Functional Characterization of 5-O-Glycosyltranferase Transforming 3-O Anthocyanins into 3,5-O Anthocyanins in Freesia hybrida
Abstract
1. Introduction
2. Results
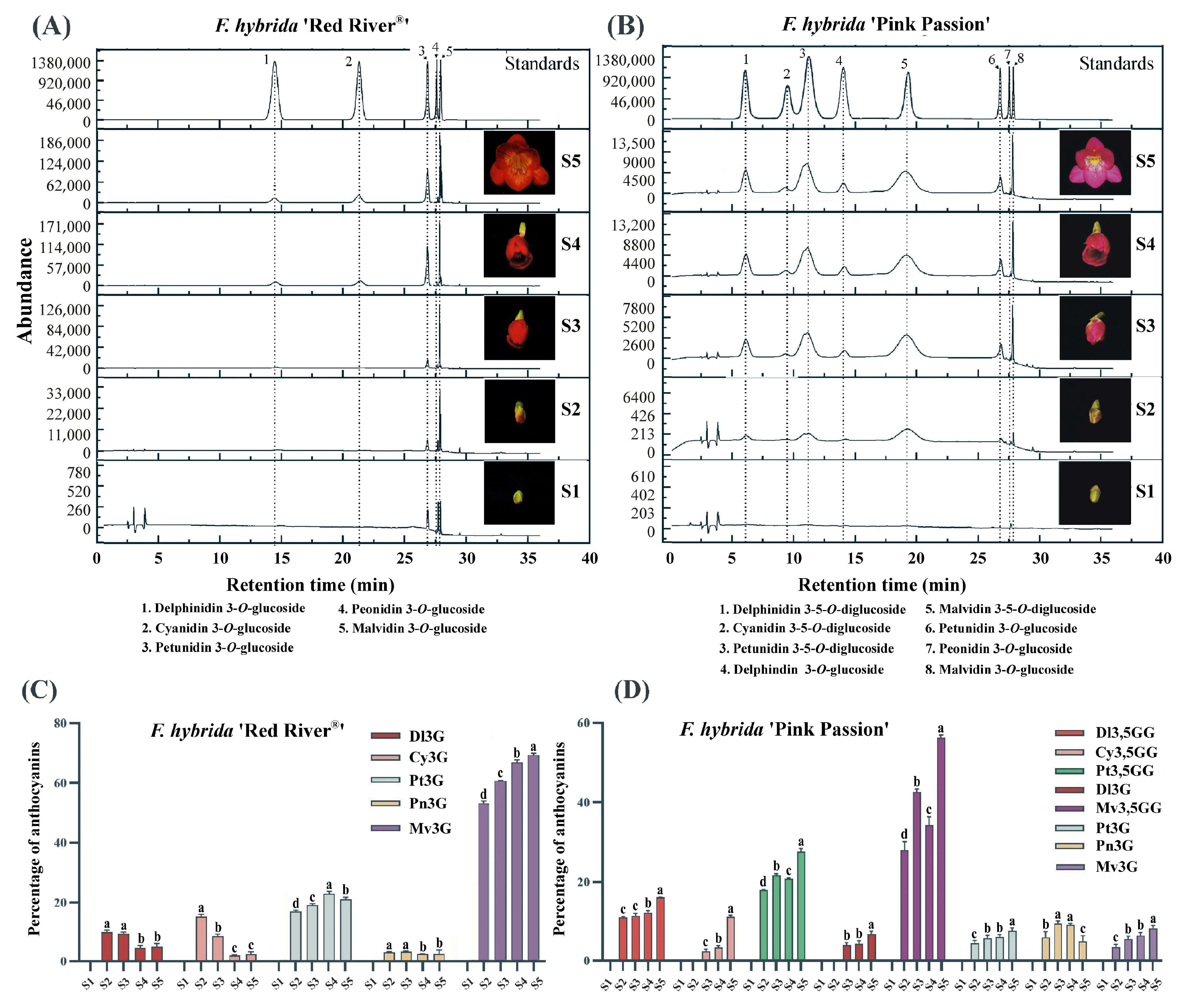
2.1. Different Colored F. hybrida Cultivars Accumulate Varied Anthocyanins
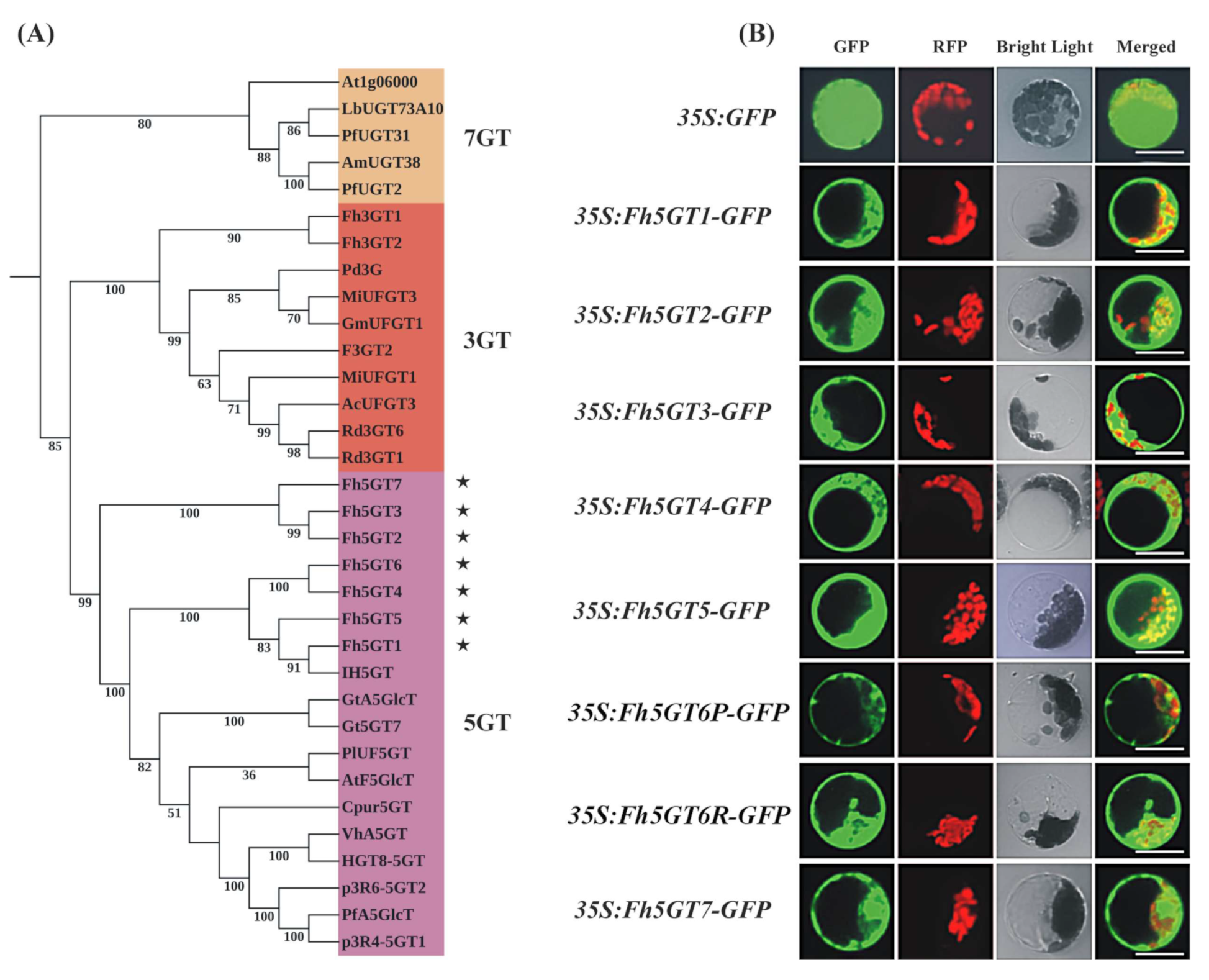
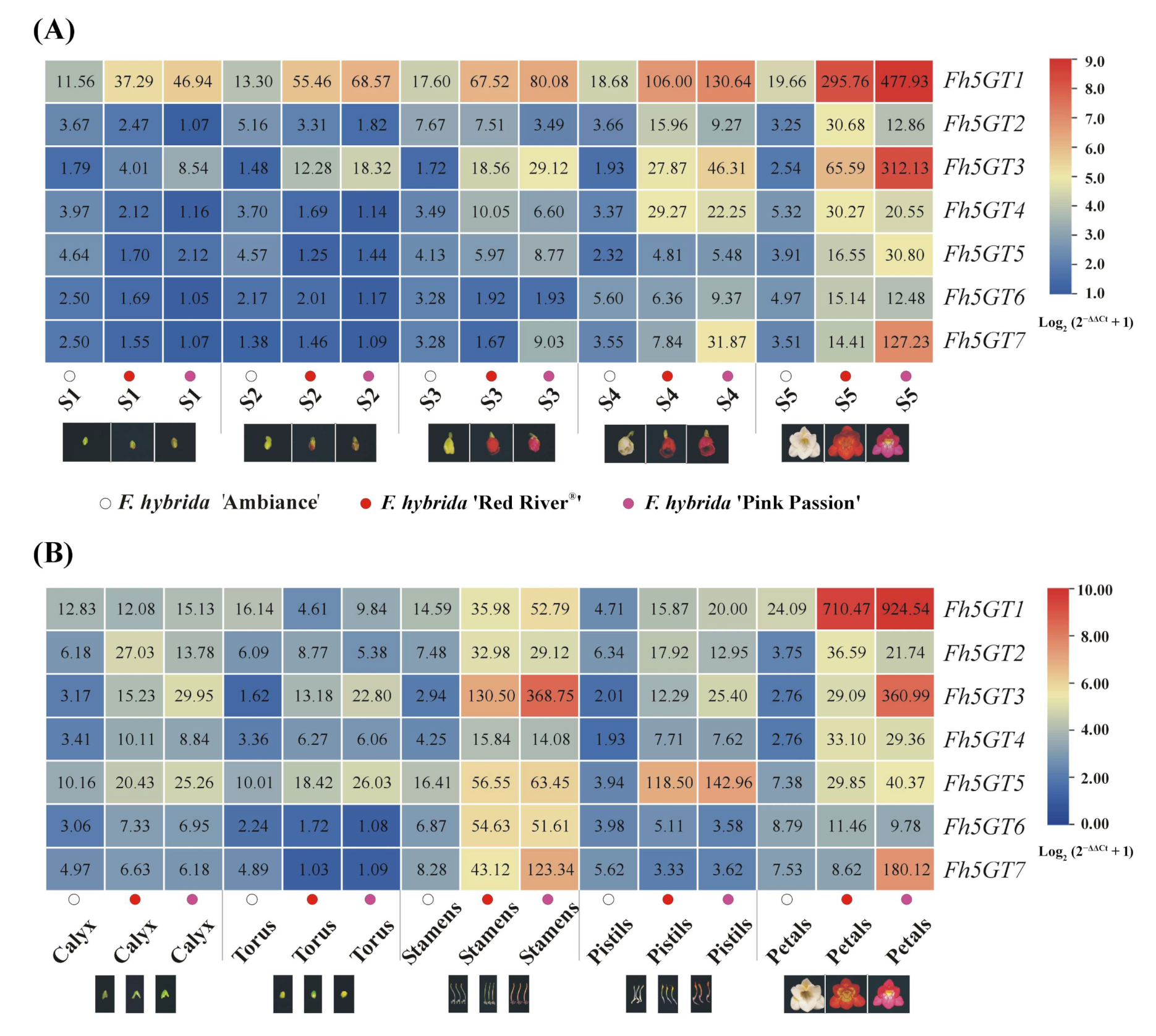
2.2. The Difference in Expression Levels, Rather Than Sequence Variations of 5GTs, May Account for the Differing Accumulation of 3,5-O-Diglucosidic Anthocyanins Between F. hybrida ‘Red River®’ and ‘Pink Passion’
2.3. Fh5GTs Prefer 3-O-Glucosidic Anthocyanins Instead of Anthocyanidins as Acceptors
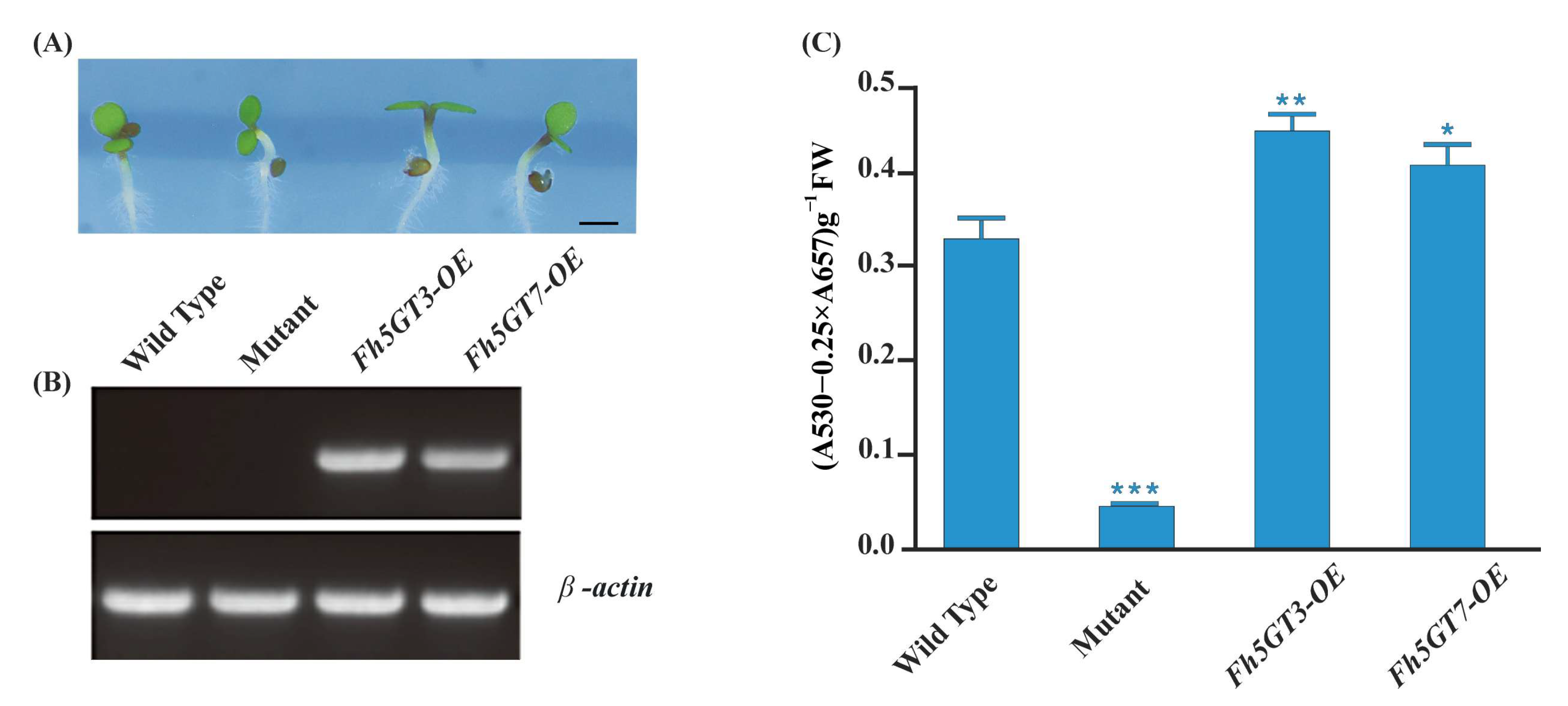
2.4. Fh5GT3 and Fh5GT7 Complement the Anthocyanin Deficiency in Arabidopsis 5gt Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Anthocyanin Extraction and HPLC Analysis
4.3. RNA-Seq Analysis
4.4. Gene Cloning and Sequence Analysis
4.5. Subcellular Localization Analysis
4.6. Heterologous Expression of Fh5GTs in E. coli BL21
4.7. Enzymatic Assay
4.8. Heterologous Expression of Fh5GTs in Arabidopsis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef] [PubMed]
