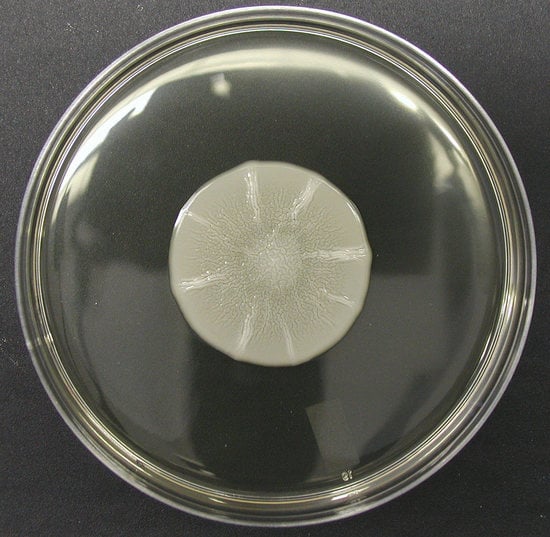
Going with the Flo: The Role of Flo11-Dependent and Independent Interactions in Yeast Mat Formation
Abstract
:1. Introduction
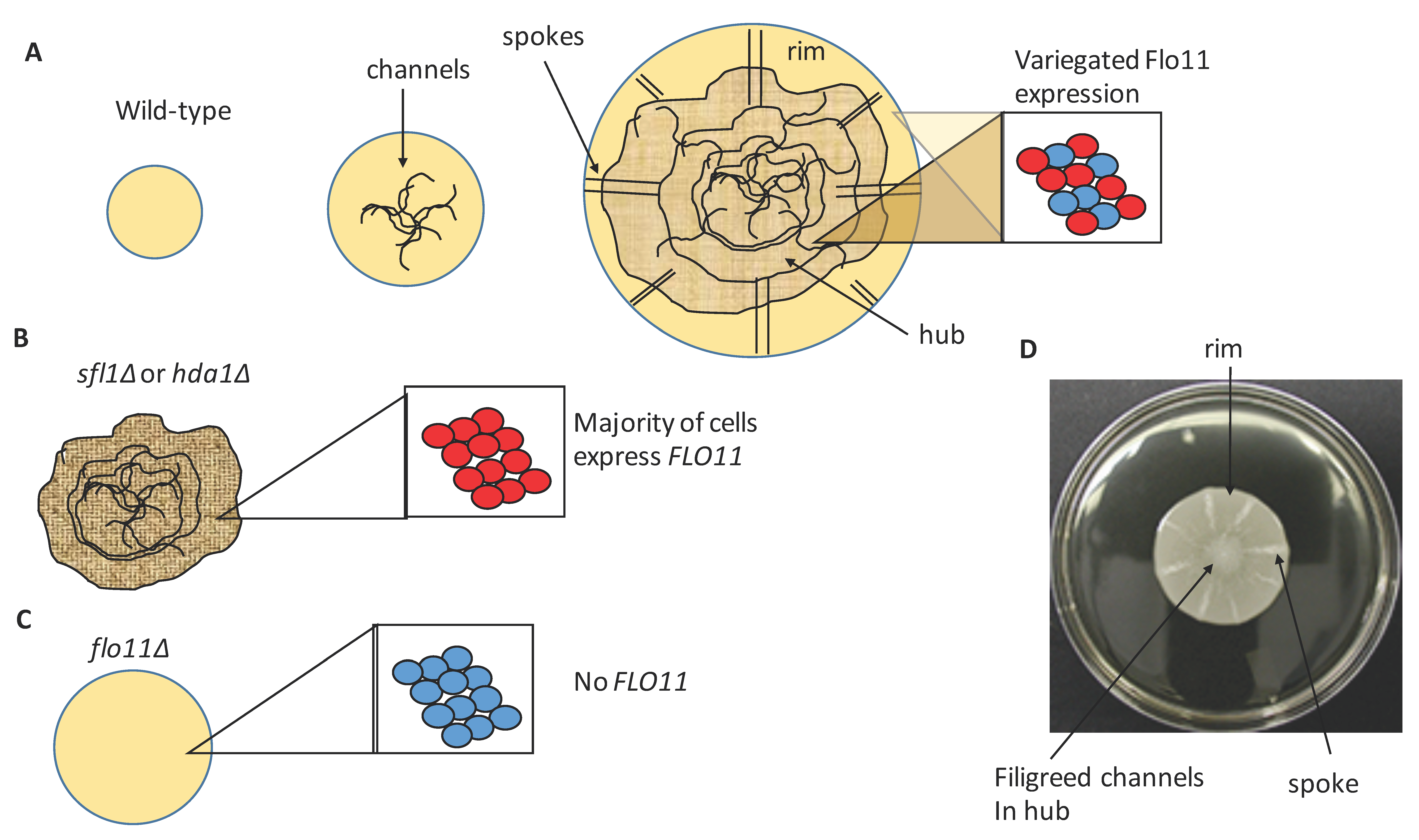
2. Mat Formation Is Dependent on Flo11
3. Environmental Conditions Control Mat Formation
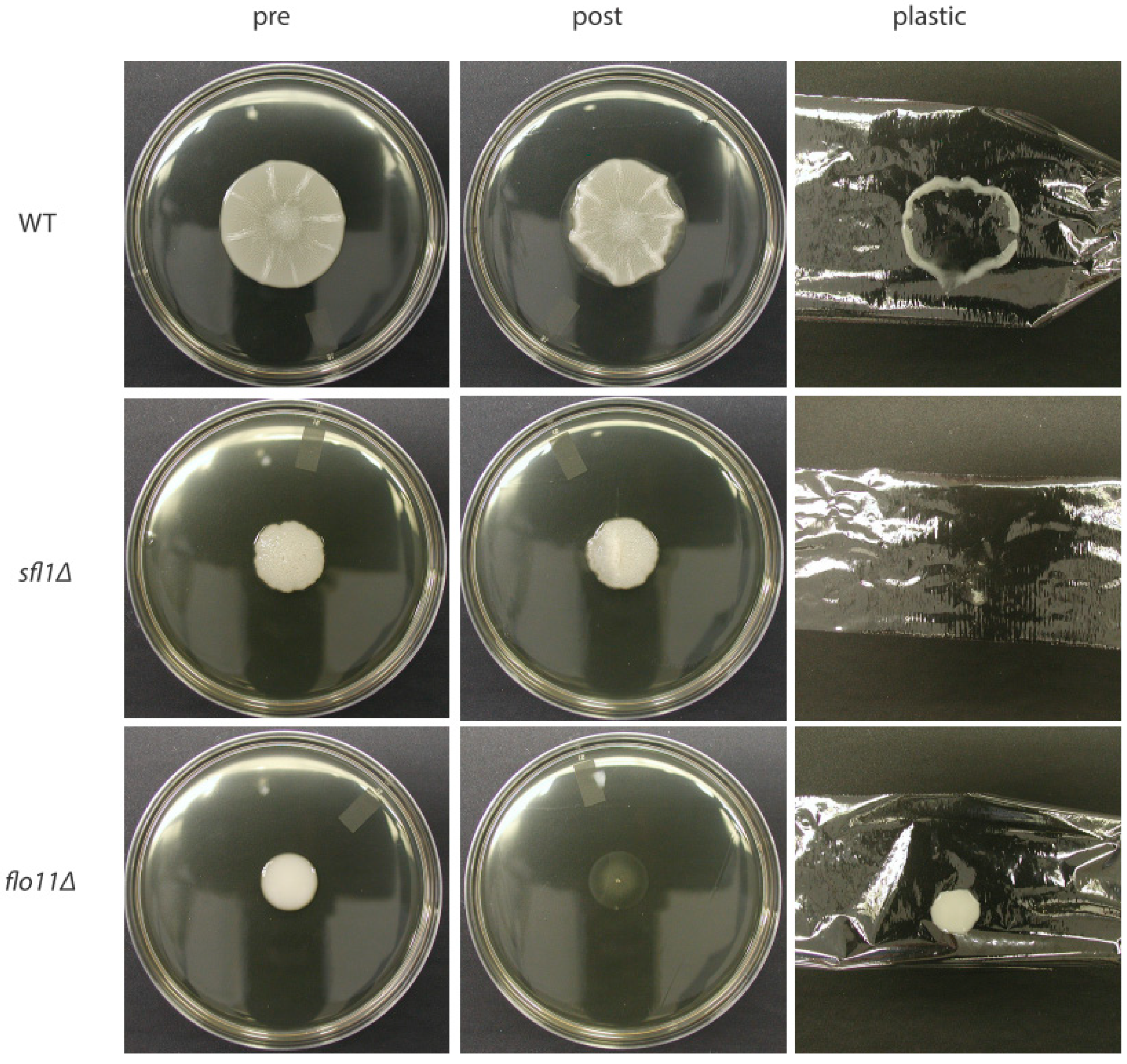
4. Cellular Flo11 Distribution Influences Mat Formation and Fitness in Low Agar Medium
5. Flo11 Expression Impacts Mat Formationf
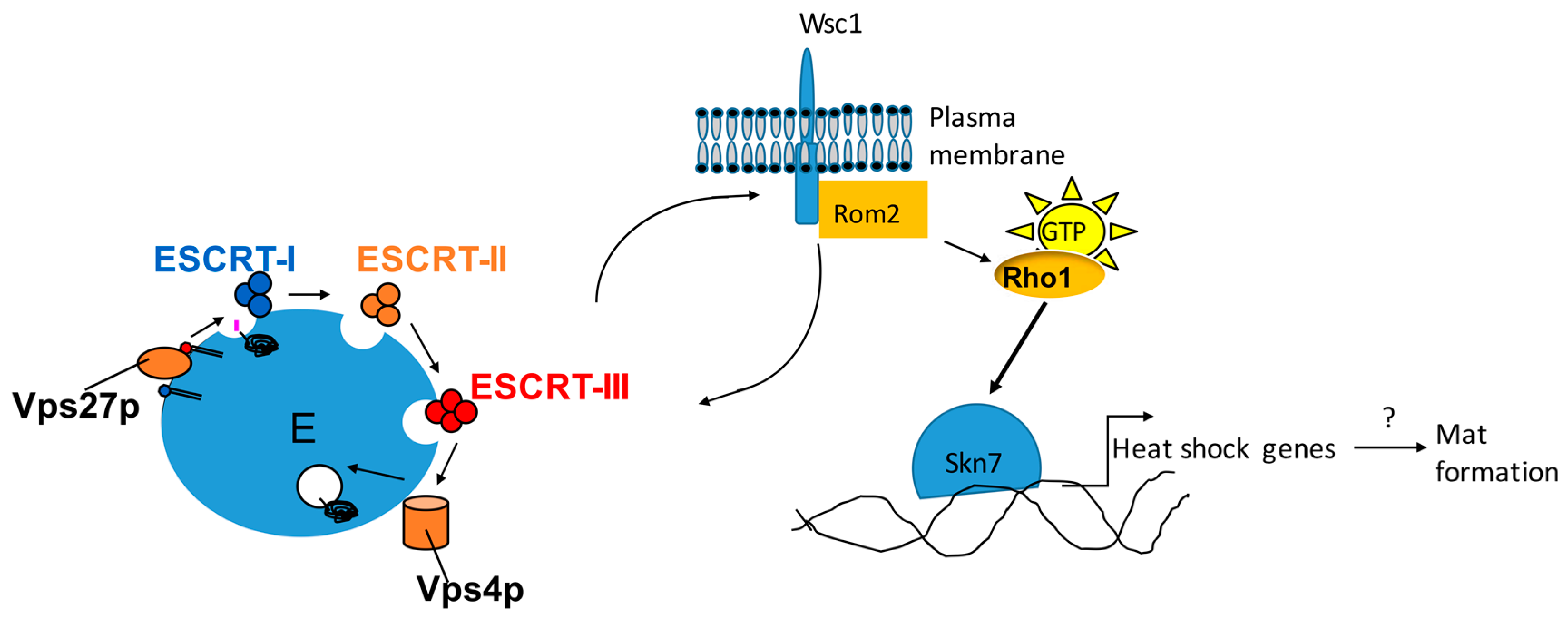
6. Flo11 Is Necessary but Not Sufficient for Mat Formation
7. Conclusions
Funding
Conflicts of Interest
References
- Verstrepen, K.J.; Reynolds, T.B.; Fink, G.R. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2004, 2, 533–540. [Google Scholar] [CrossRef] [Green Version]
- Verstrepen, K.J.; Fink, G.R. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu. Rev. Genet. 2009, 43, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Styles, C.A.; Fink, G.R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 1996, 144, 967–978. [Google Scholar] [PubMed]
- Regenberg, B.; Hanghoj, K.E.; Andersen, K.S.; Boomsma, J.J. Clonal yeast biofilms can reap competitive advantages through cell differentiation without being obligatorily multicellular. Proc. R. Soc. B. 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Pitoniak, A.; Birkaya, B.; Dionne, H.M.; Vadaie, N.; Cullen, P.J. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol. Biol. Cell 2009, 20, 3101–3114. [Google Scholar] [CrossRef] [PubMed]
- Hope, E.A.; Dunham, M.J. Ploidy-regulated variation in biofilm-related phenotypes in natural isolates of Saccharomyces cerevisiae. G3 2014, 4, 1773–1786. [Google Scholar] [CrossRef] [PubMed]
- Tek, E.L.; Sundstrom, J.F.; Gardner, J.M.; Oliver, S.G.; Jiranek, V. Evaluation of the ability of commercial wine yeasts to form biofilms (mats) and adhere to plastic: Implications for the microbiota of the winery environment. FEMS Microbiol. Ecol. 2018, 94, fix188. [Google Scholar] [CrossRef]
