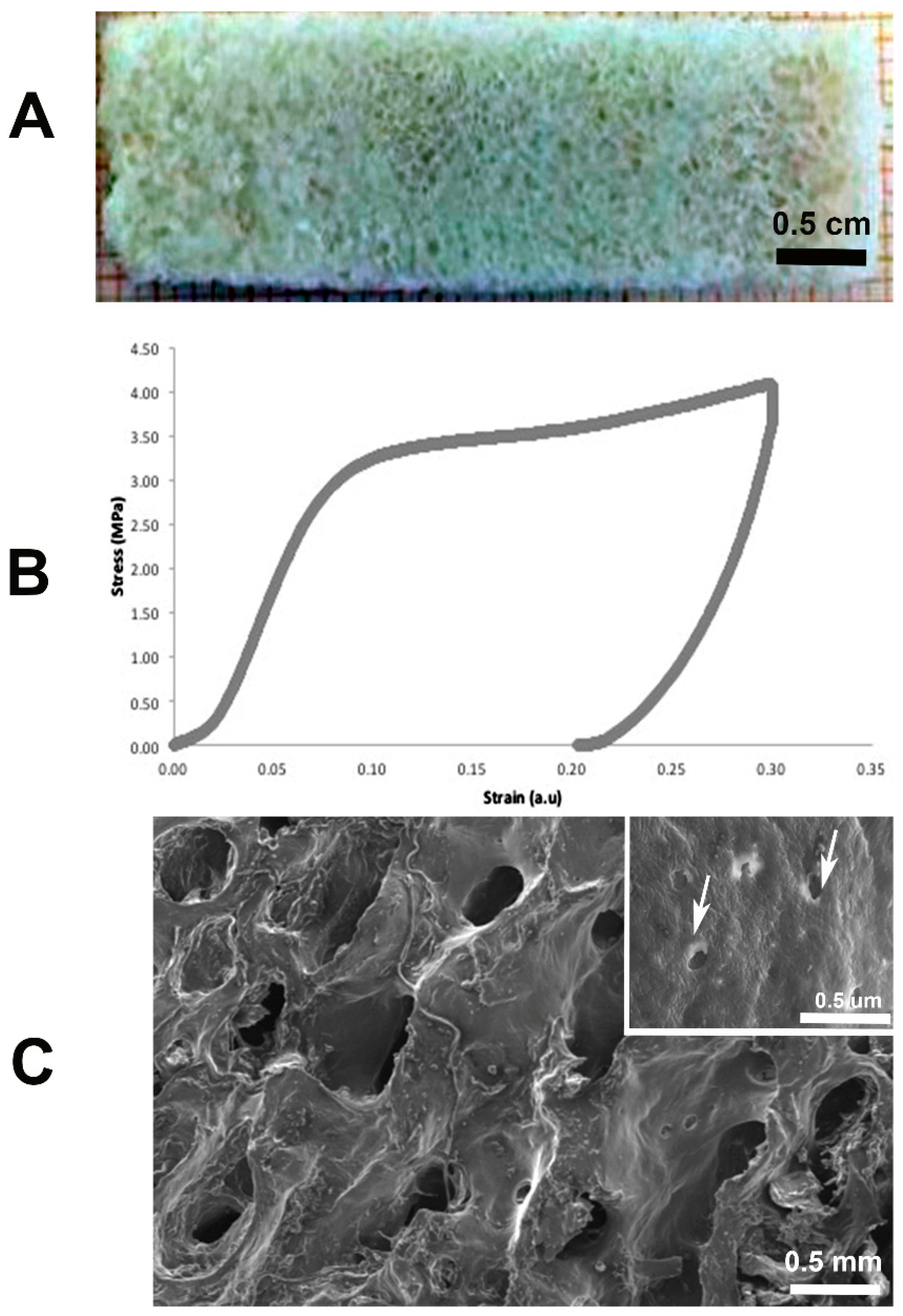
2.1. Bone Demineralization and Mechanical Tests
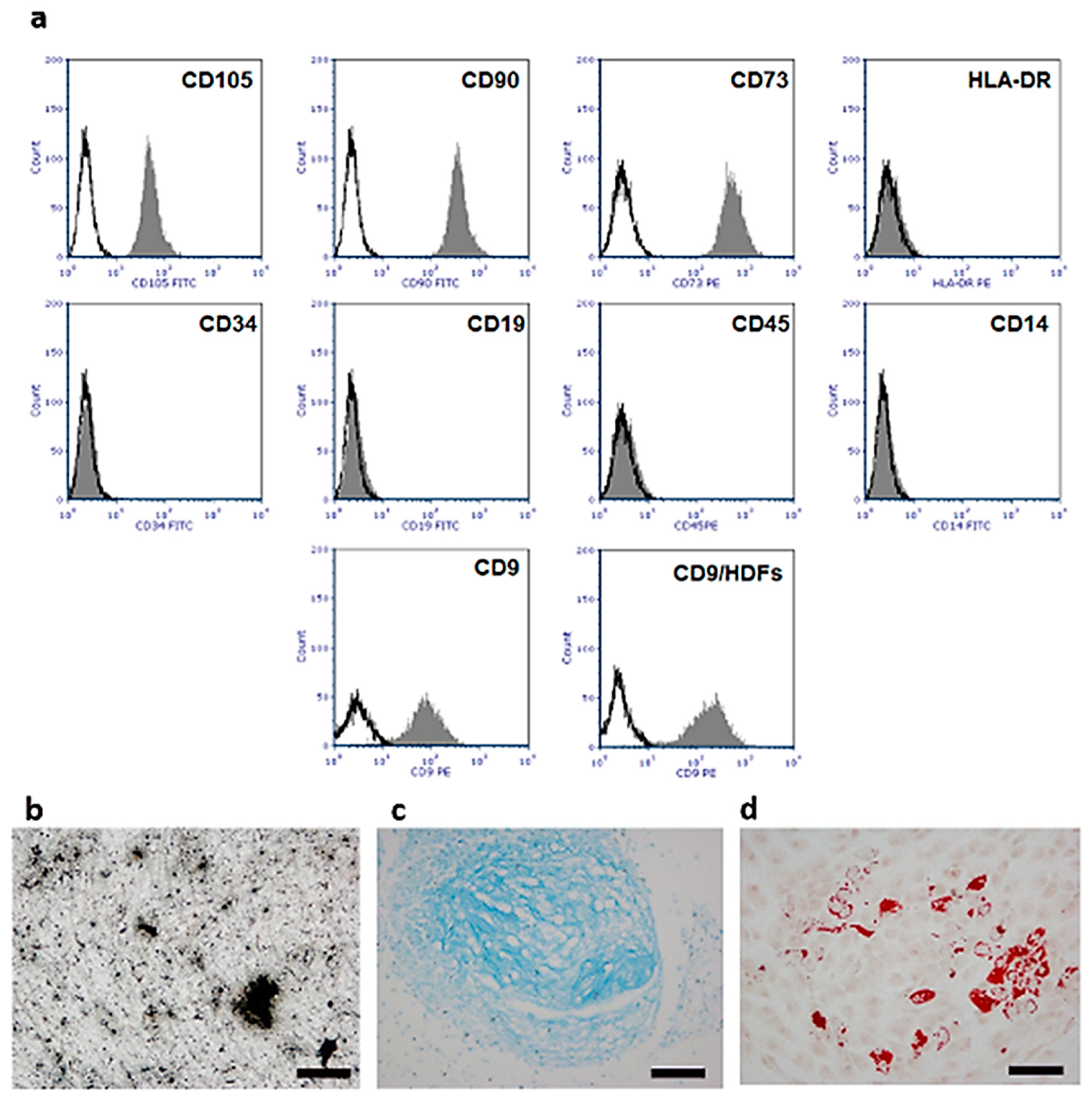
2.2. Isolation and Characterization of Human Umbilical Cord Stem Cells (hUC-SCs)
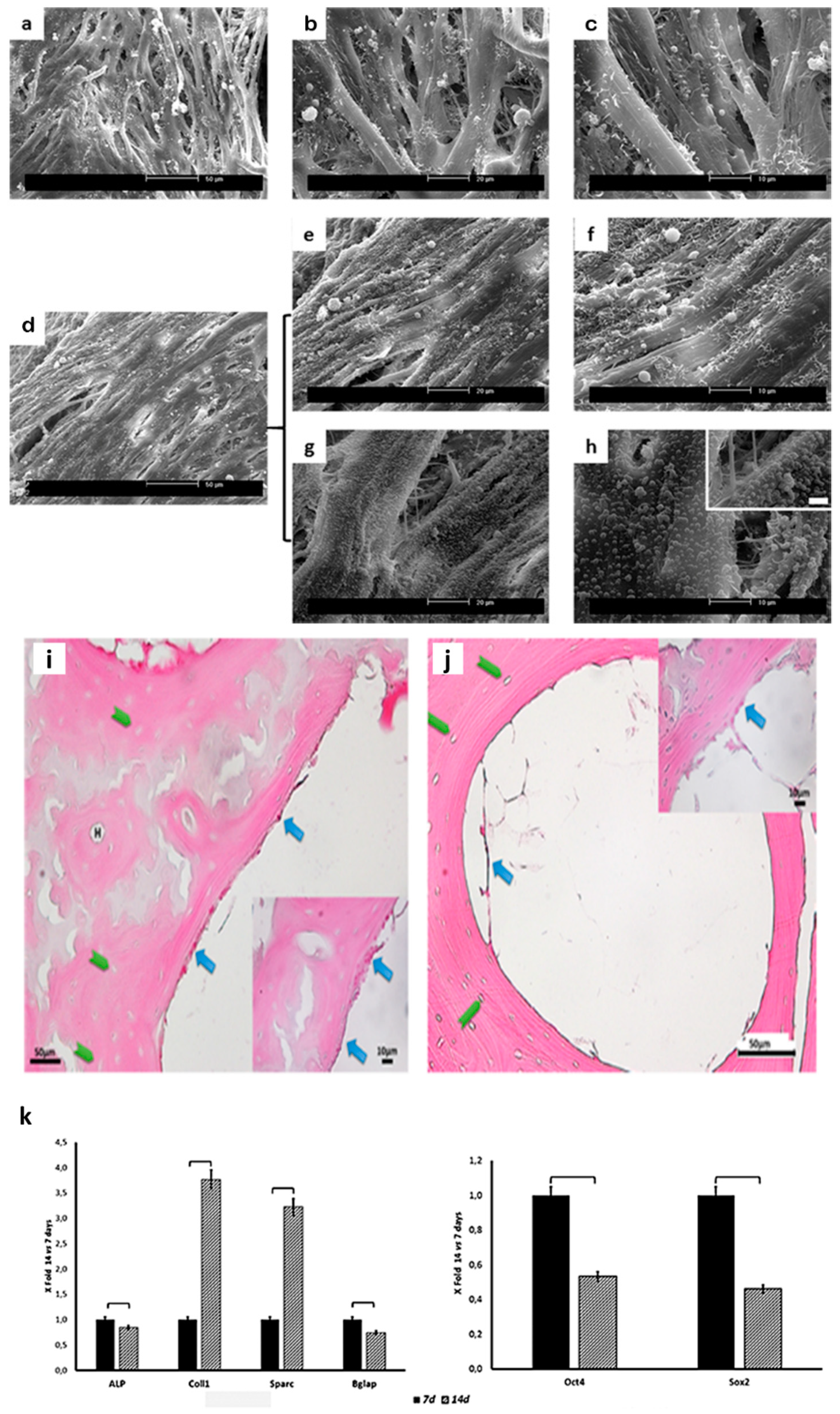
2.3. Scaffold Colonization
2.4. Gene Expression
3. Discussion
4. Materials and Methods
4.1. Bone Demineralization and Mechanical Tests
4.2. Isolation and Culture of Human Umbilical Cord Mesenchymal Stem Cells (hUC-SCs)
4.3. hUC-SC In Vitro Differentiation
4.4. Scaffold Seeding and Culture
4.5. Scanning Electron Microscopy (SEM)
4.6. Light Microscopy (LM)
4.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
4.8. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murphy, W.L.; McDevitt, T.C.; Engler, A.J. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547–557. [Google Scholar] [CrossRef]
- Mattioli-Belmonte, M.; Vozzi, G.; Kyriakidou, K.; Pulieri, E.; Lucarini, G.; Vinci, B.; Pugnaloni, A.; Biagini, G.; Ahluwalia, A. Rapid-prototyped and salt-leached PLGA scaffolds condition cell morpho-functional behavior. J. Biomed. Mater. Res. A 2008, 85, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Santoro, M.; Perale, G. Polymeric scaffolds as stem cell carriers in bone repair. J. Tissue Eng. Regen. Med. 2015, 9, 1093–1119. [Google Scholar] [PubMed]
- Ciardelli, G.; Gentile, P.; Chiono, V.; Mattioli-Belmonte, M.; Vozzi, G.; Barbani, N.; Giusti, P. Enzymatically crosslinked porous composite matrices for bone tissue regeneration. J. Biomed. Mater. Res. A 2010, 92, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Jelen, C.; Mattei, G.; Montemurro, F.; De Maria, C.; Mattioli-Belmonte, M.; Vozzi, G. Bone scaffolds with homogeneous and discrete gradient mechanical properties. Mat. Sci. Eng. C. 2013, 3, 28–36. [Google Scholar] [CrossRef]
