Adhesins of Yeasts: Protein Structure and Interactions
Abstract
1. Introduction
2. The Structure of Yeast Adhesins
2.1. Architectures of Yeast Adhesins
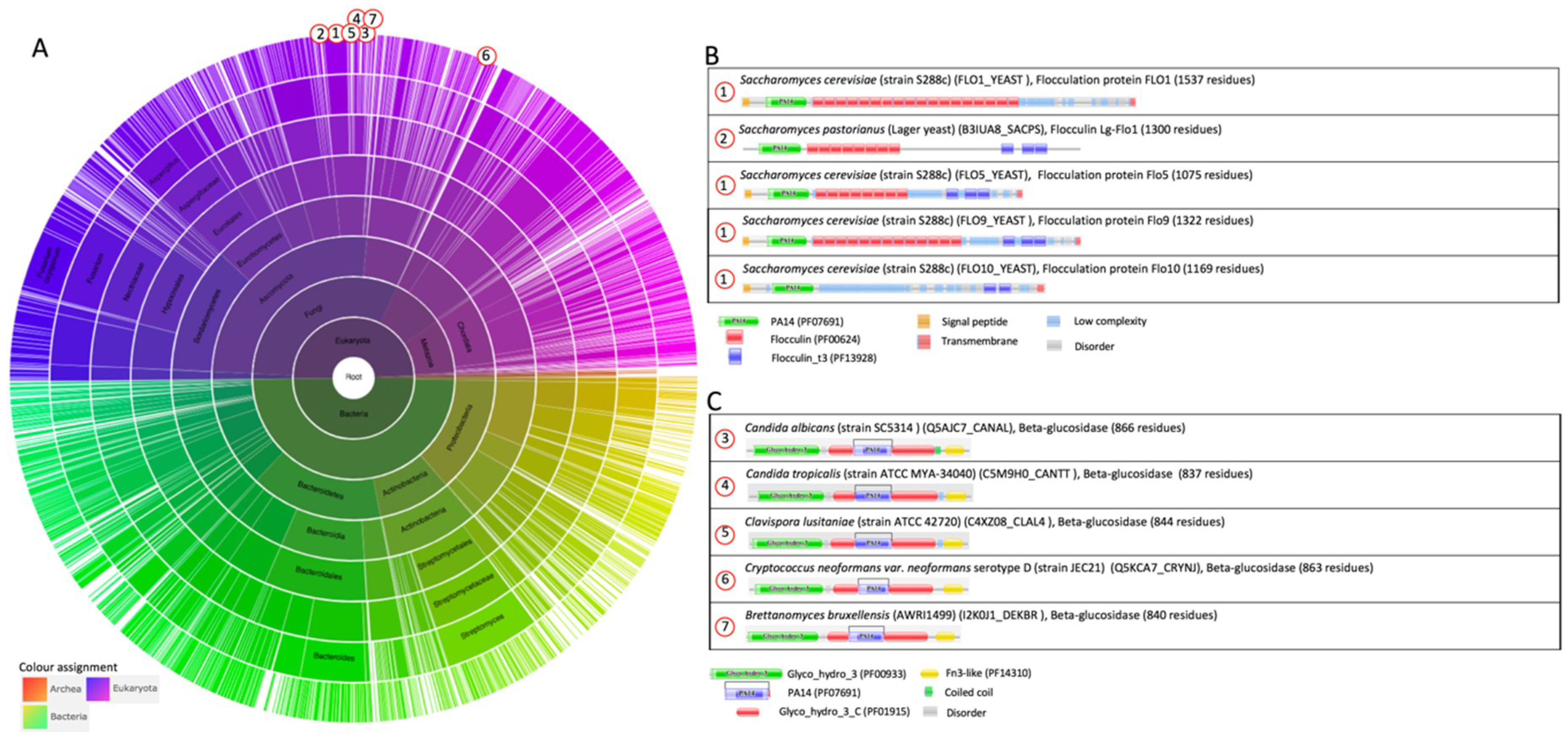
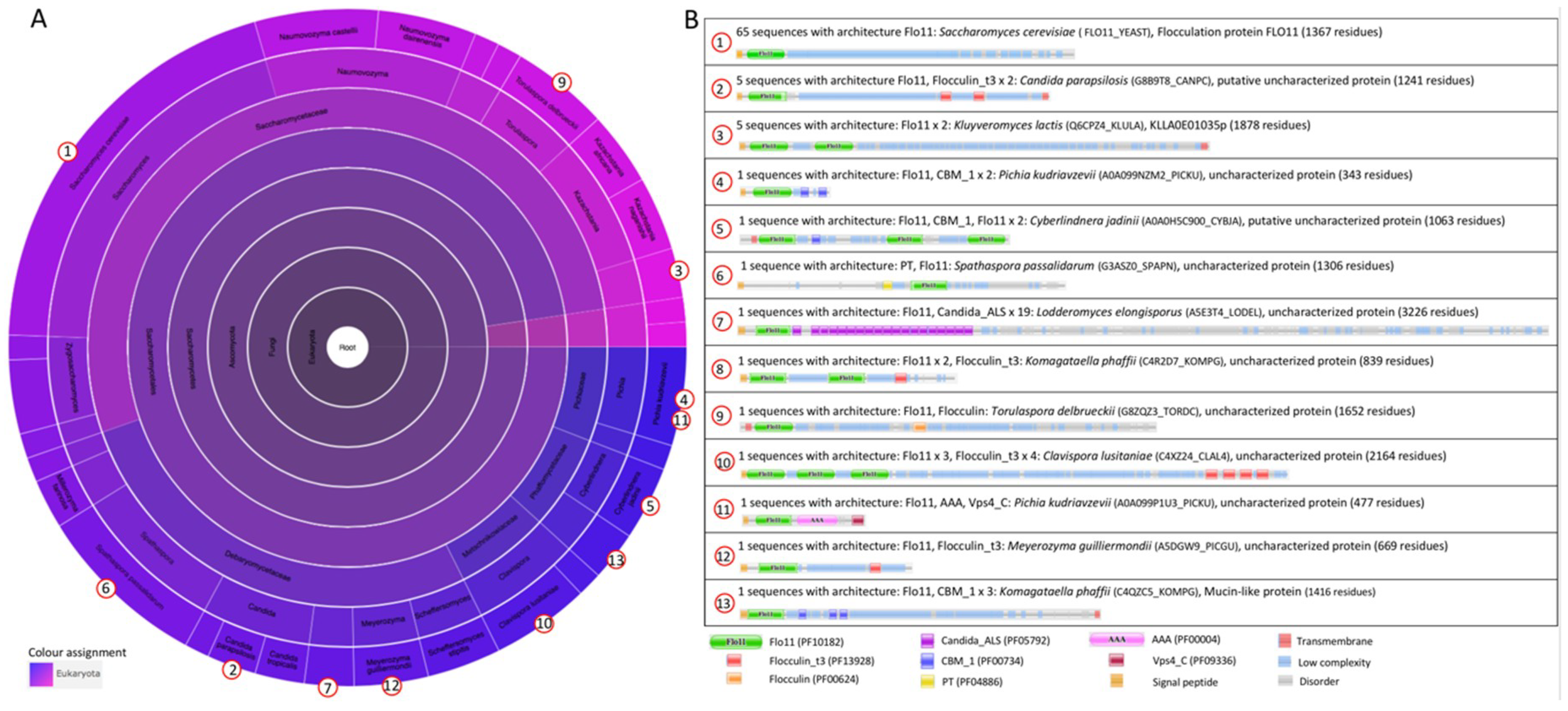
2.1.1. The S. cerevisiae Flocculation Protein Family
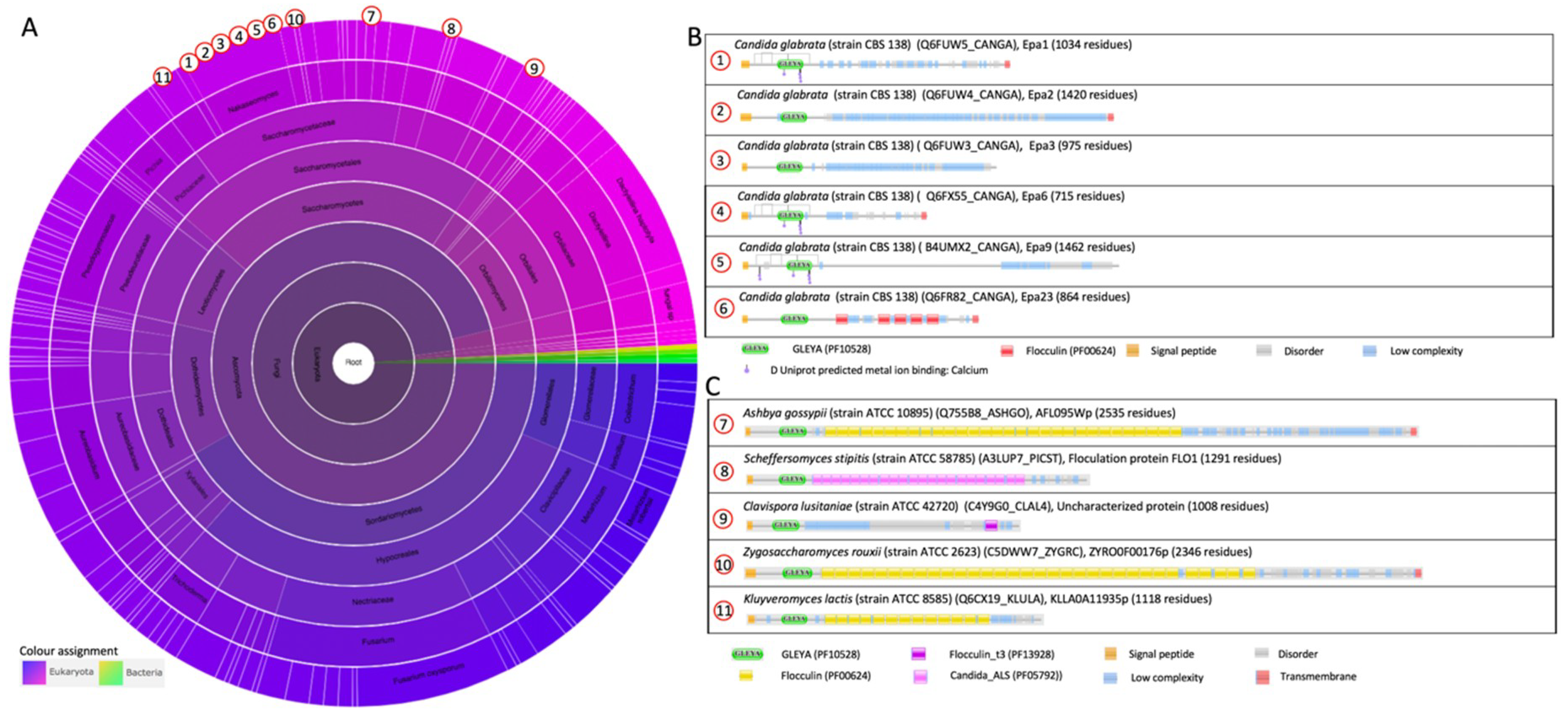
2.1.2. The Epithelial Adhesin Family
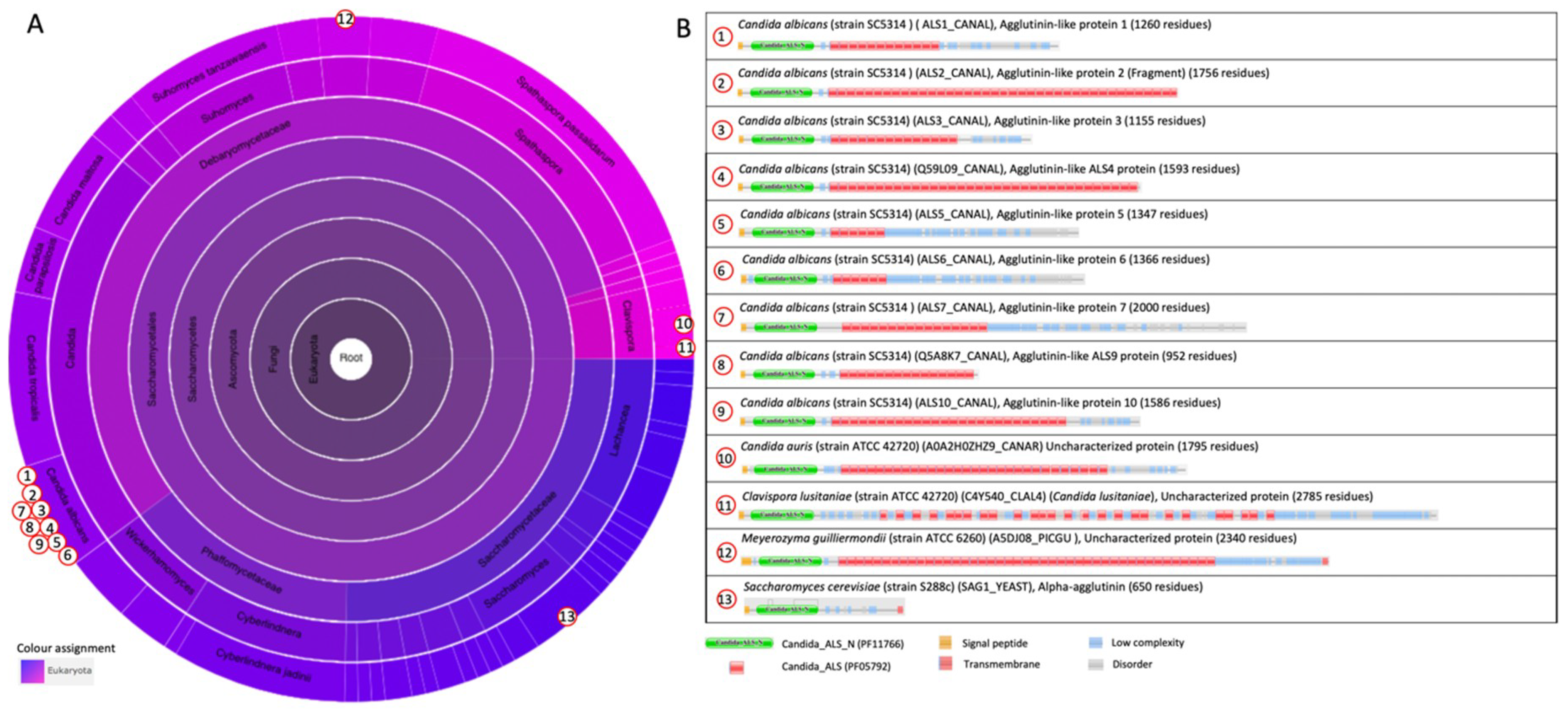
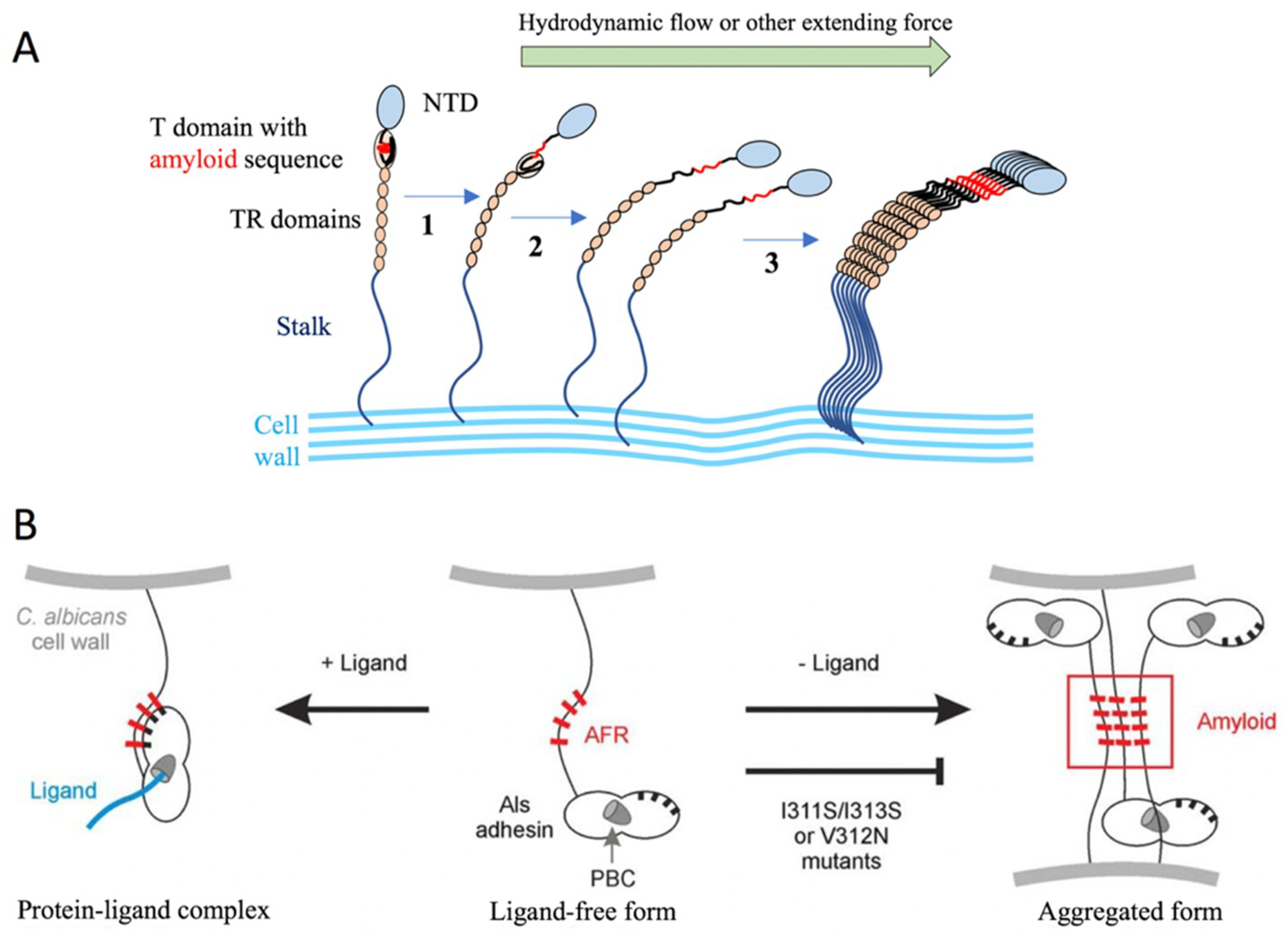
2.1.3. The Agglutinin-like Sequence Protein Family
2.2. Protein Structures of Yeast Adhesins
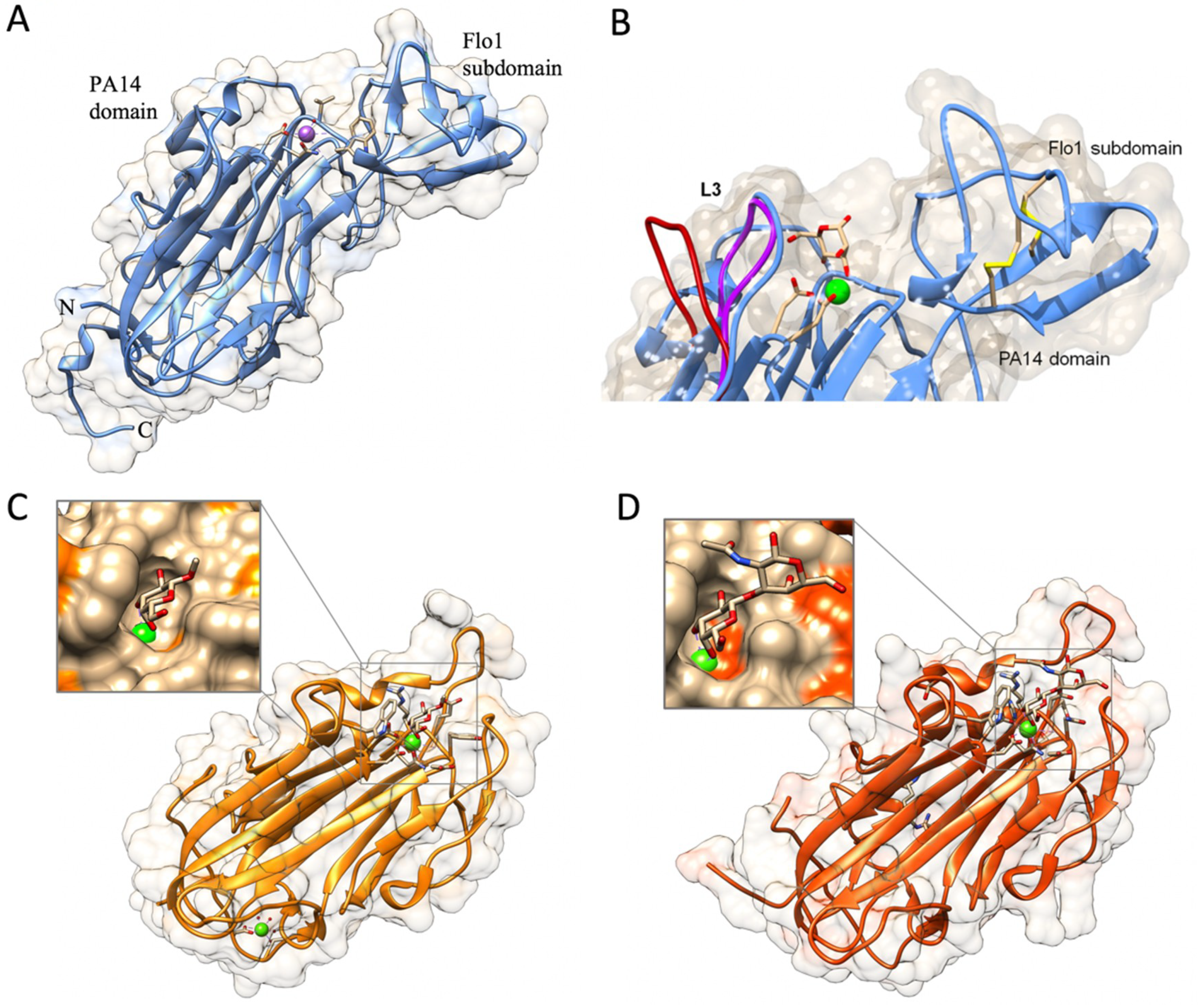
2.2.1. The PA14 Fold in Flocculation Protein and Epithelial Adhesin Family
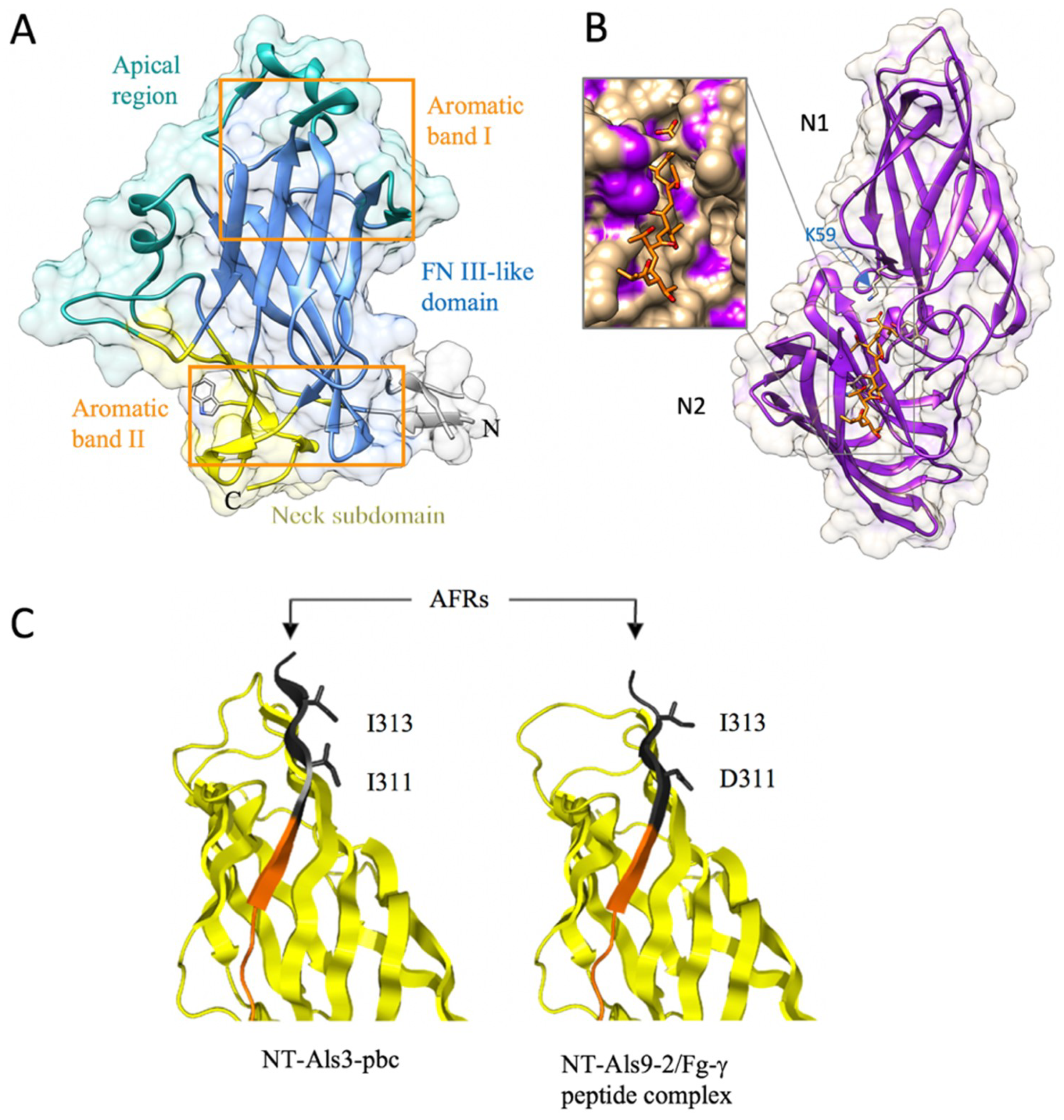
2.2.2. The Flo11 Fold
2.2.3. The Ig-like Fold in Agglutinin-like Sequence Protein (Als) Family
3. Yeast-Yeast and Yeast-Host Cell Interactions
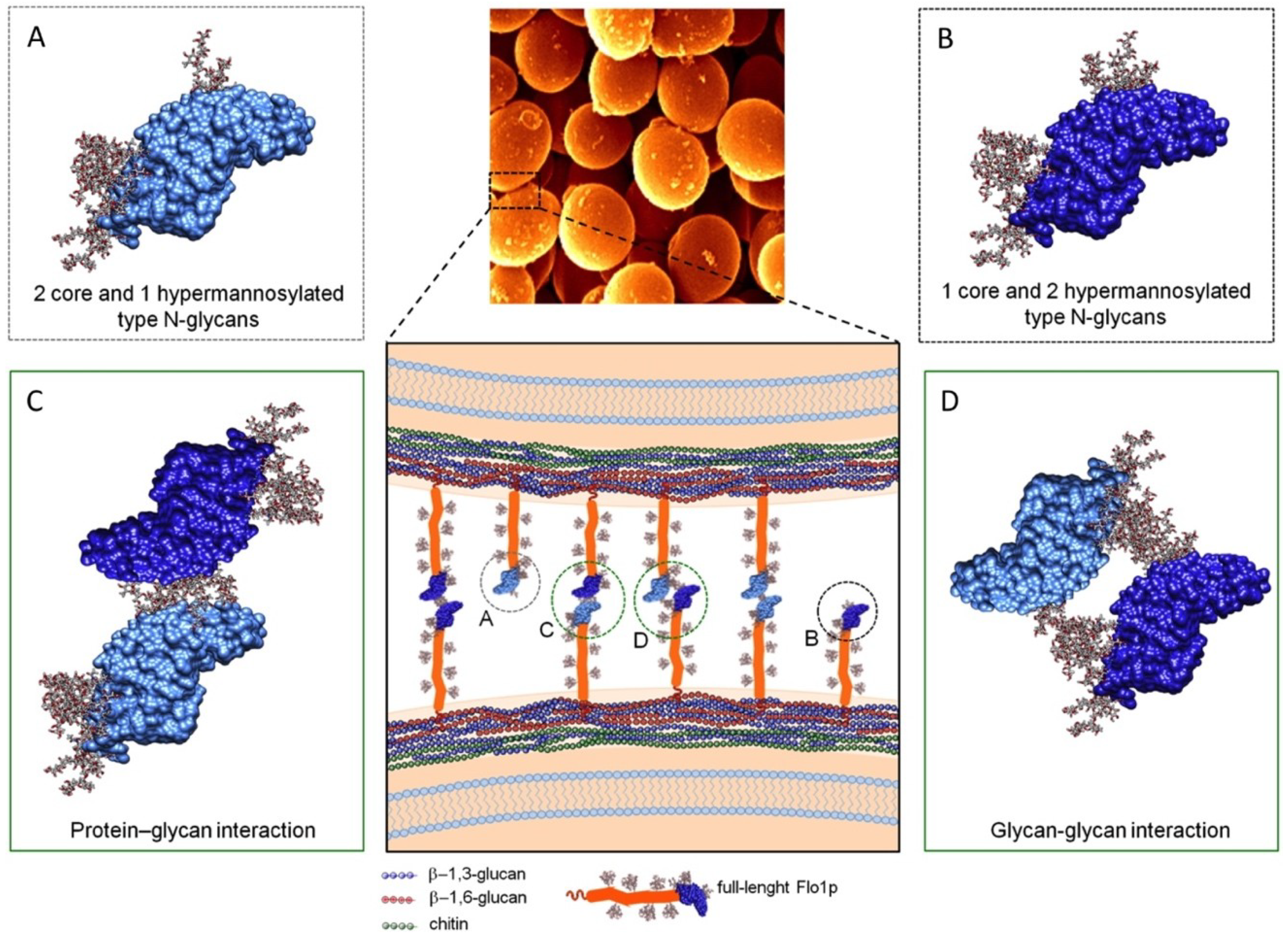
3.1. Flo Proteins Interactions
3.1.1. Cell-Cell Binding Based on S. cerevisiae-Lectin-Flocculin Interaction
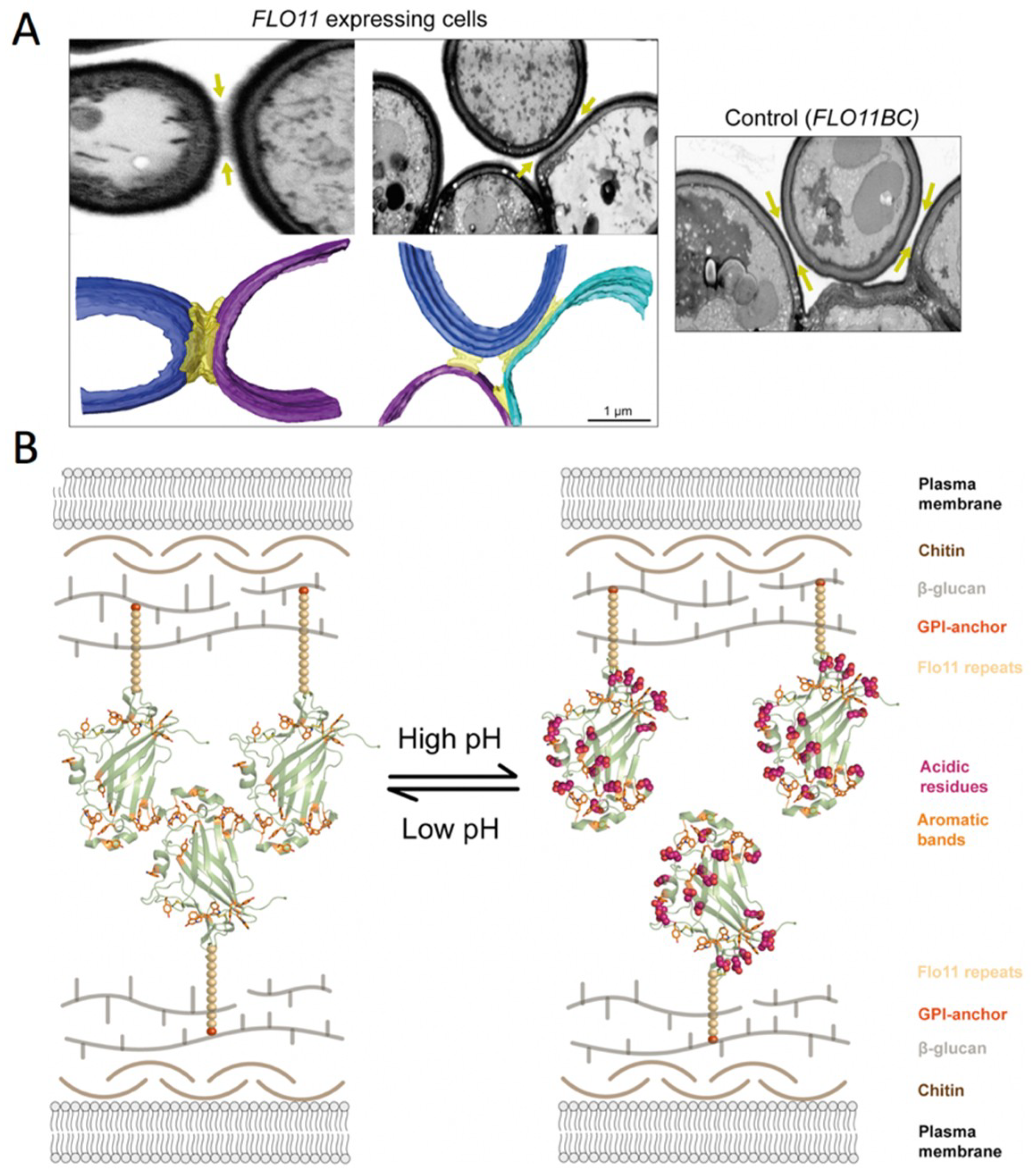
3.1.2. Cell-Cell Binding Based on S. cerevisiae-Flo11 Protein Interaction
3.2. C. albicans Als Protein Interactions
3.3. C. glabrata Epa Protein Interactions
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Teunissen, A.W.R.H.; Steensma, H.Y. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 1995, 11, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.W.J.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in Human Fungal Pathogens: Glue with Plenty of Stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; van Zandvoort, M.A.M.J.; oude Egbrink, M.G.A. The endothelial glycocalyx: Composition, functions, and visualization. Pflugers Arch. 2007, 454, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Critchley, I.A.; Douglas, L.J. Role of Glycosides as Epithelial Cell Receptors for Candida albicans. Microbiology 1987, 133, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Vardar-Ünlü, G.; McSharry, C.; Douglas, L.J. Fucose-specific adhesins on germ tubes of Candida albicans. FEMS Immunol. Med. Microbiol. 2006, 20, 55–67. [Google Scholar] [CrossRef]
- Brückner, S.; Mösch, H.-U. Choosing the right lifestyle: Adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2012, 36, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F.J. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13619–13624. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z. Multicellular microorganisms: Laboratory versus nature. EMBO Rep. 2004, 5, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling Human Fungal Infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; de Onis, M.; Ezzati, M.; Grantham-McGregor, S.; Katz, J.; Martorell, R.; et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef]
- Rahman, S.A.; Adjeroh, D. Surface-Based Body Shape Index and Its Relationship with All-Cause Mortality. PLoS ONE 2015, 10, e0144639. [Google Scholar] [CrossRef] [PubMed]
- Bourke, C.D.; Berkley, J.A.; Prendergast, A.J. Immune Dysfunction as a Cause and Consequence of Malnutrition. Trends Immunol. 2016, 37, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Katona, P.; Katona Apte, J. The Interaction between Nutrition and Infection. Clin. Infect. Dis. 2008, 46, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Olofin, I.; McDonald, C.M.; Ezzati, M.; Flaxman, S.; Black, R.E.; Fawzi, W.W.; Caulfield, L.E.; Danaei, G.; Nutrition Impact Model Study (anthropometry cohort pooling). Associations of Suboptimal Growth with All-Cause and Cause-Specific Mortality in Children under Five Years: A Pooled Analysis of Ten Prospective Studies. PLoS ONE 2013, 8, e64636. [Google Scholar] [CrossRef] [PubMed]
- Rytter, M.J.H.; Kolte, L.; Briend, A.; Friis, H.; Christensen, V.B. The Immune System in Children with Malnutrition—A Systematic Review. PLoS ONE 2014, 9, e105017. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.K.; Zambruni, M.; Melby, C.L.; Melby, P.C. Impact of Childhood Malnutrition on Host Defense and Infection. Clin. Microbiol. Rev. 2017, 30, 919–971. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, R.; Syrjänen, J. Obesity and the risk and outcome of infection. Int. J. Obes. 2012, 37, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Lamps, L.W.; Lai, K.K.T.; Milner, D.A. Fungal Infections of the Gastrointestinal Tract in the Immunocompromised Host. Adv. Anat. Pathol. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Antinori, S.; Milazzo, L.; Sollima, S.; Galli, M.; Corbellino, M. Candidemia and invasive candidiasis in adults: A narrative review. Eur. J. Intern. Med. 2016, 34, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. North. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, V.; Barnes, A.J. Non-albicans Candida spp. causing fungaemia: Pathogenicity and antifungal resistance. J. Hosp. Infect. 2002, 50, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar] [CrossRef]
- Perlroth, J.; Choi, B.; Spellberg, B. Nosocomial fungal infections: Epidemiology, diagnosis, and treatment. Med. Mycol. 2007, 45, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Dougherty, S.D.; Erdman, S.E. Conserved WCPL and CX4C Domains Mediate Several Mating Adhesin Interactions in Saccharomyces cerevisiae. Genetics 2009, 182, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Wojciechowicz, D.; Kurjan, J. AG alpha 1 is the structural gene for the Saccharomyces cerevisiae alpha-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 1989, 9, 3155–3165. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Hauser, K.; Mrśa, V.; Watzele, M.; Watzele, G.; Gruber, C.; Tanner, W. Saccharomyces cerevisiae a- and alpha-agglutinin: Characterization of their molecular interaction. EMBO J. 1991, 10, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Baldermann, C.; Rachel, R.; Tanner, W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: Cell wall attachment and active sites of a- and alpha-agglutinin. EMBO J. 1994, 13, 4737–4744. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Lu, C.F.; Marykwas, D.L.; Lipke, P.N.; Kurjan, J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 1991, 11, 4196–4206. [Google Scholar] [CrossRef] [PubMed]
- Erdman, S.; Lin, L.; Malczynski, M.; Snyder, M. Pheromone-regulated Genes Required for Yeast Mating Differentiation. J. Cell. Biol. 1998, 140, 461–483. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.; Willaert, R. Flocculation protein structure and cell–cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 2010, 32, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; van Eijsden, R.G.E.; Siewers, V.; et al. Molecular Mechanism of Flocculation Self-Recognition in Yeast and Its Role in Mating and Survival. MBio 2015, 6, e00427-15. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.S.; Dranginis, A.M. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 1996, 178, 7144–7151. [Google Scholar] [CrossRef] [PubMed]
- Vajjhala, P.R.; Wong, J.S.; To, H.-Y.; Munn, A.L. The β domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 2006, 273, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Linder, T.; Gustafsson, C.M. Molecular phylogenetics of ascomycotal adhesins—A novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2008, 45, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Decanniere, K.; Willaert, R.G. The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: Structural and functional study of the carbohydrate-binding domain. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Reyna, M.; Diderrich, R.; Veelders, M.S.; Eulenburg, G.; Kalugin, V.; Brückner, S.; Keller, P.; Rupp, S.; Mösch, H.-U.; Essen, L.-O. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. USA 2012, 109, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Bidaud, A.L.; Chowdhary, A.; Dannaoui, E. Candida auris: An emerging drug resistant yeast—A mini-review. J. Mycol. Med. 2018, 28, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida aurison 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Sears, D.; Schwartz, B.S. Candida auris: An emerging multidrug-resistant pathogen. Int. J. Infect. Dis. 2017, 63, 95–98. [Google Scholar] [CrossRef] [PubMed]
- de Cássia Orlandi Sardi, J.; Silva, D.R.; Soares Mendes-Giannini, M.J.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018, 125, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabhaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2018, 64, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Hecht, J.E. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast 2000, 18, 49–60. [Google Scholar] [CrossRef]
- Diderrich, R.; Kock, M.; Maestre-Reyna, M.; Keller, P.; Steuber, H.; Rupp, S.; Essen, L.-O.; Mösch, H.-U. Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family. J. Biol. Chem. 2015, 290, 19597–19613. [Google Scholar] [CrossRef] [PubMed]
- Veelders, M.; Brückner, S.; Ott, D.; Unverzagt, C.; Mösch, H.-U.; Essen, L.-O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.; Groes, M.; Olesen, K.; Henriksen, A. Structural and biochemical characterization of the N-terminal domain of flocculin Lg-Flo1p from Saccharomyces pastorianus reveals a unique specificity for phosphorylated mannose. FEBS J. 2013, 280, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Verhaeghe, T.; Desmet, T.; Willaert, R.G. Engineering the carbohydrate-binding site of Epa1p from Candida glabrata: Generation of adhesin mutants with different carbohydrate specificity. Glycobiology 2014, 24, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, M.G.; Bauer, F.F.; Marmur, J.; Pretorius, I.S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 8419–8424. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.-S.; Dranginis, A.M. The Cell Surface Flocculin Flo11 Is Required for Pseudohyphae Formation and Invasion by Saccharomyces cerevisiae. Mol. Biol. Cell. 1998, 9, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Rupp, S. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999, 18, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.L.; Fink, G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994, 8, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.G.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Jeffery-Smith, A.; Taori, S.K.; Schelenz, S.; Jeffery, K.; Johnson, E.M.; Borman, A.; Manuel, R.; Brown, C.S. Candida auris: A Review of the Literature. Clin. Microbiol. Rev. 2018, 31, e00029-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Oh, S.-H.; Jones, R.; Garnett, J.A.; Salgado, P.S.; Rusnakova, S.; Matthews, S.J.; Hoyer, L.L.; Cota, E. The Peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J. Biol. Chem. 2014, 289, 18401–18412. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.S.; Yan, R.; Taylor, J.D.; Burchell, L.; Jones, R.; Hoyer, L.L.; Matthews, S.J.; Simpson, P.J.; Cota, E. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 15775–15779. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, T.; Brückner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mösch, H.-U.; Essen, L.-O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2004, 237, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Kron, S.J.; Styles, C.A.; Fink, G.R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1994, 5, 1003–1022. [Google Scholar] [CrossRef] [PubMed]
- Mösch, H.U. Pseudohyphal Development of Saccharomyces cerevisiae. Contrib. Microbiol. 2000, 5, 185–200. [Google Scholar]
- Reynolds, T.B.; Fink, G.R. Bakers’ Yeast, a Model for Fungal Biofilm Formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Jansen, A.; Peng, X.; Fink, G.R. Mat Formation in Saccharomyces cerevisiae Requires Nutrient and pH Gradients. Eukaryot. Cell 2008, 7, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11-Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 Is the Primary Factor in Flor Formation Caused by Cell Surface Hydrophobicity in Wild-Type Flor Yeast. Biosci. Biotechnol. Biochem. 2014, 70, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, H.D.; Dupont, K.; Jespersen, L.; Willats, W.G.T.; Arneborg, N. Identification of amino acids involved in the Flo11p-mediated adhesion of Saccharomyces cerevisiaeto a polystyrene surface using phage display with competitive elution. J. Appl. Microbiol. 2007, 103, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Bayly, J.; Douglas, L.; Pretorius, I.; Bauer, F.; Dranginis, A. Characteristics of Flo11-dependent flocculation in. FEMS Yeast Res. 2005, 5, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and Characterization of the Flocculin Flo11/Muc1, a Saccharomyces cerevisiae Mannoprotein with Homotypic Properties of Adhesion. Eukaryot. Cell 2007, 6, 2214–2221. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Willaert, R.G. The N-terminal domain of the Flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2011, 12, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Smit, G.; Straver, M.H.; Lugtenberg, B.J.; Kijne, J.W. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 1992, 58, 3709–3714. [Google Scholar] [PubMed]
- Straver, M.H.; Aar, P.C.V.D.; Smit, G.; Kijne, J.W. Determinants of flocculence of brewer’s yeast during fermentation in wort. Yeast 1993, 9, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Wilcocks, K. The importance of surface charge and hydrophobicity for the flocculation of chain-forming brewing yeast strains and resistance of these parameters to acid washing. FEMS Microbiol. Lett. 1995, 134, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Li, L.; Lipke, P.N.; Dranginis, A.M. Molecular Basis for Strain Variation in the Saccharomyces cerevisiaeAdhesin Flo11p. mSphere 2016, 1, e00129-16. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Simpson, P.J.; Matthews, S.J.; Cota, E. Backbone 1H, 15N, 13C and Ile, Leu, Val methyl chemical shift assignments for the 33.5 kDa N-terminal domain of Candida albicans ALS1. Biomol. NMR Assign. 2010, 4, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.S.; Yan, R.; Rowan, F.; Cota, E. Expression, crystallization and preliminary X-ray data analysis of NT-Als9-2, a fungal adhesin from Candida albicans. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2011, 67, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Ponnuraj, K.; Bowden, M.G.; Davis, S.; Gurusiddappa, S.; Moore, D.; Choe, D.; Xu, Y.; Höök, M.; Narayana, S.V.L. A “dock, lock, and latch” Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen. Cell 2003, 115, 217–228. [Google Scholar] [CrossRef]
- Bowden, M.G.; Heuck, A.P.; Ponnuraj, K.; Kolosova, E.; Choe, D.; Gurusiddappa, S.; Narayana, S.V.L.; Johnson, A.E.; Höök, M. Evidence for the “Dock, Lock, and Latch” Ligand Binding Mechanism of the Staphylococcal Microbial Surface Component Recognizing Adhesive Matrix Molecules (MSCRAMM) SdrG. J. Biol. Chem. 2008, 283, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, V.K.; Rivera, J.J.; Smeds, E.; Ko, Y.-P.; Bowden, M.G.; Wann, E.R.; Gurusiddappa, S.; Fitzgerald, J.R.; Höök, M. A Structural Model of the Staphylococcus aureus ClfA–Fibrinogen Interaction Opens New Avenues for the Design of Anti-Staphylococcal Therapeutics. PLoS Pathog. 2008, 4, e1000226. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Green, C.B.; Oh, S.-H.; Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 2008, 46, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, J.E., Jr. Functional and Structural Diversity in the Als Protein Family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489. [Google Scholar] [CrossRef] [PubMed]
- Loza, L.; Fu, Y.; Ibrahim, A.S.; Sheppard, D.C.; Filler, S.G.; Edwards, J.E. Functional analysis of theCandida albicans ALS1 gene product. Yeast 2004, 21, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 2004, 150, 2415–2428. [Google Scholar] [CrossRef] [PubMed]
- Otoo, H.N.; Lee, K.G.; Qiu, W.; Lipke, P.N. Candida albicans Als Adhesins Have Conserved Amyloid-Forming Sequences. Eukaryot. Cell 2008, 7, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrêne, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Garcia, M.C.; Alsteens, D.; Ramsook, C.B.; Klotz, S.A.; Dufrêne, Y.F. Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012, 20, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and Mammalian Transglutaminase Substrate Properties of Candida albicans Hwp1. Science 1999, 283, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Tsuchimori, N.; Sharkey, L.L.; Fonzi, W.A.; French, S.W.; Edwards, J.E.; Filler, S.G. Reduced Virulence of HWP1-Deficient Mutants of Candida albicans and Their Interactions with Host Cells. Infect. Immun. 2000, 68, 1997–2002. [Google Scholar] [CrossRef] [PubMed]
- Staab, J.F.; Bahn, Y.-S.; Tai, C.-H.; Cook, P.F.; Sundstrom, P. Expression of Transglutaminase Substrate Activity on Candida albicans Germ Tubes through a Coiled, Disulfide-bonded N-terminal Domain of Hwp1 Requires C-terminal Glycosylphosphatidylinositol Modification. J. Biol. Chem. 2004, 279, 40737–40747. [Google Scholar] [CrossRef] [PubMed]
- Dalle, F.; Wächtler, B.; L’Ollivier, C.; Holland, G.; Bannert, N.; Wilson, D.; Labruère, C.; Bonnin, A.; Hube, B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 2010, 12, 248–271. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Moyes, D.L.; Wächtler, B.; Hube, B. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011, 13, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; He, H.; Dong, Y.; Pan, H. Hyphae-Specific Genes HGC1, ALS3, HWP1, and ECE1 and Relevant Signaling Pathways in Candida albicans. Mycopathologia 2013, 176, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Nguyen, Q.B.; Wolyniak, M.J.; Frechette, G.; Lehman, C.R.; Fox, B.K.; Sundstrom, P. Release of transcriptional repression through the HCR promoter region confers uniform expression of HWP1 on surfaces of Candida albicans germ tubes. PLoS ONE 2018, 13, e0192260-24. [Google Scholar] [CrossRef] [PubMed]
- Hayek, P.; Dib, L.; Yazbeck, P.; Beyrouthy, B.; Khalaf, R.A. Characterization of Hwp2, a Candida albicans putative GPI-anchored cell wall protein necessary for invasive growth. Microbiol. Res. 2010, 165, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Schneider, H.A.; Nett, J.E.; Sheppard, D.C.; Filler, S.G.; Andes, D.R.; Mitchell, A.P. Complementary Adhesin Function in C. albicans Biofilm Formation. Curr. Biol. 2008, 18, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Andes, D.R.; Nett, J.E.; Smith, F.J.; Yue, F.; Phan, Q.-T.; Edwards, J.E.; Filler, S.G.; Mitchell, A.P. Critical Role of Bcr1-Dependent Adhesins in C. albicans Biofilm Formation In Vitro and In Vivo. PLoS Pathog. 2006, 2, e63. [Google Scholar] [CrossRef] [PubMed]
- Ene, I.V.; Bennett, R.J. Hwp1 and Related Adhesins Contribute to both Mating and Biofilm Formation in Candida albicans. Eukaryot. Cell 2009, 8, 1909–1913. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Vickerman, M.M.; Jenkinson, H.F. Heterologous Expression of Candida albicans Cell Wall-Associated Adhesins in Saccharomyces cerevisiae Reveals Differential Specificities in Adherence and Biofilm Formation and in Binding Oral Streptococcus gordonii. Eukaryot. Cell 2010, 9, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.; Thompson, A.; Xie, Z.; Kashleva, H.; Ganguly, S.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Role of Bcr1-Activated Genes Hwp1 and Hyr1 in Candida Albicans Oral Mucosal Biofilms and Neutrophil Evasion. PLoS ONE 2011, 6, e16218. [Google Scholar] [CrossRef] [PubMed]
- Orsi, C.F.; Borghi, E.; Colombari, B.; Neglia, R.G.; Quaglino, D.; Ardizzoni, A.; Morace, G.; Blasi, E. Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb. Pathog. 2014, 69–70, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ponniah, G.; Rollenhagen, C.; Bahn, Y.-S.; Staab, J.F.; Sundstrom, P. State of differentiation defines buccal epithelial cell affinity for cross-linking to Candida albicans Hwp1. J. Oral Patho.l Med. 2007, 36, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Stassen, C.; Stals, I.; Donohue, D.S.; Devreese, B.; De Greve, H.; Willaert, R.G. The N-Terminal Domain of the Flo1 Flocculation Protein from Saccharomyces cerevisiae Binds Specifically to Mannose Carbohydrates. Eukaryot. Cell 2011, 10, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Donohue, D.S.; Ielasi, F.S.; Goossens, K.V.Y.; Willaert, R.G. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol. Microbiol. 2011, 80, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. MBio 2016, 7, e00584-16. [Google Scholar] [CrossRef] [PubMed]
- Bony, M.; Thines-Sempoux, D.; Barre, P.; Blondin, B. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J. Bacteriol. 1997, 179, 4929–4936. [Google Scholar] [CrossRef] [PubMed]
- Spillmann, D.; Burger, M.M. Carbohydrate-carbohydrate interactions in adhesion. J. Cell. Biochem. 1996, 61, 562–568. [Google Scholar] [CrossRef]
- Haseley, S.R.; Vermeer, H.J.; Kamerling, J.P.; Vliegenthart, J.F.G. Carbohydrate self-recognition mediates marine sponge cellular adhesion. Proc. Natl. Acad. Sci. USA 2001, 98, 9419–9424. [Google Scholar] [CrossRef] [PubMed]
- Bucior, I.; Burger, M.M. Carbohydrate–carbohydrate interactions in cell recognition. Curr. Opin. Struct. Biol. 2004, 14, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Busquets, X.; Kornig, A.; Bucior, I.; Burger, M.M.; Anselmetti, D. Self-Recognition and Ca2+-Dependent Carbohydrate-Carbohydrate Cell Adhesion Provide Clues to the Cambrian Explosion. Mol. Biol. Evol. 2009, 26, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Mill, P.J. The Nature of the Interactions between Flocculent Cells in the Flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1964, 35, 61–68. [Google Scholar] [CrossRef] [PubMed]
- JAYATISSA, P.M.; ROSE, A.H. Role of Wall Phosphomannan in Flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1976, 96, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Miki, B.L.A.; Poon, N.H.; James, A.P.; Seligy, V.L. Possible Mechanism for Flocculation Interactions Governed by Gene FLO1 in Saccharomyces cerevisiae. J. Bacteriol. 1982, 150, 878–889. [Google Scholar] [PubMed]
- Willaert, R.; Kasas, S.; Devreese, B.; Dietler, G. Yeast Nanobiotechnology. Fermentation 2016, 2, 18. [Google Scholar] [CrossRef]
- Alsteens, D.; Beaussart, A.; Derclaye, S.; El-Kirat-Chatel, S.; Park, H.R.; Lipke, P.N.; Dufrêne, Y.F. Single-cell force spectroscopy of Als-mediated fungal adhesion. Anal. Methods 2013, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- El-Kirat-Chatel, S.; Beaussart, A.; Derclaye, S.; Alsteens, D.; Kucharíková, S.; Van Dijck, P.; Dufrêne, Y.F. Force Nanoscopy of Hydrophobic Interactions in the Fungal Pathogen Candida glabrata. ACS Nano 2015, 9, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Touhami, A. Aggregation of yeast cells: Direct measurement of discrete lectin-carbohydrate interactions. Microbiology 2003, 149, 2873–2878. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Šťovíček, V.; Hlaváček, O.; Chernyavskiy, O.; Štěpánek, L.; Kubínová, L.; Palková, Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell. Biol. 2011, 194, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Gaur, N.K.; Lake, D.F.; Chan, V.; Rauceo, J.; Lipke, P.N. Degenerate Peptide Recognition by Candida albicans Adhesins Als5p and Als1p. Infect. Immun. 2004, 72, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Wishart, J.A.; Leng, R.; Macaskill, S.; Mavor, A.; Alexandris, T.; Nicholls, S.; Knight, A.W.; Enjalbert, B.; Walmsley, R.; et al. Developmental Regulation of an Adhesin Gene during Cellular Morphogenesis in the Fungal Pathogen Candida albicans. Eukaryot. Cell 2007, 6, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a Multifunctional Adhesin and Invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Issa, S.; Moran, A.P.; Ustinov, S.N.; Lin, J.H.-H.; Ligtenberg, A.J.; Karlsson, N.G. O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine. Glycobiology 2010, 20, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.P.; Haidaris, C.G. Analysis of Candida albicans Adhesion to Salivary Mucin. Infect. Immun. 1993, 61, 1940–1949. [Google Scholar] [PubMed]
- de Repentigny, L.; Aumont, F.; Bernard, K.; Belhumeur, P. Characterization of Binding of Candida albicans to Small Intestinal Mucin and Its Role in Adherence to Mucosal Epithelial Cells. Infect. Immun. 2000, 68, 3172–3179. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.C.; Lee, J.T.; Ramsook, C.B.; Alsteens, D.; Dufrêne, Y.F.; Lipke, P.N. A Role for Amyloid in Cell Aggregation and Biofilm Formation. PLoS ONE 2011, 6, e17632. [Google Scholar] [CrossRef] [PubMed]
- de Nobel, H.; Lipke, P.N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell. Biol. 1994, 4, 42–45. [Google Scholar] [CrossRef]
- Lipke, P.N.; Klotz, S.A.; Dufrêne, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microb. Mol. Biol. Rev. 2018, 82, 237. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans Agglutinin-Like Sequence (Als) Family Vignettes: A Review of Als Protein Structure and Function. Front. Microbiol. 2016, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and Characterization of an In Vivo Central Venous Catheter Candida albicans Biofilm Model. Infect. Immun. 2004, 72, 6023–6031. [Google Scholar] [CrossRef] [PubMed]
- Shuford, J.A.; Rouse, M.S.; Piper, K.E.; Steckelberg, J.M.; Patel, R. Evaluation of Caspofungin and Amphotericin B Deoxycholate against Candida albicans Biofilms in an Experimental Intravascular Catheter Infection Model. J. Infect. Dis. 2006, 194, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Lazzell, A.L.; Chaturvedi, A.K.; Pierce, C.G.; Prasad, D.; Uppuluri, P.; Lopez-Ribot, J.L. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J. Antimicrob. Chemother. 2009, 64, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Řičicová, M.; Kucharíková, S.; Tournu, H.; Hendrix, J.; Bujdáková, H.; Van Eldere, J.; Lagrou, K.; Van Dijck, P. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 2010, 156, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Brooks, E.G.; Cabezas-Olcoz, J.; Sanchez, H.; Zarnowski, R.; Marchillo, K.; Andes, D.R. Rat Indwelling Urinary Catheter Model of Candida albicans Biofilm Infection. Infect. Immun. 2014, 82, 4931–4940. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Bui, C.K.; Nett, J.E.; Hartooni, N.; Mui, M.C.; Andes, D.R.; Nobile, C.J.; Johnson, A.D. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol. Microbiol. 2015, 96, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.B.; Gulati, M.; Johnson, A.D.; Nobile, C.J. Development and regulation of single- and multi-species Candida albicans biofilms. Nat. Rev. Microbiol. 2017, 16, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A Biochemical Guide to Yeast Adhesins: Glycoproteins for Social and Antisocial Occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Palecek, S.P. Distinct domains of the Candida albicans adhesin Eap1p mediate cell–cell and cell–substrate interactions. Microbiology 2008, 154, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Frade, J.P.; Arthington-Skaggs, B.A. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses 2010, 54, e154–e162. [Google Scholar] [CrossRef] [PubMed]
- Sandini, S.; Stringaro, A.; Arancia, S.; Colone, M.; Mondello, F.; Murtas, S.; Girolamo, A.; Mastrangelo, N.; De Bernardis, F. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.T.; Silva, S.; Pereira, L.; Williams, D.W.; Azeredo, J.; Henriques, M. Effect of progesterone on Candida albicans vaginal pathogenicity. Int J. Med. Microbiol. 2014, 304, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; Mitchell, A.P. Candida albicans Biofilm Development and Its Genetic Control. Microbiol. Spectr. 2015, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Zupancic, M.L.; Frieman, M.; Smith, D.; Alvarez, R.A.; Cummings, R.D.; Cormack, B.P. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 2008, 68, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Wolf, M. Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Hospenthal, M.K.; Zyla, D.; Costa, T.R.D.; Redzej, A.; Giese, C.; Lillington, J.; Glockshuber, R.; Waksman, G. The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod. Structure 2017, 25, 1829–1838. [Google Scholar] [CrossRef] [PubMed]

| Micro-Organism | Adhesin | Ligand in the Structure | Mutations | PDB Code | Interacting Substrate/Function Properties | Refs |
|---|---|---|---|---|---|---|
| C. albicans | NT-Als3p (residues1-313) | Heptathreonine | - | 4LEB | Endothelial and epithelial cells; fibronectin, laminin, type-IV collagen; abiotic surfaces (glass, plastics) | [62] |
| Apo | K59M, A116V, Y301F | 4LEE | [62] | |||
| NT-Als3p (residues 1-299) | Apo | - | 4LE8 | * | ||
| NT-Als9-2p | Apo | - | 2Y7N | [63] | ||
| Apo | G299W | 2Y7O, 2YLH | ||||
| Fg-γ 1 | - | 2Y7L | ||||
| C. glabrata | N-Epa1p | Gal | - | 4A3X | Epithelial cells, fibronectin, mucin | [43] |
| Galβ-1,3-Glc | - | 4AF9 | [44] | |||
| Galβ-1,3-Glc | E227D, Y228N | 4AFC | [44] | |||
| Galβ-1,3-GalNAc (T antigen) | - | 4ASL | [44] | |||
| 4D3W | [52] | |||||
| Galβ-1,4-Glc (lactose) | 4COU | [52] | ||||
| Glycerol | E227D, Y228N, D229N | 4AFA | [44] | |||
| Glycerol | R226I, E227G, Y228K | 4AFB | [44] | |||
| N-Epa6p | Galβ-1,4-Glc (lactose) | 4COU | [52] | |||
| Galβ-1,3-GalNAc (T-antigen) | 4COW | [52] | ||||
| N-acetyl-D-lactosamine | 4COY | [52] | ||||
| Lacto-N-biose | 4COZ | [52] | ||||
| α1-3-galactobiose | 4COV | [52] | ||||
| N-Epa9p | Galβ-1,4-Glc | - | 4CP0 | * | ||
| Galβ-1,3-GlcNAc | - | 4CP1 | * | |||
| Galβ-1,4-GlcNAc | - | 4CP2 | * | |||
| S. cerevisiae | N-Flo1p | Apo | - | 4LHL | Cell-cell interaction via cell surface mannans | [39] |
| Man | - | 4LHK | [39] | |||
| N-Lg-Flo1p | Apo | - | 4GQ7 | Cell-cell interaction via cell surface mannans and phosphomannans | [5] | |
| Manα-1,2-Man | - | 4LHK | [39] | |||
| N-Lg-Flo5p | Apo | - | 2XJQ | Cell-cell interaction via cell surface mannans | [6] | |
| Man | - | 2XJP | [6] | |||
| Man3(D1) | - | 2XJT | [6] | |||
| Man5(D2-3) | - | 2XJR | [6] | |||
| Manα-1,2-Man | - | 2XJS | [6] | |||
| Manα-1,2-Man | S277A | 2XJU | [6] | |||
| Glc | - | 2XJV | [6] | |||
| N-Flo11p | Apo | - | 4UYR | Cell-cell and cell-hydrophobic plastic adhesion via hydrophobic interactions | [64] | |
| - | 4UYS | [64] | ||||
| - | 4UYT | [64] |
| Microorganism | Adhesin | Ligand | Dissociation Constant KD (mM) | Refs |
|---|---|---|---|---|
| C. albicans | N-Als1p | Fibronectin | 0.0016 ± 0.0006 | [111] |
| Laminin | 0.013 ± 0.002 | [111] | ||
| Fucose | 0.21 ± 0.03 | [111] | ||
| N-Als1p | 0.020 ± 0.001 | [111] | ||
| N-Als3p | Fibronectin | 0.41 ± 0.04 | [112] | |
| Laminin | 0.01 ± 0.001 | [112] | ||
| GlcNAc | 0.034 ± 0.004 | [112] | ||
| N-Als3p | 0.014 ± 0.002 | [112] | ||
| C. glabrata | N-Epa1p | Galβ | 0.115 ± 0.011 | [43] |
| Galβ-1,4-Glc 1 | 0.031 ± 0.004 | [43] | ||
| 0.035 ± 0.006 | [3] | |||
| Galα-1,3-Gal 2 | 0.0050 ± 0.0009 | [7] | ||
| Galβ-1,3-Gal 3 | 0.0009 ± 0.00005 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0035 ± 0.0004 | [43] | ||
| 0.0021 ± 0.0003 | [3] | |||
| 0.0009 ± 0.0001 | [7] | |||
| Galβ-1,3-GlcNAc 5 | 0.0049 ± 0.003 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0020 ± 0.0002 | [7] | ||
| Mucin | 0.0047 ± 0.0009 | [112] | ||
| Fibronectin | 0.911 10−3 ± 0.122 10−3 | [8] | ||
| N-Epa1p E227A | Fibronectin | 0.317 10−3 ± 0.026 10−3 | [8] | |
| N-Epa1p Y228W | Fibronectin | 0.545 10−3 ± 0.058 10−3 | [8] | |
| N-Epa6p | Galα-1,3-Gal 2 | 0.0004 ± 0.0007 | [7] | |
| Galβ-1,3-Gal 3 | 0.0003 ± 0.0002 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0005 ± 0.0011 | [7] | ||
| Galβ-1,3-GlcNAc 5 | 0.0148 ± 0.0001 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0094 ± 0.0013 | [7] | ||
| N-Epa7p | Galα-1,3-Gal 2 | 0.0124 ± 0.0009 | [7] | |
| Galβ-1,3-Gal 3 | 0.0013 ± 0.0001 | [7] | ||
| Galβ-1,3-GalNAc 4 | 0.0015 ± 0.00001 | [7] | ||
| Galβ-1,3-GlcNAc 5 | 0.0147 ± 0.0004 | [7] | ||
| Galβ-1,4-GlcNAc 6 | 0.0048 ± 0.0004 | [7] | ||
| S. cerevisiae | N-Flo1p | Man | 8.7 ± 0.4 | [39] |
| Manα-1,2-Man | 0.6 ± 0.1 | [39] | ||
| Manα-1,3-Man | 3.3 ± 0.3 | [39] | ||
| Manα-1,6-Man | 6.9 ± 0.6 | [39] | ||
| Glc | >100 | [39] | ||
| N-Lg-Flo1p | Man | 0.8 ± 0.03 | [54] | |
| Man | 0.5 ± 0.05 | [39] | ||
| Man1P 7 | 0.06 ± 0.002 | [54] | ||
| Manα-1,2-Man | 4.5 ± 0.38 | [54] | ||
| Manα-1,3-Man | 3.9 ± 0.98 | [54] | ||
| Manα-1,6-Man | 3.0 ± 0.33 | [54] | ||
| Glc | 5.8 ± 0.80 | [54] | ||
| Glc1P 8 | 0.41 ± 0.03 | [54] | ||
| N-Flo5p (S227A) | Man | 29.3 ± 3.6 (9.7 ± 0.6) | [6] | |
| Manα-1,2-Man | 3.5 ± 0.3 (1.6 ± 0.1) | [6] | ||
| Manα-1,3-Man | No binding | [6] | ||
| Manα-1,6-Man | No binding | [6] | ||
| Man3(D1) | 2.8 ± 0.2 | [6] | ||
| Man5(D2-3) | 2.2 ± 0.5 | [6] | ||
| Glc | >1000 | [6] | ||
| N-Flo11p | N-Flo11 | 0.0195 ± 0.0023 | [64] |
| Cell type | Adhesin | Ligand | Rupture force | Comments | Refs |
|---|---|---|---|---|---|
| C. albicans | Als5p | Fibronectin | 2800 ± 600 pN | SMFS 1 in vitro | [122] |
| C. glabrata | Epa6p | Hydrophobic surface (dodecane-thiol) | 31–52 nN | SCFC 2: cell immobilization on the AFM probe using dopamine | [123] |
| S. cerevisiae | Flo1p | Flo1p | 300 (100–600) | SMFS | [39] |
| S. pastorianus | Lg-Flo1p | Glucose | 121 ± 53 pN | Amylose-coated AFM probe interact with cell surface | [124] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willaert, R.G. Adhesins of Yeasts: Protein Structure and Interactions. J. Fungi 2018, 4, 119. https://doi.org/10.3390/jof4040119
Willaert RG. Adhesins of Yeasts: Protein Structure and Interactions. Journal of Fungi. 2018; 4(4):119. https://doi.org/10.3390/jof4040119
Chicago/Turabian StyleWillaert, Ronnie G. 2018. "Adhesins of Yeasts: Protein Structure and Interactions" Journal of Fungi 4, no. 4: 119. https://doi.org/10.3390/jof4040119
APA StyleWillaert, R. G. (2018). Adhesins of Yeasts: Protein Structure and Interactions. Journal of Fungi, 4(4), 119. https://doi.org/10.3390/jof4040119