- Bou Zeidan, M.; Carmona, L.; Zara, S.; Marcos, J.F. FLO11 Gene Is Involved in the Interaction of Flor Strains of Saccharomyces cerevisiae with a Biofilm-Promoting Synthetic Hexapeptide. Appl. Environ. Microbiol. 2013, 79, 6023–6032. [Google Scholar] [CrossRef]
- Zara, G.; Goffrini, P.; Lodi, T.; Zara, S.; Mannazzu, I.; Budroni, M. FLO11 expression and lipid biosynthesis are required for air-liquid biofilm formation in a Saccharomyces cerevisiae flor strain. FEMS Yeast Res. 2012, 12, 864–866. [Google Scholar] [CrossRef]
- Zara, G.; Zara, S.; Pinna, C.; Marceddu, S.; Budroni, M. FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae. Microbiology 2009, 155, 3838–3846. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Jansen, A.; Peng, X.; Fink, G.R. Mat formation in Saccharomyces cerevisiae requires nutrient and pH gradients. Eukaryot. Cell 2008, 7, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, S.; Vadaie, N.; Chavel, C.A.; Birkaya, B.; Joshi, J.; Grell, L.; Cullen, P.J. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol. 2010, 20, 1389–1395. [Google Scholar] [CrossRef] [PubMed]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans Agglutinin-Like Sequence (Als) Family Vignettes: A Review of Als Protein Structure and Function. Front. Microbiol. 2016, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Domergue, R.; Zupancic, M.L.; Cormack, B.P. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 2005, 8, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P.; Menon, A.K. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar] [CrossRef]
- De Nobel, H.; Lipke, P.N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 1994, 4, 42–45. [Google Scholar] [CrossRef]
- Lu, C.F.; Kurjan, J.; Lipke, P.N. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 1994, 14, 4825–4833. [Google Scholar] [CrossRef]