- Rajangam, T.; An, S.S. Fibrinogen and fibrin-based micro and nano scaffolds incorporated with drugs, proteins, cells and genes for therapeutic biomedical applications. Int. J. Nanomed. 2013, 8, 3641–3662. [Google Scholar]
- Kruger, T.E.; Miller, A.H.; Wang, J. Collagen scaffolds in bone sialoprotein-mediated bone regeneration. Sci. World J. 2013. [Google Scholar] [CrossRef]
- Vindigni, V.; Cortivo, R.; Iacobellis, L.; Abatangelo, G.; Zavan, B. Hyaluronan benzyl ester as a scaffold for tissue engineering. Int. J. Mol. Sci. 2009, 10, 2972–2985. [Google Scholar] [CrossRef]
- Gloria, A.; De Santis, R.; Ambrosio, L. Polymer-based composite scaffolds for tissue engineering. J. Appl. Biomater. Biomech. 2010, 8, 57–67. [Google Scholar] [PubMed]
- Boccaccini, A.R.; Blaker, J.J. Bioactive composite materials for tissue engineering scaffolds. Expert. Rev. Med. Devices 2005, 2, 303–317. [Google Scholar] [CrossRef]
- Shi, X.; Sitharaman, B.; Pham, Q.P.; Liang, F.; Wu, K.; Edward Billups, W.; Wilson, L.J.; Mikos, A.G. Fabrication of porous ultra-short single-walled carbon nanotube nanocomposite scaffolds for bone tissue engineering. Biomaterials 2007, 28, 4078–4090. [Google Scholar] [CrossRef] [PubMed]
- Veetil, J.V.; Ye, K. Tailored carbon nanotubes for tissue engineering applications. Biotechnol. Prog. 2009, 25, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Mattioli-Belmonte, M.; Vozzi, G.; Whulanza, Y.; Seggiani, M.; Fantauzzi, V.; Orsini, G.; Ahluwalia, A. Tuning polycaprolactone–carbon nanotube composites for bone tissue engineering scaffolds. Mat. Sci. Eng. C 2012, 32, 152–159. [Google Scholar] [CrossRef]
