Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Phylogenetic Reconstruction
2.3. ITS2 Consensus Secondary Structure Modelling
2.4. Compensatory Base Changes (CBCs)
3. Results
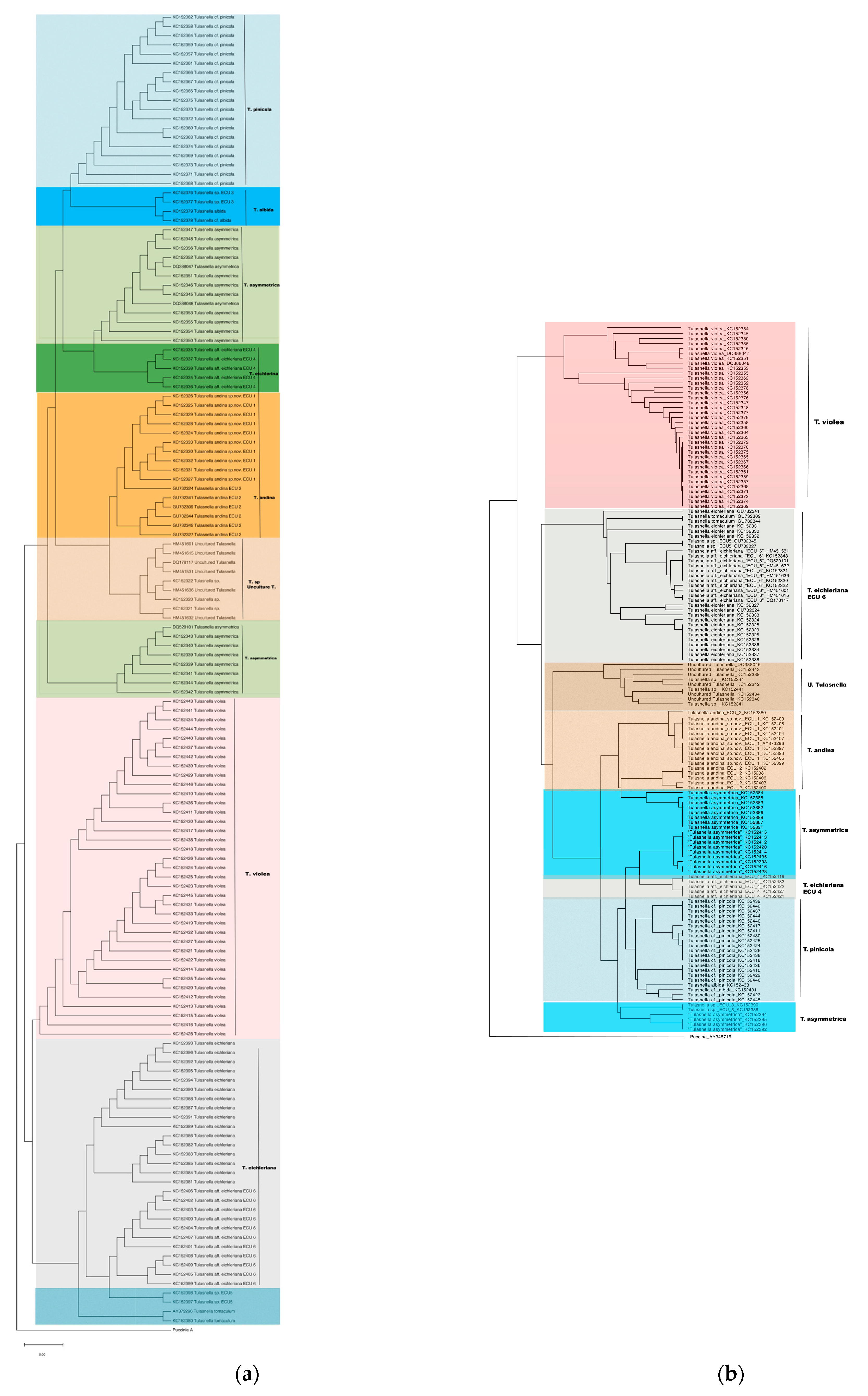
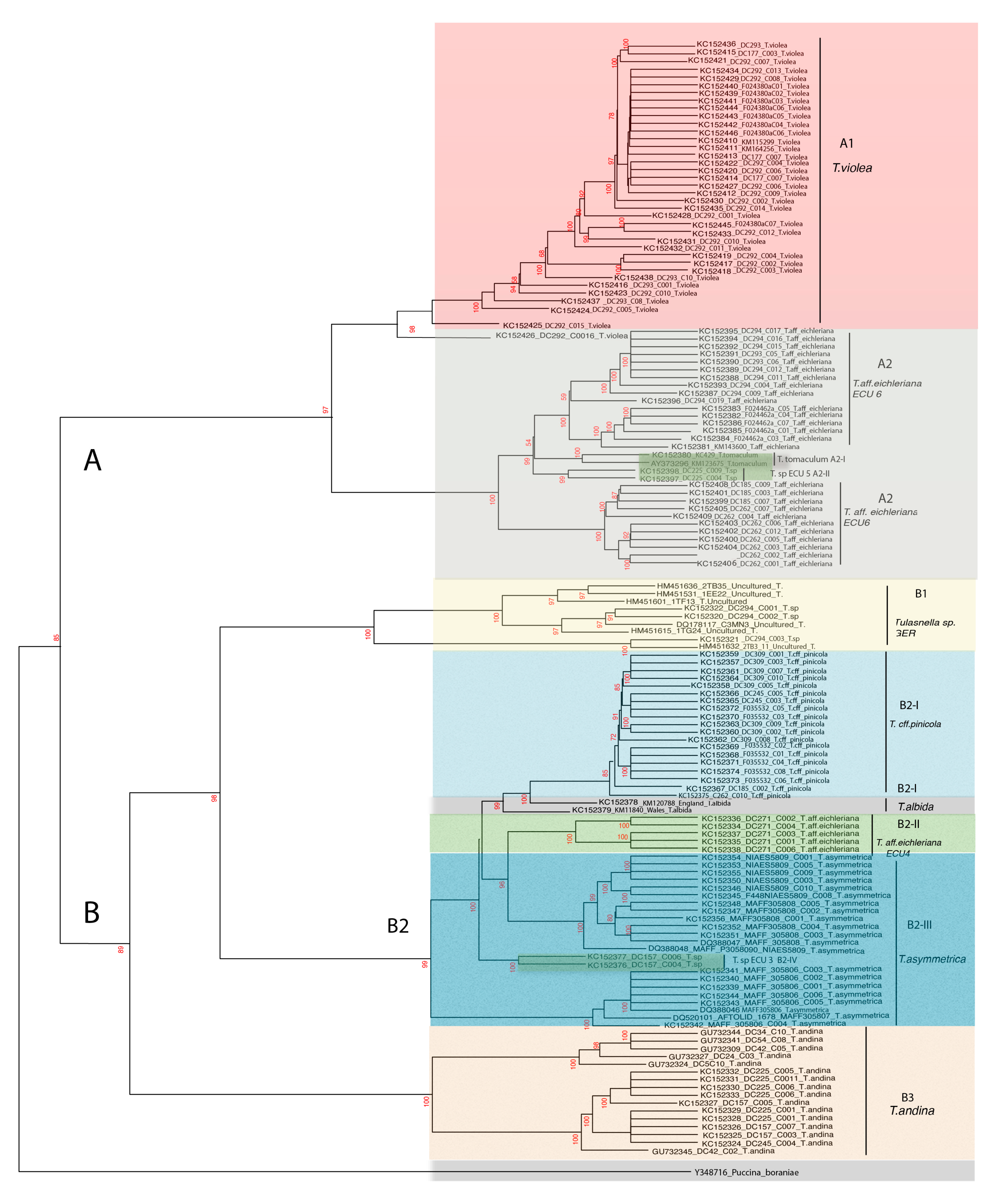
3.1. AB and AF Phylogenetic Reconstruction
3.2. AB and AF Distance Integration with DistatisR
3.3. Consensus Secondary Structure Modelling for Intra-Specific Differentiation
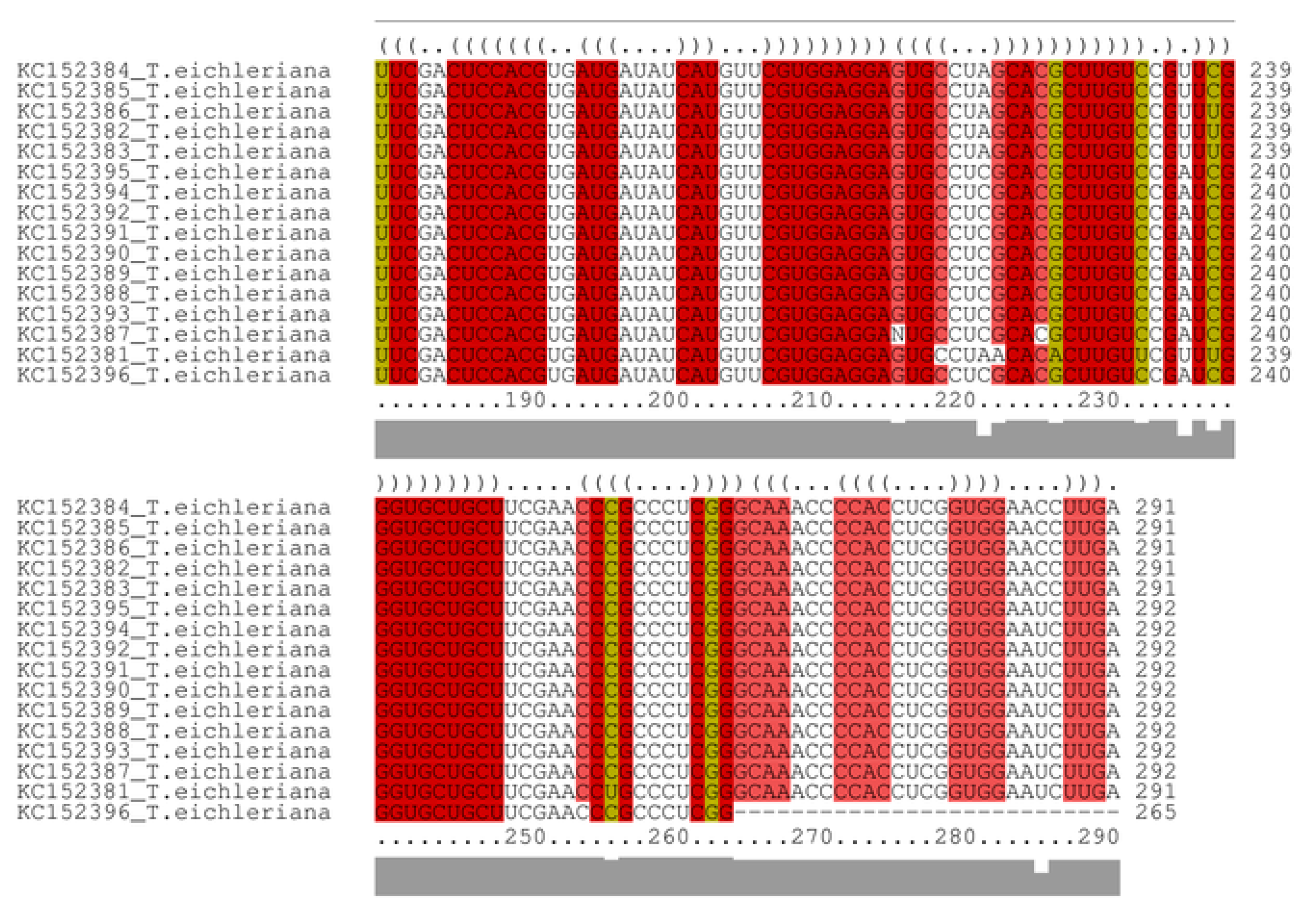
3.4. CBCs in the ITS2 Secondary Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BOLD | Barcode of Life |
| CBC | Compensatory base change |
| GMYC | Generalized mixed Yule-coalescent |
| ITS | Internal transcribed spacers |
| -InL | The negative log-likelihood |
| LRT | Likelihood ratio test |
| MFE | Minimum free energy |
| MSA | Multiple sequence alignments |
| ML | Maximum likelihood |
| MDS | Metric multidimensional scaling |
| NCBI | National Center for Biotechnology Information |
| NJ | Neighbor Joining |
Appendix A. List of Software and Packages to Tulasnella Species Delimitation
| Software | Description | Link (accessed on 1 February 2022) |
|---|---|---|
| Genbank | Database National Center for Biotechnology Information (NCBI), to download sequences. | http://www.ncbi.nlm.nih.gov/ |
| ITSx v1.1.3 | ITS2 region extraction | https://microbiology.se/software/itsx/ |
| Maff-add v7 | Sequences alignment | http://mafft.cbrc.jp/alignment/software |
| Mfold v 3.6 | To infer 2D structures by a MFE (free energy minimization) parameter. | http://www.unafold.org/mfold/applications/rna-folding-form.php |
| RNA distance V2.6.2 | Distance matrix from predicted structures was obtained with RNA distance fromVienna package. | https://www.tbi.univie.ac.at/RNA/ |
| MEGA6.0 | An integrated tool for conducting automatic and manual sequence alignment and inferring phylogenetic trees. | https://www.megasoftware.net |
| Clustal Omega v1.2.1 | Sequences alignment | http://www.clustal.org/omega/ |
| OPAL v2.1.3 | Sequences alignment | http:/opal.cs.arizona.edu |
| RaxML-ng v.1.2.0 | Phylogenetic tree inference. | https://github.com/amkozlov/raxml-ng |