- Narbona, E.; del Valle, J.C.; Whittall, J.B. Painting the green canvas: How pigments produce flower colours. Biochem 2021, 43, 6–12. [Google Scholar] [CrossRef]
- Liu, H.; Jin, Y.; Huang, L.; Miao, C.; Tang, J.; Zhang, H.; Yin, H.; Lu, X.; Li, N.; Dai, S. Transcriptomics and metabolomics reveal the underlying mechanism of drought treatment on anthocyanin accumulation in postharvest blood orange fruit. BMC Plant Biol. 2024, 24, 160. [Google Scholar] [CrossRef]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From mechanisms of regulation in plants to health benefits in foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef]
- Salehi, B.; Sharifi-Rad, J.; Cappellini, F.; Reiner, Ž.; Zorzan, D.; Imran, M.; Sener, B.; Kilic, M.; El-Shazly, M.; Fahmy, N.M. The therapeutic potential of anthocyanins: Current approaches based on their molecular mechanism of action. Front. Pharmacol. 2020, 11, 1300. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, P.; Kumar, A.; Chawla, N.; Dhatt, A.S. Multifaceted regulation of anthocyanin biosynthesis in plants: A comprehensive review. J. Plant Growth Regul. 2024, 43, 3048–3062. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Muskała, M.; Merecz-Sadowska, A.; Sikora, J.; Picot, L.; Sitarek, P. Anti-Inflammatory and Anticancer Effects of Anthocyanins in In Vitro and In Vivo Studies. Antioxidants 2024, 13, 1143. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Quideau, S. Flavonoids. Chemistry, Biochemistry and Applications; Andersen, Ø.M., Markham, K.R., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2006. [Google Scholar]
- Trapero, A.; Ahrazem, O.; Rubio-Moraga, A.; Jimeno, M.L.; Gómez, M.D.; Gómez-Gómez, L. Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol. 2012, 159, 1335–1354. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin, J.; Cheng, T.; Liu, Y.; Han, F. Methylation, hydroxylation, glycosylation and acylation affect the transport of wine anthocyanins in Caco-2 cells. Foods 2022, 11, 3793. [Google Scholar] [CrossRef]
- Nie, S.; Ma, H.-Y.; Shi, T.-L.; Tian, X.-C.; El-Kassaby, Y.A.; Porth, I.; Yang, F.-S.; Mao, J.-F. Progress in phylogenetics, multi-omics and flower coloration studies in Rhododendron. Ornam. Plant Res. 2024, 4, e003. [Google Scholar] [CrossRef]