- Whulanza, Y.; Battini, E.; Vannozzi, L.; Vomero, M.; Ahluwalia, A.; Vozzi, G. Electrical and Mechanical Characterisation of single wall carbon nanotubes-based composites for tissue engineering applications. J. Nanosci. Nanotechnol. 2013, 13, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Mattioli-Belmonte, M.; De Maria, C.; Vitale-Brovarone, C.; Baino, F.; Dicarlo, M.; Vozzi, G. Pressure-activated microsyringe (PAM) fabrication of bioactive glass-poly (lactic-co-glycolic acid) composite scaffolds for bone tissue regeneration. J. Tissue Eng. Regen. Med. 2017, 11, 1986–1997. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.A., Jr.; Hoehn, J.G. Use of commercial porcine skin for wound dressings. Plast. Reconstr. Surg. 1973, 52, 401–405. [Google Scholar] [CrossRef]
- Edgar, L.; McNamara, K.; Wong, T.; Tamburrini, R.; Katari, R.; Orlando, G. Heterogeneity of Scaffold Biomaterials in Tissue Engineering. Materials 2016, 9, 332. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hall, H.G.; Parry, G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 1982, 99, 31–68. [Google Scholar] [CrossRef]
- Lin, X.; Fang, X.; Wang, Q.; Hu, Z.; Chen, K.; Shan, Z.; Chen, S.; Wang, J.; Mo, J.; Ma, J.; et al. Decellularized allogeneic intervertebral disc: Natural biomaterials for regenerating disc degeneration. Oncotarget 2016, 7, 12121–12136. [Google Scholar] [CrossRef][Green Version]
- Saldin, L.T.; Cramer, M.C.; Velankar, S.S.; White, L.J.; Badylak, S.F. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater. 2017, 49, 1–15. [Google Scholar] [CrossRef]
- Cheng, C.W.; Solorio, L.D.; Alsberg, E. Decellularized tissue and cell-derived extracellular matrices as scaffolds for orthopaedic tissue engineering. Biotechn. Adv. 2014, 32, 462–484. [Google Scholar] [CrossRef]
- Roberts, T.T.; Rosenbaum, A.J. Bone grafts, bone substitutes and orthobiologics. The bridge between basic science and clinical advancements in fracture healing. Organogenesis 2012, 8, 114–124. [Google Scholar] [CrossRef]
- Mattei, G.; Ferretti, C.; Tirella, A.; Ahluwalia, A.; Mattioli-Belmonte, M. Decoupling the role of stiffness from other hydroxyapatite signalling cues in periosteal derived stem cell differentiation. Sci. Rep. 2015, 5, 10778. [Google Scholar] [CrossRef]
- Even-Ram, S.; Artym, V.; Yamada, K.M. Matrix control of stem cell fate. Cell 2006, 126, 645–647. [Google Scholar] [CrossRef]
- Eagle, M.J.; Rooney, P.; Kearney, J.N. Optimized demineralization of human cancellous bone by application of a vacuum. J. Biomed. Mater. Res. B 2015, 103, 1023–1029. [Google Scholar] [CrossRef]
- Chen, P.Y.; McKittric, J. Compressive mechanical properties of demineralized and deproteinized cancellous bone. J. Mech. Behav. Biomed. Mater. 2011, 4, 961–973. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for defining multipotent mesenchymal stem cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Halfon, S.; Abramov, N.; Grinblat, B.; Ginis, I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011, 20, 53–66. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Keane, T.J.; Swinehart, I.T.; Badylak, S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods 2015, 84, 25–34. [Google Scholar] [CrossRef]
- Xing, Q.; Yates, K.; Tahtinen, M.; Shearier, E.; Qian, Z.; Zhao, F. Decellularization of fibroblast cell sheets for natural extracellular matrix scaffold preparation. Tissue Eng. Part C Methods. 2014, 21, 77–87. [Google Scholar] [CrossRef]
- Voytik-Harbin, S.L.; Brightman, A.O. Small intestinal submucosa: A tissue derived extracellular matrix that promotes tissue-specific growth and differentiation of cells in vitro. Tissue Eng. 1998, 4, 157–174. [Google Scholar] [CrossRef]
- Badylak, S.F.; Freytes, D.O.; Gilbert, T.W. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009, 5, 1–13. [Google Scholar] [CrossRef]
- Porzionato, A.; Stocco, E.; Barbon, S.; Grandi, F.; Macchi, V.; De Caro, R. Tissue-Engineered Grafts from Human Decellularized Extracellular Matrices: A Systematic Review and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 4117. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, W.; Zhou, G.; Cao, Y.; Liu, W.; Zhang, Z.Y. Demineralized bone matrix-based microcarrier scaffold favors vascularized large bone regeneration in vivo in a rat model. J. Biomater. Appl. 2018, 33, 182–195. [Google Scholar] [CrossRef]
- Li, T.; Xia, M.; Gao, Y.; Chen, Y.; Xu, Y. Human umbilical cord mesenchymal stem cells: An overview of their potential in cell-based therapy. Expert Opin. Biol. Ther. 2015, 15, 1293–1306. [Google Scholar] [CrossRef]
- Wang, W.T.; Gu, P.; Qiu, F.C.; Zhang, L.N.; Zhang, Z.X.; Xie, B.C.; Dong, C.; Han, R.; Liu, H.M.; Yan, B.Y. Intravenous transplantation of Allograft hUC-MSC was more effective than subarachnoid transplantation of BM-MSCs in patients with Parkinson’s Syndrome and Secondary Parkinson’s Syndrome. J. Biomater. Tiss. Eng. 2016, 6, 158–164. [Google Scholar] [CrossRef]
- Moroncini, G.; Paolini, C.; Orlando, F.; Capelli, C.; Grieco, A.; Tonnini, C.; Agarbati, S.; Mondini, E.; Saccomanno, S.; Goteri, G.; et al. Mesenchymal stromal cells from human umbilical cord prevent the development of lung fibrosis in immunocompetent mice. PLoS ONE 2018, 13, e0196048. [Google Scholar] [CrossRef]
- Merlo, B.; Teti, G.; Mazzotti, E.; Ingrà, L.; Salvatore, V.; Buzzi, M.; Cerqueni, G.; Dicarlo, M.; Lanci, A.; Castagnetti, C.; et al. Wharton’s Jelly Derived Mesenchymal Stem Cells: Comparing Human and Horse. Stem Cell Rev. 2018, 14, 574–584. [Google Scholar] [CrossRef]
- Xu, J.; Sun, M.; Tan, Y.; Wang, H.; Wang, H.; Li, P.; Xu, Z.; Xia, Y.; Li, L.; Li, Y. Effect of matrix stiffness on the proliferation and differentiation of umbilical cord mesenchymal stem cells. Differentiation 2017, 96, 30–39. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Witkowska-Zimny, M.; Walenko, K.; Walkiewicz, A.E.; Pojda, Z.; Przybylski, J.; Lewandowska-Szumiel, M. Effect of substrate stiffness on differentiation of umbilical cord stem cells. Acta Biochim. Pol. 2016, 59, 261–264. [Google Scholar] [CrossRef]
- Duchamp de Lageneste, O.; Julien, A.; Abou-Khalil, R.; Frangi, G.; Carvalho, C.; Cagnard, N.; Cordier, C.; Conway, S.J.; Colnot, C. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun. 2018, 9, 773. [Google Scholar] [CrossRef]
- Manilay, Z.; Novitskaya, E.; Sadovnikov, E.; McKittrick, J. A comparative study of young and mature bovine cortical bone. Acta Biomater. 2013, 9, 5280–5288. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Sulochana, KN. Manual of Medical Laboratory Techniques; JP Medical Ltd.: London, UK, 2012; p. 388. [Google Scholar]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef]
| Genes | 7 Days | 14 Days | |||
|---|---|---|---|---|---|
| Mean | DS | Mean | DS | p | |
| ALP | 2.93 × 10−4 | 1.47 × 10−5 | 2.46 × 10−4 | 1.23 × 10−5 | 0.039 |
| COL1 | 1.33 × 10−1 | 6.64 × 10−3 | 4.99 × 10−1 | 2.50 × 10−2 | 0.0096 |
| SPARC | 1.52 × 10−1 | 7.59 × 10−3 | 4.89 × 10−1 | 2.44 × 10−2 | 0.013 |
| BGLAP | 4.96 × 10−6 | 2.48 × 10−7 | 3.64 × 10−6 | 1.82 × 10−7 | 0.047 |
| Oct4 | 9.79 × 10−4 | 4.89 × 10−5 | 5.21 × 10−4 | 2.60 × 10−5 | 0.034 |
| Sox2 | 3.93 × 10−5 | 1.97 × 10−6 | 1.77 × 10−5 | 8.85 × 10−7 | 0.042 |
| Genes | Detected Transcript | Primer Forward (5′→3′) | Primer Reverse (3′→5′) | Amplicon Length (bp) |
|---|---|---|---|---|