| DNAdist v3.69 | Used to obtain the distance matrix from Which evaluates the similarity between distance matrices Opal MSA | https://evolution.genetics.washington.edu/phylip/doc/dnadist.html |
| DistatisR. | The compromise matrix was used as the input data to construct the third phylogenetic tree using pvclust library with R package v 1.10 | https://cran.r-project.org/web/packages/DistatisR/index.html |
| ITS2 database | An exhaustive dataset of internal transcribed spacer 2 sequences from NCBI GenBank, accurately reannotated. | http://its2.bioapps.biozentrum.uni-wuerzburg.de/ |
| LocARNA V2.0.0 | Software to perform structural-alignment and consensus secondary structure modelling | https://rna.informatik.uni-freiburg.de/LocARNA/Input.jsp;jsessionid=115161C5C9E9AB44CC6D3C81763A1699 |
| 4SALE tool v1.7.1 | Secondary Structure Alignment | http://4sale.bioapps.biozentrum.uni-wuerzburg.de/ |
| Figtree v1.4.4 | Tree visualization | http://tree.bio.ed.ac.uk/software/figtree/ |
| pv-clust | Hierarchical Clustering Analysis library with R package V2.2-0 | https://cran.r-project.org/web/packages/pvclust/index.html |
| Splits-gmyc | Delimiting species and automated taxonomy library with R package V1.0-20 | https://rdrr.io/rforge/splits/ |
Appendix B. Compensatory Base Changes in Tulasnella Sequences
| Sub-Clades A/B | Internal and External Sub-Clades | Evolutionary Base Pared Changes in Tulasnella spp. | ||
|---|---|---|---|---|
| CBCs among Internal Sub-Clades Threshold (1–9) | CBCs among External Sub-Clades Threshold (1–10) | |||
| CBCs = 0 | CBCs > 0 | CBCs > 0 | ||
| A (98%) | A1 T. violea | A1 → A1 A1 → A2-II | A1 → A2 (4–7) A1 → A2-I (3–5) | A1 → B1 (1–7) |
| A1 → B2-I (1–7) | ||||
| A1 → B2-II (1–9) | ||||
| A1 → B2-III (1–6) | ||||
| A1 → B2-V (1–5) | ||||
| A1 → B-IV (1–2) | ||||
| A1 → B3 | ||||
| A2 T. eichleriana ECU6 | A2 → A2 | A2 → A2-I (1–5) A2 → A2-II (1–1) | A2 → B1(1–7) | |
| A2 → B2-I (1–7) | ||||
| A2 → B2-II (1–9) | ||||
| A2 → B2-III (1–5) | ||||
| A2 → B2-V (1–4) | ||||
| A2 → B2-IV (1–2) | ||||
| A2 → B3 | ||||
| A2-I T. tomaculum | A2-I → A2-I | A2-I → A2-II(1–5) - | A2-I → B1 (1–7) | |
| A2-I → B2-I (1–7) | ||||
| A2-I → B2-II (1–9) | ||||
| A2-I → B2-III (1–5) | ||||
| A2-I → B2-V (1–4) | ||||
| A2-I → B2-IV (1–4) | ||||
| A2-I → B3 | ||||
| A2-II T. sp ECU5 | A2-II→ A2-II | A2-II→ A2-I (1–1) | A2-II → B1 (1–7) | |
| A2-II → B2-I (1–7) | ||||
| A2-II → B2-II (1–9) | ||||
| A2-II → B2-III (1–5) | ||||
| A2-II → B2-V (1–4) | ||||
| A2-II → B2-IV (1–4) | ||||
| A2-II → B3 | ||||
| B (89%) | B1 T. sp (GER) | B1 → B1 B1 → B2-II B1 → B2-II | B1 → B2-I (1–8) B1 → B2-V (1–2) B1 → B2-IV (1–3) | |
| B2-I T. pinicola | B2-I → B2-I B2-I → B4 B2-I → B2-IV | B2-I → B2-II (1–2) B2-I → B2-V (1–5) B2-I → B2-II (1–1) | ||
| B2-II T. albida | B2-II → B2-II B2-II → B2-III B2-II → B2-V | B2-II → B2-IV (1–3) B2-II → B2-II (1–2) | ||
| B2-III T. eichleriana ECU4 | B2-III → B2-III B2-III → B2-IV | B2-III→B2-V (1–4) B2-III → B2-II (1–5) | ||
| B2-V T. asymmetrica | B2-V → B2-V B2-V → B2-IV | B2-V→ B3(1–4) | ||
| B2-IV T. sp ECU3 | B2-IV → B2-IV B2-IV→ B3 | |||
| B3 T. andina | B3 → B3 | |||
| Total | ∑ CBCi = 0 21 | ∑ CBCi > 0 212 | ∑ CBCe > 0 433 | |
References
- Chethana, K.W.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Ding, X.; Xiao, J.H.; Li, L.; Conran, J.G.; Li, J. Congruent species delimitation of two controversial gold-thread nanmu tree species based on morphological and restriction site-associated DNA sequencing data. J. Syst. Evol. 2019, 57, 234–246. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Stanke, K.M.; Quattrone, A.C.; Herr, J.R. Improving Taxonomic Delimitation of Fungal Species in the Age of Genomics and Phenomics. Front. Microbiol. 2022, 13, 847067. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Elnitski, L.; Church, D.M.; Dubchak, I.; Hardison, R.C. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 2003, 13, 1–12. [Google Scholar] [CrossRef]
- DeSalle, R.; Goldstein, P. Review and interpretation of trends in DNA barcoding. Front. Ecol. Evol. 2019, 7, 302. [Google Scholar] [CrossRef]
- Cruz, D.; Suárez, J.P.; Kottke, I.; Piepenbring, M. Cryptic species revealed by molecular phylogenetic analysis of sequences obtained from basidiomata of Tulasnella. Mycologia 2014, 106, 708–722. [Google Scholar] [CrossRef]
- Edgar, R.C.; Batzoglou, S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006, 16, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Agüero-Chapin, G.; Jiménez, Y.; Sánchez-Rodríguez, A.; Molina-Ruiz, R.; Vivanco, O.; Antunes, A. DISTATIS: A Promising Framework to Integrate Distance Matrices in Molecular Phylogenetics. Curr. Top. Med. Chem. 2021, 21, 599–611. [Google Scholar] [CrossRef]
- Janies, D.; Studer, J.; Handelman, S.; Linchangco, G. A comparison of supermatrix and supertree methods for multilocus phylogenetics using organismal datasets. Cladistics 2013, 29, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Abeysundera, M.; Kenney, T.; Field, C.; Gu, H. Combining distance matrices on identical taxon sets for multigene analysis with singular value decomposition. PLoS ONE 2014, 9, e94279. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Coleman, A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. TRENDS Genet. 2003, 19, 370–375. [Google Scholar] [CrossRef]
- Coleman, A.W.; Vacquier, V.D. Exploring the phylogenetic utility of ITS sequences for animals: A test case for abalone (Haliotis). J. Mol. Evol. 2002, 54, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Koetschan, C.; Foerster, F.; Keller, A.; Schleicher, T.; Ruderisch, B.; Schwarz, R.; Müller, T.; Wolf, M.; Schultz, J. The ITS2 Database III—Sequences and structures for phylogeny. Nucleic Acids Res. 2010, 38 (Suppl. S1), D275–D279. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Caisová, L.; Marin, B.; Melkonian, M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 2013, 164, 482–496. [Google Scholar] [CrossRef]
- Schultz, J.; Wolf, M. ITS2 sequence–structure analysis in phylogenetics: A how-to manual for molecular systematics. Mol. Phylogenetics Evol. 2009, 52, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.; Jagan, E.G.; Kathamuthu, G.; Pandi, M. Internal transcribed spacer 2 (ITS2) molecular morphometric analysis based species delimitation of foliar endophytic fungi from Aglaia elaeagnoidea, Flacourtia inermis and Premna serratifolia. PLoS ONE 2019, 14, e0215024. [Google Scholar] [CrossRef]
- Cao, R.; Tong, S.; Luan, T.; Zheng, H.; Zhang, W. Compensatory Base Changes and Varying Phylogenetic Effects on Angiosperm ITS2 Genetic Distances. Plants 2022, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Müller, T. Compensatory Base Changes in ITS2 Secondary Structures Correlate with the Biological Species Concept Despite Intragenomic Variability in ITS2 Sequences—A Proof of Concept. PLoS ONE 2013, 8, e66726. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Friedrich, J.; Dandekar, T.; Muller, T. CBCAnalyzer: Inferring phylogenies based on compensatory base changes in RNA secondary structures. Silico Biol. 2005, 5, 291–294. [Google Scholar]
- Ruhl, M.; Wolf, M.; Jenkins, T.M. Compensatory base changes illuminate morphologically difficult taxonomy. Mol. Phylogenetics Evol. 2009, 54, 664–669. [Google Scholar] [CrossRef]
- Agüero-Chapin, G.; Sánchez-Rodríguez, A.; Hidalgo-Yanes, P.I.; Pérez-Castillo, Y.; Molina-Ruiz, R.; Marchal, K.; Vasconcelos, V.; Antunes, A. An alignment-free approach for eukaryotic ITS2 annotation and phylogenetic inference. PLoS ONE 2011, 6, e26638. [Google Scholar] [CrossRef] [PubMed]
- Zahin, T.; Abrar, M.H.; Rahman, M.; Tasnim, T.; Bayzid, M.S.; Rahman, A. An Alignment-free Method for Phylogeny Estimation using Maximum Likelihood. bioRxiv 2019. [Google Scholar] [CrossRef]
- Zuo, G. CVTree: A parallel alignment-free phylogeny and taxonomy tool based on composition vectors of genomes. Genom. Proteom. Bioinform. 2021, 19, 662–667. [Google Scholar] [CrossRef]
- Cheng, J.; Cao, F.; Liu, Z. AGP: A multimethods web server for alignment-free genome phylogeny. Mol. Biol. Evol. 2013, 30, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Abeysundera, M.; Field, C.; Gu, H. Phylogenetic analysis based on spectral methods. Mol. Biol. Evol. 2011, 29, 579–597. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2009, 37 (Suppl. S1), D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; Nilsson, R.H. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast folding and comparison of RNA secondary structures. Monatshefte Chem. Chem. Mon. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Anderson, C.; Strope, C.; Moriyama, E. Assessing Multiple Sequence Alignments Using Visual Tools. In Bioinformatics: Trends and Methodologies; Intechopen: London, UK, 2011. [Google Scholar]
- Wheeler, T.J.; Kececioglu, J.D. Multiple alignment by aligning alignments. Bioinformatics 2007, 23, i559–i568. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Ankenbrand, M.J.; Keller, A.; Wolf, M.; Schultz, J.; Förster, F. ITS2 database V: Twice as much. Mol. Biol. Evol. 2015, 32, 3030–3032. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.L.; Dong, Y.W.; Somero, G.N. Thermal adaptation of mRNA secondary structure: Stability versus lability. Proc. Natl. Acad. Sci. USA 2021, 118, e2113324118. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.; Backofen, R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. Rna 2012, 18, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, H.; Zhao, F.; Jiang, L.; Peng, H.; Zhang, W.; Simmons, M.P. Alternative analyses of compensatory base changes in an ITS2 phylogeny of Corydalis (Papaveraceae). Ann. Bot. 2019, 124, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Seibel, P.; Muller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7, 498. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.; Suárez, J.P.; Piepenbring, M. Morphological revision of Tulasnellaceae, with two new species of Tulasnella and new records of Tulasnella spp. for Ecuador. Nova Hedwig 2016, 102, 279–338. [Google Scholar] [CrossRef]
- Roberts, P. Spiral-spored Tulasnella species from Devon and the New Forest. Mycol. Res. 1992, 96, 233–236. [Google Scholar] [CrossRef]
- Keller, A.; Förster, F.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol. Direct 2010, 5, 4. [Google Scholar] [CrossRef]
- Freitas, E.F.S.; da Silva, M.; Cruz, E.D.S.; Mangaravite, E.; Bocayuva, M.F.; Veloso, T.G.R.; Selosse, M.-A.; Kasuya, M.C.M. Diversity of mycorrhizal Tulasnella associated with epiphytic and rupicolous orchids from the Brazilian Atlantic Forest, including four new species. Sci. Rep. 2020, 10, 7096. [Google Scholar] [CrossRef] [PubMed]
- Schulenburg, J.; Englisch, U.; Wägele, J.W. Evolution of ITS1 rDNA in the Digenea (Platyhelminthes: Trematoda): 3′ end sequence conservation and its phylogenetic utility. J. Mol. Evol. 1999, 48, 2–12. [Google Scholar] [CrossRef]
- Coleman, A.W. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3332. [Google Scholar] [CrossRef]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [PubMed]
- Hillis, D.M.; Dixon, M.J. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef] [PubMed]
- Rampersad, S.N.; Hosein, F.N.; Carrington, C.V.F. Sequence exploration reveals information bias among molecular markers used in phylogenetic reconstruction for Colletotrichum species. SpringerPlus 2014, 3, 614. [Google Scholar] [CrossRef]
- Rampersad, S.N. ITS1, 5.8 S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. SpringerPlus 2014, 3, 684. [Google Scholar] [CrossRef]
- Zhang, W.; Tian, W.; Gao, Z.; Wang, G.; Zhao, H. Phylogenetic utility of rRNA ITS2 sequence-structure under functional constraint. Int. J. Mol. Sci. 2020, 21, 6395. [Google Scholar] [CrossRef]
- Réblová, M.; Réblová, K.; Štěpánek, V. Molecular systematics of Barbatosphaeria (Sordariomycetes): Multigene phylogeny and secondary ITS structure. Persoonia-Mol. Phylogeny Evol. Fungi 2015, 35, 21–38. [Google Scholar] [CrossRef]



| Type of Tree | Method | Result of LR Test | Number of ML Clusters |
|---|---|---|---|
| AB Tree | OPAL bootstrap | 0.02277398 | 11 |
| AF Tree | VRNAdist | 0.4530083 | 9 |
| Distance tree | Distatis R | 0.01520527 * | 8 |
| Evolutionary Base Pared Changes | CBC = 0 | CBC > 0 |
|---|---|---|
| CBCs among internal sub-clades | 0 | 212 |
| CBCs among external sub-clades | 21 | 433 |
| TOTAL CBCs | 21 | 645 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Gaona, Y.; Vivanco-Galván, O.; Cruz, D.; Armijos-Carrión, A.; Suárez, J.P. Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota). J. Fungi 2023, 9, 894. https://doi.org/10.3390/jof9090894
Jiménez-Gaona Y, Vivanco-Galván O, Cruz D, Armijos-Carrión A, Suárez JP. Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota). Journal of Fungi. 2023; 9(9):894. https://doi.org/10.3390/jof9090894
Chicago/Turabian StyleJiménez-Gaona, Yuliana, Oscar Vivanco-Galván, Darío Cruz, Angelo Armijos-Carrión, and Juan Pablo Suárez. 2023. "Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota)" Journal of Fungi 9, no. 9: 894. https://doi.org/10.3390/jof9090894
APA StyleJiménez-Gaona, Y., Vivanco-Galván, O., Cruz, D., Armijos-Carrión, A., & Suárez, J. P. (2023). Compensatory Base Changes in ITS2 Secondary Structure Alignment, Modelling, and Molecular Phylogeny: An Integrated Approach to Improve Species Delimitation in Tulasnella (Basidiomycota). Journal of Fungi, 9(9), 894. https://doi.org/10.3390/jof9090894