- Lu, C.F.; Montijn, R.C.; Brown, J.L.; Klis, F.; Kurjan, J.; Bussey, H.; Lipke, P.N. Glycosyl phosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta 1,6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1995, 128, 333–340. [Google Scholar] [CrossRef]
- Sarode, N.; Davis, S.E.; Tams, R.N.; Reynolds, T.B. The Wsc1p cell wall signaling protein controls biofilm (Mat) formation independently of Flo11p in Saccharomyces cerevisiae. G3 2014, 4, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, T.; Bruckner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mosch, H.U.; Essen, L.O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Halme, A.; Bumgarner, S.; Styles, C.; Fink, G.R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 2004, 116, 405–415. [Google Scholar] [CrossRef]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barua, S.; Li, L.; Lipke, P.N.; Dranginis, A.M. Molecular Basis for Strain Variation in the Saccharomyces cerevisiae Adhesin Flo11p. mSphere 2016, 1, e00129-16. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B. The Opi1p transcription factor affects expression of FLO11, mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Sarode, N.; Miracle, B.; Peng, X.; Ryan, O.; Reynolds, T.B. Vacuolar protein sorting genes regulate mat formation in Saccharomyces cerevisiae by Flo11p-dependent and -independent mechanisms. Eukaryot. Cell 2011, 10, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—A model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, J.M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 107–123. [Google Scholar] [CrossRef]
- Kuchin, S.; Vyas, V.K.; Carlson, M. Role of the yeast Snf1 protein kinase in invasive growth. Biochem. Soc. Trans. 2003, 31, 175–177. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Derdelinckx, G.; Verachtert, H.; Delvaux, F.R. Yeast flocculation: What brewers should know. Appl. Microbiol. Biotechnol. 2003, 61, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Ryan, O.; Shapiro, R.S.; Kurat, C.F.; Mayhew, D.; Baryshnikova, A.; Chin, B.; Lin, Z.Y.; Cox, M.J.; Vizeacoumar, F.; Cheung, D.; et al. Global gene deletion analysis exploring yeast filamentous growth. Science 2012, 337, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Heitman, J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 4874–4887. [Google Scholar] [CrossRef] [PubMed]
- Mosch, H.U.; Fink, G.R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 1997, 145, 671–684. [Google Scholar] [PubMed]
- Roberts, R.L.; Fink, G.R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994, 8, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Galitski, T.; Saldanha, A.J.; Styles, C.A.; Lander, E.S.; Fink, G.R. Ploidy regulation of gene expression. Science 1999, 285, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Martineau, C.N.; Beckerich, J.M.; Kabani, M. Flo11p-independent control of “mat” formation by hsp70 molecular chaperones and nucleotide exchange factors in yeast. Genetics 2007, 177, 1679–1689. [Google Scholar] [CrossRef]
- Martineau, C.N.; Melki, R.; Kabani, M. Swa2p-dependent clathrin dynamics is critical for Flo11p processing and ‘Mat’ formation in the yeast Saccharomyces cerevisiae. FEBS Lett. 2010, 584, 1149–1155. [Google Scholar] [CrossRef]
- Babst, M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Emr, S.D. The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 277–298. [Google Scholar] [CrossRef]
- Xu, W.; Smith, F.J., Jr.; Subaran, R.; Mitchell, A.P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 2004, 15, 5528–5537. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef] [PubMed]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 2001, 21, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Vay, H.A.; Philip, B.; Levin, D.E. Mutational analysis of the cytoplasmic domain of the Wsc1 cell wall stress