| ALP | NM_007431 | GGCCAGCTACACCACAACA | CTGAGCGTTGGTGTTATATGTCTT | 96 |
| COL1 | NM_000088.3 | CCAACCCTTCCACCTTTGGAAGT | CCGGAGGTCCACAAAGCTGAA | 132 |
| SPARC | NM_003118.3 | CCTGAGGCTGTAACTGAGAGAAAG | GTGGGAGGGGAAACAAGAAGATAA | 142 |
| BGLAP | NM_199173 | GACTGTGACGAGTTGGCTGA | GCCCACAGATTCCTCTTCTG | 119 |
| Sox2 | NM_003106.3 | ACACCAATCCCATCCACACT | GCAAACTTCCTGCAAAGCTC | 198 |
| Oct4 | NM_203289.4 | AGCGAACCAGTATCGAGAAC | GCCTCAAAATCCTCTCGTTG | 199 |
| GUSB * | NM_000181.2 | AAACGATTGCAGGGTTTCAC | TCTCGTCGGTGACTGTTCA | 81 |
| GAPDH * | NM_002046.3 | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC | 200 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattioli-Belmonte, M.; Montemurro, F.; Licini, C.; Iezzi, I.; Dicarlo, M.; Cerqueni, G.; Coro, F.; Vozzi, G. Cell-Free Demineralized Bone Matrix for Mesenchymal Stem Cells Survival and Colonization. Materials 2019, 12, 1360. https://doi.org/10.3390/ma12091360
Mattioli-Belmonte M, Montemurro F, Licini C, Iezzi I, Dicarlo M, Cerqueni G, Coro F, Vozzi G. Cell-Free Demineralized Bone Matrix for Mesenchymal Stem Cells Survival and Colonization. Materials. 2019; 12(9):1360. https://doi.org/10.3390/ma12091360
Chicago/Turabian StyleMattioli-Belmonte, Monica, Francesca Montemurro, Caterina Licini, Iolanda Iezzi, Manuela Dicarlo, Giorgia Cerqueni, Florinda Coro, and Giovanni Vozzi. 2019. "Cell-Free Demineralized Bone Matrix for Mesenchymal Stem Cells Survival and Colonization" Materials 12, no. 9: 1360. https://doi.org/10.3390/ma12091360
APA StyleMattioli-Belmonte, M., Montemurro, F., Licini, C., Iezzi, I., Dicarlo, M., Cerqueni, G., Coro, F., & Vozzi, G. (2019). Cell-Free Demineralized Bone Matrix for Mesenchymal Stem Cells Survival and Colonization. Materials, 12(9), 1360. https://doi.org/10.3390/ma12091360