- Butler, N. Purple is the new orange: Anthocyanin regulation coming together in carrot. Plant Physiol. 2019, 181, 12–13. [Google Scholar] [CrossRef]
- Saleh, N.; Fritsch, H.; Witkop, P.; Grisebach, H. UDP-glucose: Cyanidin 3-O-glucosyltransferase from cell cultures of Haplopappus gracilis. Planta 1976, 133, 41–45. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Haruta, K.S.; Pitaksutheepong, C.; Abe, Y.; Kakizaki, Y.; Yamamoto, K.; Shimada, N.; Yamamura, S.; Nishihara, M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008, 49, 1818–1829. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Sato, K.; Takahashi, H.; Yamamura, S.; Nishihara, M. Cloning and characterization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J. Exp. Bot. 2008, 59, 1241–1252. [Google Scholar] [CrossRef]
- Kang, X.; Mikami, R.; Akita, Y. Characterization of 5-O-glucosyltransferase involved in anthocyanin biosynthesis in Cyclamen purpurascens. Plant Biotechnol. 2021, 38, 263–268. [Google Scholar] [CrossRef]
- Sun, W.; Liang, L.; Meng, X.; Li, Y.; Gao, F.; Liu, X.; Wang, S.; Gao, X.; Wang, L. Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida. Front. Plant Sci. 2016, 7, 410. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Tong, L.; Wei, C.; Lu, K.; Li, S.; Kimani, S.; Wang, S.; Wang, L.; Gao, X. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis. Plant Cell Physiol. 2020, 61, 1365–1380. [Google Scholar] [CrossRef]
- Ju, Z.; Sun, W.; Meng, X.; Liang, L.; Li, Y.; Zhou, T.; Shen, H.; Gao, X.; Wang, L. Isolation and functional characterization of two 5-O-glucosyltransferases related to anthocyanin biosynthesis from Freesia hybrida. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 135, 99–110. [Google Scholar] [CrossRef]
- Meng, X.; Li, Y.; Zhou, T.; Sun, W.; Shan, X.; Gao, X.; Wang, L. Functional differentiation of duplicated flavonoid 3-O-glycosyltransferases in the flavonol and anthocyanin biosynthesis of Freesia hybrida. Front. Plant Sci. 2019, 10, 1330. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhuang, D.; Gao, R.; Qiu, M.; Zhou, L.; Zhang, J.; Wang, Y.; Zhang, Q.; Zhai, N.; