Protein-Based Systems for Translational Regulation of Synthetic mRNAs in Mammalian Cells
Abstract
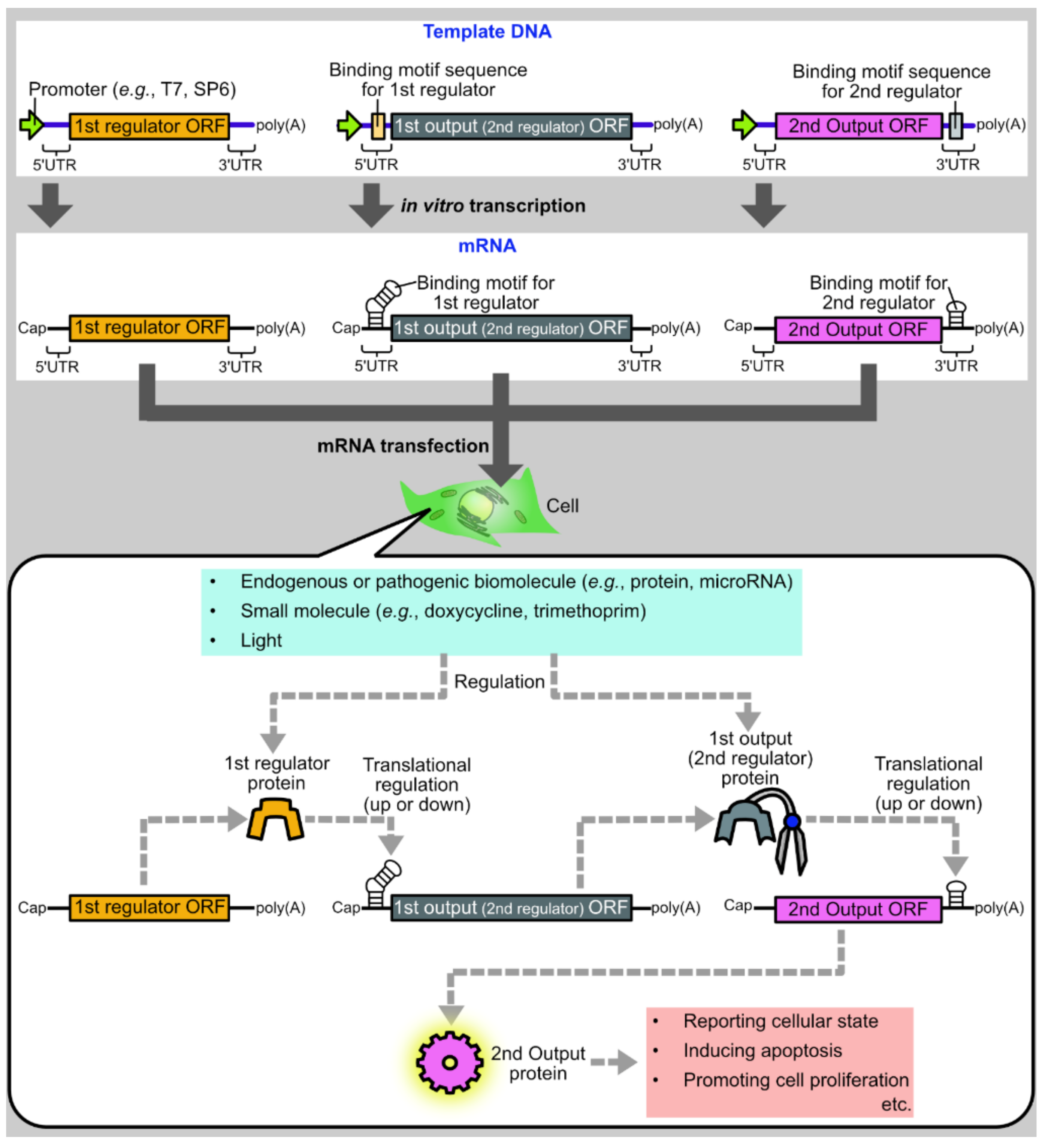
1. Introduction
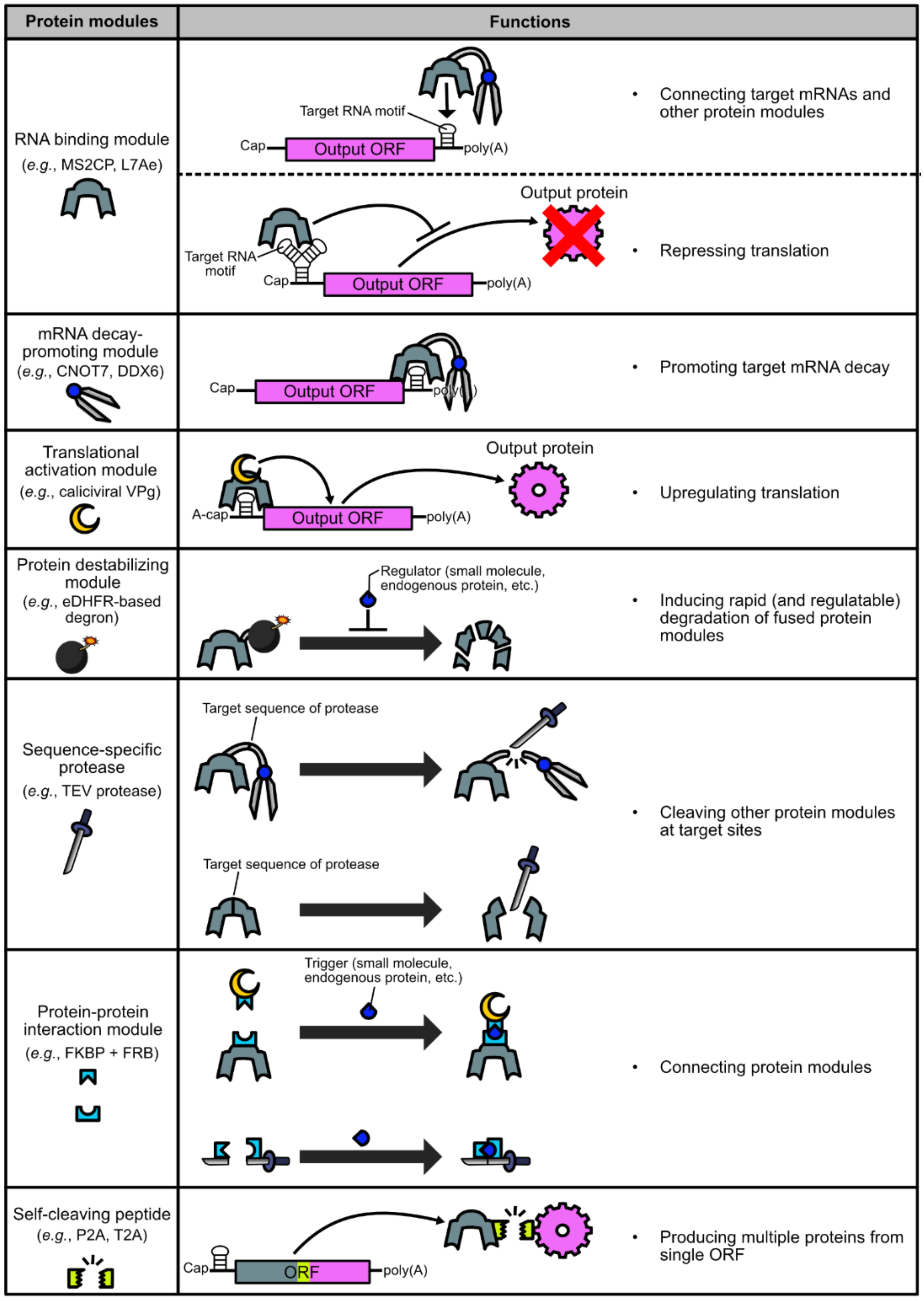
2. Basic Protein Modules for Protein-Based Translational Regulation Systems
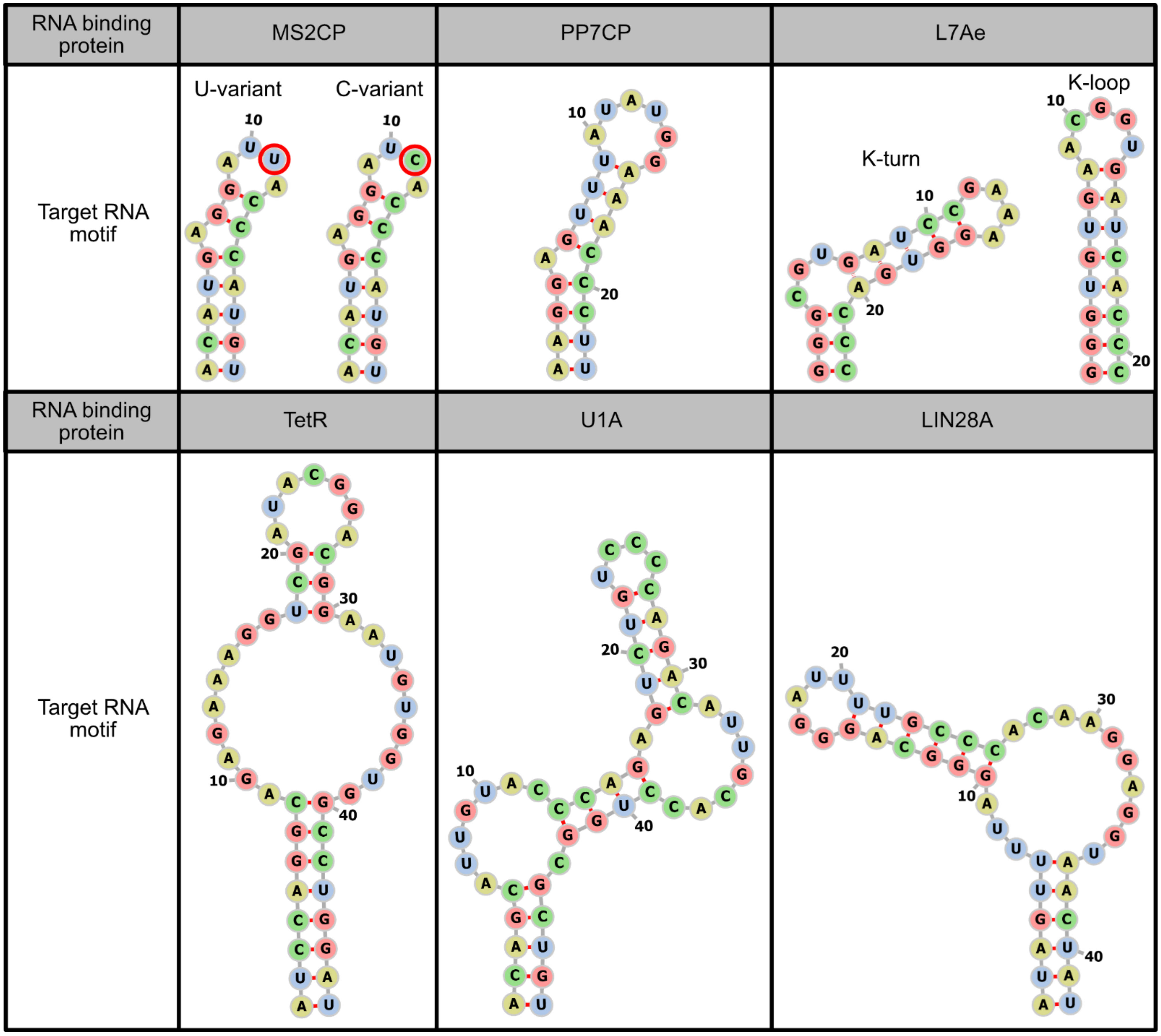
2.1. Binding to Target mRNAs
2.2. Promoting Target mRNA Decay
2.3. Activating Target mRNA Translation
2.4. Destabilizing Proteins
2.5. Cleaving Proteins
2.6. Combining Separate Proteins
2.7. Producing Multiple Proteins from a Single ORF
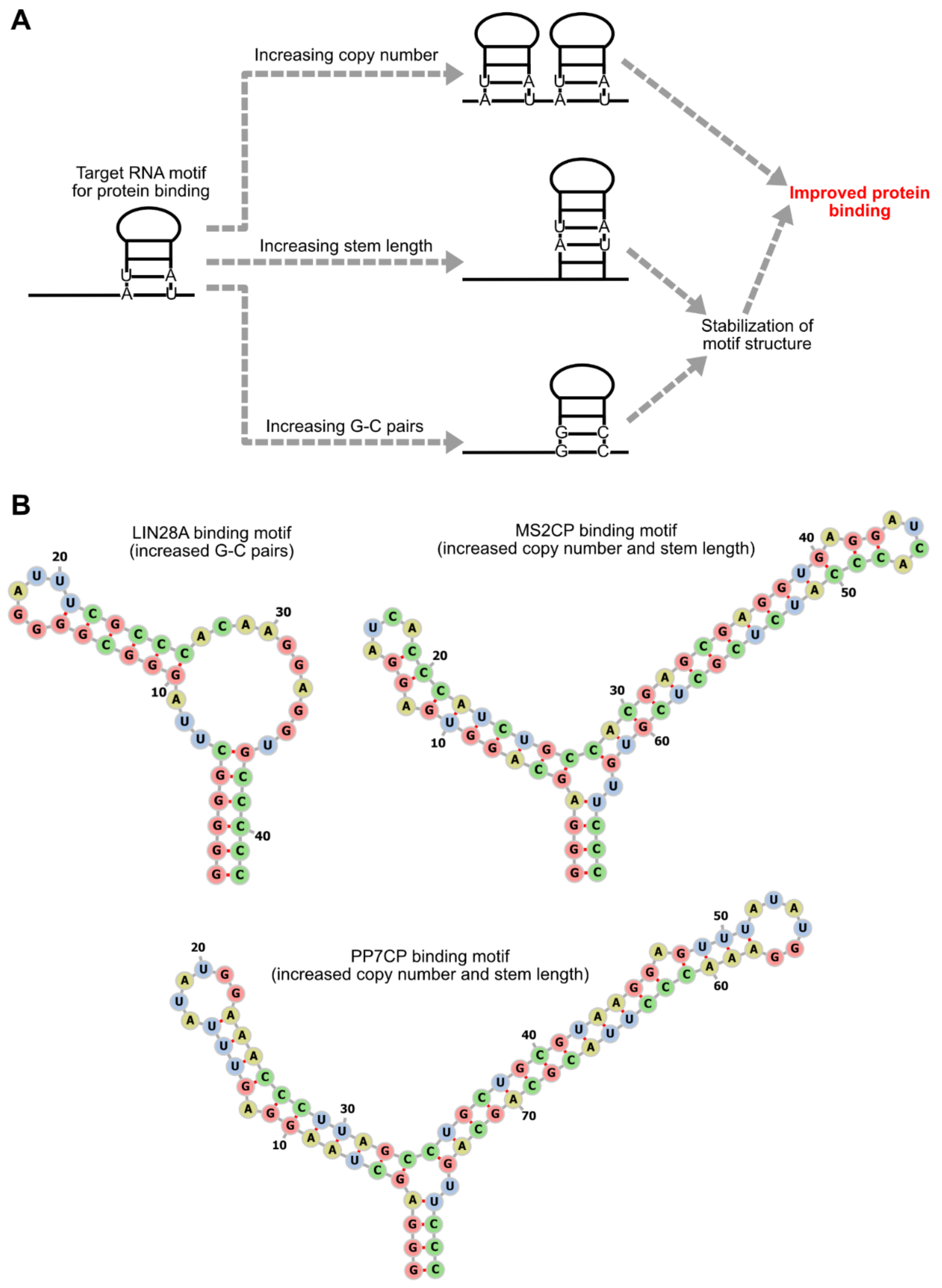
3. Optimization of RNA Motifs and Protein Modules
3.1. RNA Motif Optimization
3.2. Protein Module Optimization
4. Input Signals Utilized for Translational Regulation
4.1. Deliberate Regulation by External Cues
4.2. Endogenous or Pathogenic Protein-Responsive Autonomous Regulation
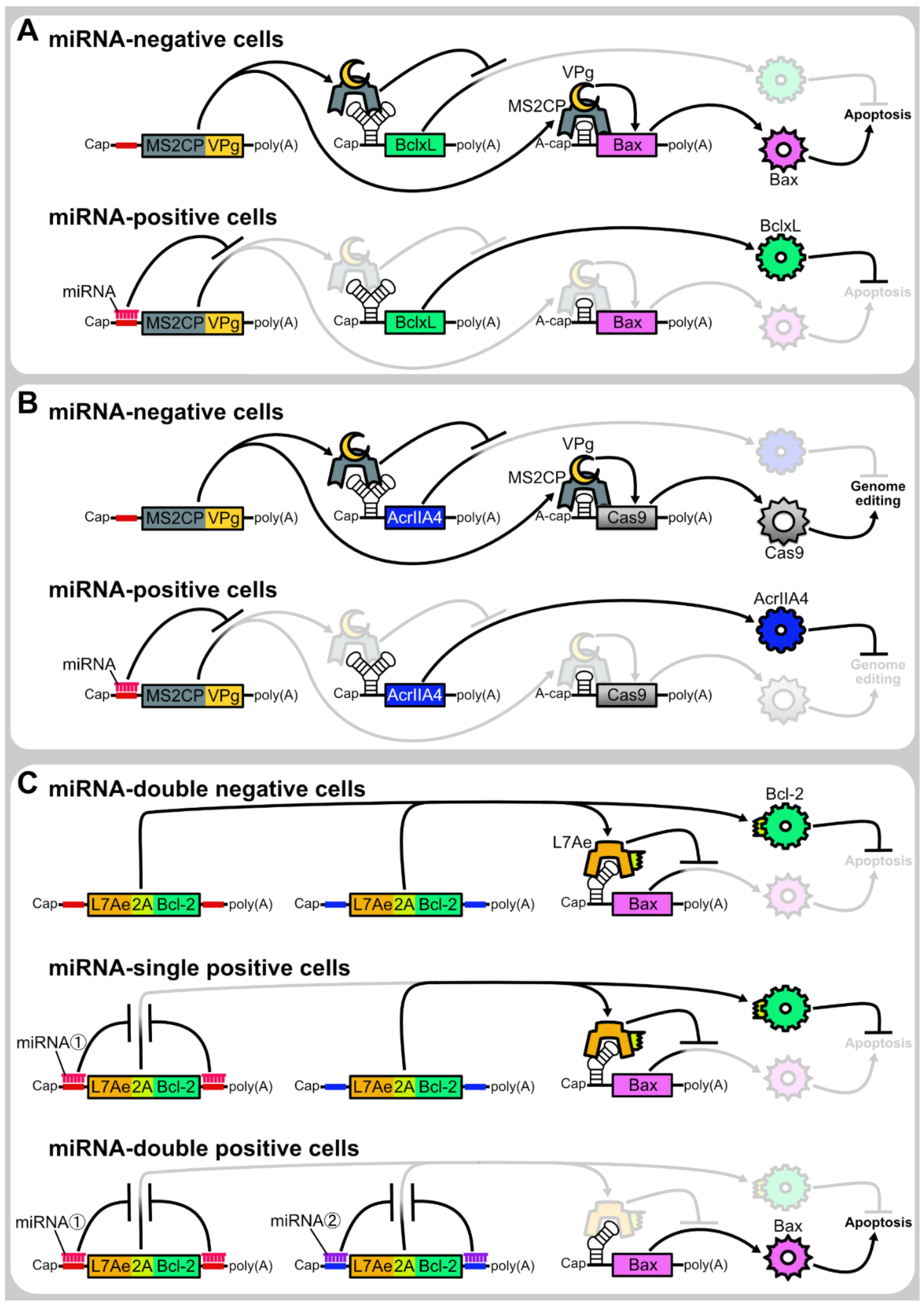
4.3. microRNA-Responsive Autonomous Regulation
5. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Solinís, M.Á.; del Pozo-Rodríguez, A. Nanomedicines to Deliver MRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Wroblewska, L.; Kitada, T.; Endo, K.; Siciliano, V.; Stillo, B.; Saito, H.; Weiss, R. Mammalian Synthetic Circuits with RNA Binding Proteins for RNA-Only Delivery. Nat. Biotechnol. 2015, 33, 839–841. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Ono, H.; Kawasaki, S.; Kuang, Y.; Fujita, Y.; Saito, H. Synthetic RNA-Based Logic Computation in Mammalian Cells. Nat. Commun. 2018, 9, 4847. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ding, S. Engineering L7Ae for RNA-Only Delivery Kill Switch Targeting CMS2 Type Colorectal Cancer Cells. ACS Synth. Biol. 2021, 10, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Saito, H. Caliciviral Protein-Based Artificial Translational Activator for Mammalian Gene Circuits with RNA-Only Delivery. Nat. Commun. 2020, 11, 1297. [Google Scholar] [CrossRef]
- Yang, J.; Ding, S. Chimeric RNA-Binding Protein-Based Killing Switch Targeting Hepatocellular Carcinoma Cells. Mol. Ther. - Nucleic Acids 2021, 25, 683–695. [Google Scholar] [CrossRef]
- Magadum, A.; Singh, N.; Kurian, A.A.; Munir, I.; Mehmood, T.; Brown, K.; Sharkar, M.T.K.; Chepurko, E.; Sassi, Y.; Oh, J.G.; et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020, 141, 1249–1265. [Google Scholar] [CrossRef]