sensor. Microbiology 2004, 150, 3281–3288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piao, H.L.; Machado, I.M.; Payne, G.S. NPFXD-mediated endocytosis is required for polarity and function of a yeast cell wall stress sensor. Mol. Biol. Cell 2007, 18, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar] [CrossRef]
- Raitt, D.C.; Johnson, A.L.; Erkine, A.M.; Makino, K.; Morgan, B.; Gross, D.S.; Johnston, L.H. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 2000, 11, 2335–2347. [Google Scholar] [CrossRef]
- Li, X.S.; Reddy, M.S.; Baev, D.; Edgerton, M. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J. Biol. Chem. 2003, 278, 28553–28561. [Google Scholar] [CrossRef]
- Li, X.S.; Sun, J.N.; Okamoto-Shibayama, K.; Edgerton, M. Candida albicans cell wall ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J. Biol. Chem. 2006, 281, 22453–22463. [Google Scholar] [CrossRef]
- Sun, J.N.; Solis, N.V.; Phan, Q.T.; Bajwa, J.S.; Kashleva, H.; Thompson, A.; Liu, Y.; Dongari-Bagtzoglou, A.; Edgerton, M.; Filler, S.G. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathog. 2010, 6, e1001181. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynolds, T.B. Going with the Flo: The Role of Flo11-Dependent and Independent Interactions in Yeast Mat Formation. J. Fungi 2018, 4, 132. https://doi.org/10.3390/jof4040132
Reynolds TB. Going with the Flo: The Role of Flo11-Dependent and Independent Interactions in Yeast Mat Formation. Journal of Fungi. 2018; 4(4):132. https://doi.org/10.3390/jof4040132
Chicago/Turabian StyleReynolds, Todd B. 2018. "Going with the Flo: The Role of Flo11-Dependent and Independent Interactions in Yeast Mat Formation" Journal of Fungi 4, no. 4: 132. https://doi.org/10.3390/jof4040132
APA StyleReynolds, T. B. (2018). Going with the Flo: The Role of Flo11-Dependent and Independent Interactions in Yeast Mat Formation. Journal of Fungi, 4(4), 132. https://doi.org/10.3390/jof4040132