- Yokobayashi, Y. Aptamer-Based and Aptazyme-Based Riboswitches in Mammalian Cells. Curr. Opin. Chem. Biol. 2019, 52, 72–78. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, C.Y.; Busby, K.N.; Alexander, S.C.; Devaraj, N.K. Light-Activated Control of Translation by Enzymatic Covalent MRNA Labeling. Angew. Chem. 2018, 130, 2872–2876. [Google Scholar] [CrossRef]
- Stripecke, R.; Oliveira, C.C.; McCarthy, J.E.; Hentze, M.W. Proteins Binding to 5’ Untranslated Region Sites: A General Mechanism for Translational Regulation of MRNAs in Human and Yeast Cells. Mol. Cell. Biol. 1994, 14, 5898–5909. [Google Scholar] [CrossRef]
- Endo, K.; Stapleton, J.A.; Hayashi, K.; Saito, H.; Inoue, T. Quantitative and Simultaneous Translational Control of Distinct Mammalian MRNAs. Nucleic Acids Res. 2013, 41, e135. [Google Scholar] [CrossRef]
- Cella, F.; Wroblewska, L.; Weiss, R.; Siciliano, V. Engineering Protein-Protein Devices for Multilayered Regulation of MRNA Translation Using Orthogonal Proteases in Mammalian Cells. Nat. Commun. 2018, 9, 4392. [Google Scholar] [CrossRef]
- Parr, C.J.C.; Wada, S.; Kotake, K.; Kameda, S.; Matsuura, S.; Sakashita, S.; Park, S.; Sugiyama, H.; Kuang, Y.; Saito, H. N1-Methylpseudouridine Substitution Enhances the Performance of Synthetic MRNA Switches in Cells. Nucleic Acids Res. 2020, 48, e35. [Google Scholar] [CrossRef]
- Liu, K.; Yang, J.; Ding, S.; Gao, Y. Daisy Chain Topology Based Mammalian Synthetic Circuits for RNA-Only Delivery. ACS Synth. Biol. 2020, 9, 269–282. [Google Scholar] [CrossRef]
- Paek, K.Y.; Hong, K.Y.; Ryu, I.; Park, S.M.; Keum, S.J.; Kwon, O.S.; Jang, S.K. Translation Initiation Mediated by RNA Looping. Proc. Natl. Acad. Sci. 2015, 112, 1041–1046. [Google Scholar] [CrossRef]
- Hosoda, N.; Kim, Y.K.; Lejeune, F.; Maquat, L.E. CBP80 Promotes Interaction of Upf1 with Upf2 during Nonsense-Mediated MRNA Decay in Mammalian Cells. Nat. Struct. Mol. Biol. 2005, 12, 893–901. [Google Scholar] [CrossRef]
- Kim, Y.K.; Furic, L.; DesGroseillers, L.; Maquat, L.E. Mammalian Staufen1 Recruits Upf1 to Specific MRNA 3′UTRs so as to Elicit MRNA Decay. Cell 2005, 120, 195–208. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.-D.; Steitz, J.A. Communication of the Position of Exon-Exon Junctions to the MRNA Surveillance Machinery by the Protein RNPS1. Science 2001, 293, 1836–1839. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.-D.; Steitz, J.A. Human Upf Proteins Target an MRNA for Nonsense-Mediated Decay When Bound Downstream of a Termination Codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef]
- Nakanishi, H.; Yoshii, T.; Kawasaki, S.; Hayashi, K.; Tsutsui, K.; Oki, C.; Tsukiji, S.; Saito, H. Light-Controllable RNA-Protein Devices for Translational Regulation of Synthetic MRNAs in Mammalian Cells. Cell Chem. Biol. 2021, 28, 662–674.e5. [Google Scholar] [CrossRef]
- Ono, H.; Kawasaki, S.; Saito, H. Orthogonal Protein-Responsive MRNA Switches for Mammalian Synthetic Biology. ACS Synth. Biol. 2020, 9, 169–174. [Google Scholar] [CrossRef]
- Stapleton, J.A.; Endo, K.; Fujita, Y.; Hayashi, K.; Takinoue, M.; Saito, H.; Inoue, T. Feedback Control of Protein Expression in Mammalian Cells by Tunable Synthetic Translational Inhibition. ACS Synth. Biol. 2012, 1, 83–88. [Google Scholar] [CrossRef]
- Wagner, T.E.; Becraft, J.R.; Bodner, K.; Teague, B.; Zhang, X.; Woo, A.; Porter, E.; Alburquerque, B.; Dobosh, B.; Andries, O.; et al. Small-Molecule-Based Regulation of RNA-Delivered Circuits in Mammalian Cells. Nat. Chem. Biol. 2018, 14, 1043–1050. [Google Scholar] [CrossRef]
- Shen, C.-C.; Hsu, M.-N.; Chang, C.-W.; Lin, M.-W.; Hwu, J.-R.; Tu, Y.; Hu, Y.-C. Synthetic Switch to Minimize CRISPR Off-Target Effects by Self-Restricting Cas9 Transcription and Translation. Nucleic Acids Res. 2019, 47, e13. [Google Scholar] [CrossRef]
- Mc Cafferty, S.; De Temmerman, J.; Kitada, T.; Becraft, J.R.; Weiss, R.; Irvine, D.J.; Devreese, M.; De Baere, S.; Combes, F.; Sanders, N.N. In Vivo Validation of a Reversible Small Molecule-Based Switch for Synthetic Self-Amplifying MRNA Regulation. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 1164–1173. [Google Scholar] [CrossRef]
- Goldfless, S.J.; Belmont, B.J.; de Paz, A.M.; Liu, J.F.; Niles, J.C. Direct and Specific Chemical Control of Eukaryotic Translation with a Synthetic RNA–Protein Interaction. Nucleic Acids Res. 2012, 40, e64. [Google Scholar] [CrossRef][Green Version]
- Kawasaki, S.; Fujita, Y.; Nagaike, T.; Tomita, K.; Saito, H. Synthetic MRNA Devices That Detect Endogenous Proteins and Distinguish Mammalian Cells. Nucleic Acids Res. 2017, 45, e117. [Google Scholar] [CrossRef]
- Lowary, P.T.; Uhlenbeck, O.C. An RNA Mutation That Increases the Affinity of an RNA-Protein Interaction. Nucleic Acids Res. 1987, 15, 10483–10493. [Google Scholar] [CrossRef]
- Nagai, K.; Oubridge, C.; Ito, N.; Avis, J.; Evans, P. The RNP Domain: A Sequence-Specific RNA-Binding Domain Involved in Processing and Transport of RNA. Trends Biochem. Sci. 1995, 20, 235–240. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (Force-Directed RNA): Simple and Effective Online RNA Secondary Structure Diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Mugridge, J.S.; Coller, J.; Gross, J.D. Structural and Molecular Mechanisms for the Control of Eukaryotic 5′–3′ MRNA Decay. Nat. Struct. Mol. Biol. 2018, 25, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Inada, T.; Makino, S. Novel Roles of the Multi-Functional CCR4-NOT Complex in Post-Transcriptional Regulation. Front. Genet. 2014, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Artus, C.G.; Filipowicz, W. Tethering of Human Ago Proteins to MRNA Mimics the MiRNA-Mediated Repression of Protein Synthesis. RNA 2004, 10, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Babendure, J.R.; Babendure, J.L.; Ding, J.-H.; Tsien, R.Y. Control of Mammalian Translation by MRNA Structure near Caps. RNA 2006, 12, 851–861. [Google Scholar] [CrossRef]
- Petrakova, O.; Volkova, E.; Gorchakov, R.; Paessler, S.; Kinney, R.M.; Frolov, I. Noncytopathic Replication of Venezuelan Equine Encephalitis Virus and Eastern Equine Encephalitis Virus Replicons in Mammalian Cells. J. Virol. 2005, 79, 7597–7608. [Google Scholar] [CrossRef]
- Yoshioka, N.; Gros, E.; Li, H.-R.; Kumar, S.; Deacon, D.C.; Maron, C.; Muotri, A.R.; Chi, N.C.; Fu, X.-D.; Yu, B.D.; et al. Efficient Generation of Human IPSCs by a Synthetic Self-Replicative RNA. Cell Stem Cell 2013, 13, 246–254. [Google Scholar] [CrossRef]
- Fukao, A.; Tomohiro, T.; Fujiwara, T. Translation Initiation Regulated by RNA-Binding Protein in Mammals: The Modulation of Translation Initiation Complex by Trans-Acting Factors. Cells 2021, 10, 1711. [Google Scholar] [CrossRef]
- Herbert, T.P.; Brierley, I.; Brown, T.D. Identification of a Protein Linked to the Genomic and Subgenomic MRNAs of Feline Calicivirus and Its Role in Translation. J. Gen. Virol. 1997, 78, 1033–1040. [Google Scholar] [CrossRef]
- Goodfellow, I.; Chaudhry, Y.; Gioldasi, I.; Gerondopoulos, A.; Natoni, A.; Labrie, L.; Laliberté, J.-F.; Roberts, L. Calicivirus Translation Initiation Requires an Interaction between VPg and EIF4E. EMBO Rep. 2005, 6, 968–972. [Google Scholar] [CrossRef]
- Chaudhry, Y.; Nayak, A.; Bordeleau, M.-E.; Tanaka, J.; Pelletier, J.; Belsham, G.J.; Roberts, L.O.; Goodfellow, I.G. Caliciviruses Differ in Their Functional Requirements for EIF4F Components. J. Biol. Chem. 2006, 281, 25315–25325. [Google Scholar] [CrossRef]
- Matsui, A.; Uchida, S.; Ishii, T.; Itaka, K.; Kataoka, K. Messenger RNA-Based Therapeutics for the Treatment of Apoptosis-Associated Diseases. Sci. Rep. 2015, 5, 15810. [Google Scholar] [CrossRef]
- De Gregorio, E.; Baron, J.; Preiss, T.; Hentze, M.W. Tethered-Function Analysis Reveals That ElF4E Can Recruit Ribosomes Independent of Its Binding to the Cap Structure. RNA 2001, 7, 106–113. [Google Scholar] [CrossRef]
- Banaszynski, L.A.; Chen, L.; Maynard-Smith, L.A.; Lisa Ooi, A.G.; Wandless, T.J. A Rapid, Reversible, and Tunable Method to Regulate Protein Function in Living Cells Using Synthetic Small Molecules. Cell 2006, 126, 995–1004. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An Auxin-Based Degron System for the Rapid Depletion of Proteins in Nonplant Cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Iwamoto, M.; Björklund, T.; Lundberg, C.; Kirik, D.; Wandless, T.J. A General Chemical Method to Regulate Protein Stability in the Mammalian Central Nervous System. Chem. Biol. 2010, 17, 981–988. [Google Scholar] [CrossRef]
- Taxis, C.; Stier, G.; Spadaccini, R.; Knop, M. Efficient Protein Depletion by Genetically Controlled Deprotection of a Dormant N-Degron. Mol. Syst. Biol. 2009, 5, 267. [Google Scholar] [CrossRef]
- Tözsér, J.; Tropea, J.E.; Cherry, S.; Bagossi, P.; Copeland, T.D.; Wlodawer, A.; Waugh, D.S. Comparison of the Substrate Specificity of Two Potyvirus Proteases. FEBS J. 2005, 272, 514–523. [Google Scholar] [CrossRef]
- Pethe, M.A.; Rubenstein, A.B.; Khare, S.D. Data-Driven Supervised Learning of a Viral Protease Specificity Landscape from Deep Sequencing and Molecular Simulations. Proc. Natl. Acad. Sci. USA 2019, 116, 168–176. [Google Scholar] [CrossRef]
- Bayle, J.H.; Grimley, J.S.; Stankunas, K.; Gestwicki, J.E.; Wandless, T.J.; Crabtree, G.R. Rapamycin Analogs with Differential Binding Specificity Permit Orthogonal Control of Protein Activity. Chem. Biol. 2006, 13, 99–107. [Google Scholar] [CrossRef]
- Liberles, S.D.; Diver, S.T.; Austin, D.J.; Schreiber, S.L. Inducible Gene Expression and Protein Translocation Using Nontoxic Ligands Identified by a Mammalian Three-Hybrid Screen. Proc. Natl. Acad. Sci. USA 1997, 94, 7825–7830. [Google Scholar] [CrossRef]
- Kennedy, M.J.; Hughes, R.M.; Peteya, L.A.; Schwartz, J.W.; Ehlers, M.D.; Tucker, C.L. Rapid Blue-Light–Mediated Induction of Protein Interactions in Living Cells. Nat. Methods 2010, 7, 973–975. [Google Scholar] [CrossRef] [PubMed]
- Dagliyan, O.; Krokhotin, A.; Ozkan-Dagliyan, I.; Deiters, A.; Der, C.J.; Hahn, K.M.; Dokholyan, N.V. Computational Design of Chemogenetic and Optogenetic Split Proteins. Nat. Commun. 2018, 9, 4042. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.-R.; Li, L.-H.; Park, H.-J.; Park, J.-H.; Lee, K.Y.; Kim, M.-K.; Shin, B.A.; Choi, S.-Y. High Cleavage Efficiency of a 2A Peptide Derived from Porcine Teschovirus-1 in Human Cell Lines, Zebrafish and Mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into MRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-Methylpseudouridine-Incorporated MRNA Outperforms Pseudouridine-Incorporated MRNA by Providing Enhanced Protein Expression and Reduced Immunogenicity in Mammalian Cell Lines and Mice. J. Controlled Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Fukunaga, K.; Yokobayashi, Y. Directed Evolution of Orthogonal RNA–RBP Pairs through Library-vs-Library in Vitro Selection. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef]
- Peabody, D.S.; Ely, K.R. Control of Translational Repression by Protein-Protein Interactions. Nucleic Acids Res. 1992, 20, 1649–1655. [Google Scholar] [CrossRef]
- Lim, F.; Peabody, D.S. Mutations That Increase the Affinity of a Translational Repressor for RNA. Nucleic Acids Res. 1994, 22, 3748–3752. [Google Scholar] [CrossRef]
- Podrazký, O.; Peterka, P.; Kašík, I.; Vytykáčová, S.; Proboštová, J.; Mrázek, J.; Kuneš, M.; Závalová, V.; Radochová, V.; Lyutakov, O.; et al. In Vivo Testing of a Bioresorbable Phosphate-Based Optical Fiber. J. Biophotonics 2019, 12, e201800397. [Google Scholar] [CrossRef]
- Weissleder, R. A Clearer Vision for in Vivo Imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef]
- Ryu, M.-H.; Gomelsky, M. Near-Infrared Light Responsive Synthetic c-Di-GMP Module for Optogenetic Applications. ACS Synth. Biol. 2014, 3, 802–810. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Huq, E.; Tepperman, J.M.; Quail, P.H. A Light-Switchable Gene Promoter System. Nat. Biotechnol. 2002, 20, 1041–1044. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef]
- Kawano, F.; Suzuki, H.; Furuya, A.; Sato, M. Engineered Pairs of Distinct Photoswitches for Optogenetic Control of Cellular Proteins. Nat. Commun. 2015, 6, 6256. [Google Scholar] [CrossRef]
- Weber, A.M.; Kaiser, J.; Ziegler, T.; Pilsl, S.; Renzl, C.; Sixt, L.; Pietruschka, G.; Moniot, S.; Kakoti, A.; Juraschitz, M.; et al. A Blue Light Receptor That Mediates RNA Binding and Translational Regulation. Nat. Chem. Biol. 2019, 15, 1085–1092. [Google Scholar] [CrossRef]
- Guntas, G.; Hallett, R.A.; Zimmerman, S.P.; Williams, T.; Yumerefendi, H.; Bear, J.E.; Kuhlman, B. Engineering an Improved Light-Induced Dimer (ILID) for Controlling the Localization and Activity of Signaling Proteins. Proc. Natl. Acad. Sci. 2015, 112, 112–117. [Google Scholar] [CrossRef]
- Quejada, J.R.; Park, S.-H.E.; Awari, D.W.; Shi, F.; Yamamoto, H.E.; Kawano, F.; Jung, J.C.; Yazawa, M. Optimized Light-Inducible Transcription in Mammalian Cells Using Flavin Kelch-Repeat F-Box1/GIGANTEA and CRY2/CIB1. Nucleic Acids Res. 2017, 45, e172. [Google Scholar] [CrossRef]
- Bonger, K.M.; Rakhit, R.; Payumo, A.Y.; Chen, J.K.; Wandless, T.J. General Method for Regulating Protein Stability with Light. ACS Chem. Biol. 2014, 9, 111–115. [Google Scholar] [CrossRef]
- Renicke, C.; Schuster, D.; Usherenko, S.; Essen, L.-O.; Taxis, C. A LOV2 Domain-Based Optogenetic Tool to Control Protein Degradation and Cellular Function. Chem. Biol. 2013, 20, 619–626. [Google Scholar] [CrossRef]
- Wang, Z.; Thang, D.C.; Han, Q.; Zhao, X.; Xie, X.; Wang, Z.; Lin, J.; Xing, B. Near-Infrared Photocontrolled Therapeutic Release via Upconversion Nanocomposites. J. Controlled Release 2020, 324, 104–123. [Google Scholar] [CrossRef]
- Wu, X.; Zhu, X.; Chong, P.; Liu, J.; Andre, L.N.; Ong, K.S.; Brinson, K.; Mahdi, A.I.; Li, J.; Fenno, L.E.; et al. Sono-Optogenetics Facilitated by a Circulation-Delivered Rechargeable Light Source for Minimally Invasive Optogenetics. Proc. Natl. Acad. Sci. 2019, 116, 26332–26342. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and Small Interfering RNAs Can Inhibit MRNA Expression by Similar Mechanisms. Proc. Natl. Acad. Sci. 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating MicroRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Miki, K.; Endo, K.; Takahashi, S.; Funakoshi, S.; Takei, I.; Katayama, S.; Toyoda, T.; Kotaka, M.; Takaki, T.; Umeda, M.; et al. Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 2015, 16, 699–711. [Google Scholar] [CrossRef]
- Nakanishi, H.; Miki, K.; Komatsu, K.R.; Umeda, M.; Mochizuki, M.; Inagaki, A.; Yoshida, Y.; Saito, H. Monitoring and Visualizing MicroRNA Dynamics during Live Cell Differentiation Using MicroRNA-Responsive Non-Viral Reporter Vectors. Biomaterials 2017, 128, 121–135. [Google Scholar] [CrossRef]
- Endo, K.; Hayashi, K.; Saito, H. Numerical Operations in Living Cells by Programmable RNA Devices. Sci. Adv. 2019, 5, eaax0835. [Google Scholar] [CrossRef]
- Mauger, D.M.; Cabral, B.J.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. MRNA Structure Regulates Protein Expression through Changes in Functional Half-Life. Proc. Natl. Acad. Sci. 2019, 116, 24075–24083. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Cheng, Y.M.; Chakraborty, T.; Presnyak, V.; John, M.; Sonenberg, N. N1-Methyl-Pseudouridine in MRNA Enhances Translation through EIF2α-Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res. 2017, 45, 6023–6036. [Google Scholar] [CrossRef]
- Uchida, S.; Kataoka, K.; Itaka, K. Screening of MRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics 2015, 7, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Thess, A.; Grund, S.; Mui, B.L.; Hope, M.J.; Baumhof, P.; Fotin-Mleczek, M.; Schlake, T. Sequence-Engineered MRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015, 23, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.K.M.A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 MRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. - Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Itaka, K.; Uchida, H.; Hayakawa, K.; Ogata, T.; Ishii, T.; Fukushima, S.; Osada, K.; Kataoka, K. In Vivo Messenger RNA Introduction into the Central Nervous System Using Polyplex Nanomicelle. PLOS ONE 2013, 8, e56220. [Google Scholar] [CrossRef]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving MRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering Circular RNA for Potent and Stable Translation in Eukaryotic Cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef]
- Kojima, R.; Aubel, D.; Fussenegger, M. Building Sophisticated Sensors of Extracellular Cues That Enable Mammalian Cells to Work as “Doctors” in the Body. Cell. Mol. Life Sci. 2020, 77, 3567–3581. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakanishi, H. Protein-Based Systems for Translational Regulation of Synthetic mRNAs in Mammalian Cells. Life 2021, 11, 1192. https://doi.org/10.3390/life11111192
Nakanishi H. Protein-Based Systems for Translational Regulation of Synthetic mRNAs in Mammalian Cells. Life. 2021; 11(11):1192. https://doi.org/10.3390/life11111192
Chicago/Turabian StyleNakanishi, Hideyuki. 2021. "Protein-Based Systems for Translational Regulation of Synthetic mRNAs in Mammalian Cells" Life 11, no. 11: 1192. https://doi.org/10.3390/life11111192
APA StyleNakanishi, H. (2021). Protein-Based Systems for Translational Regulation of Synthetic mRNAs in Mammalian Cells. Life, 11(11), 1192. https://doi.org/10.3390/life11111192





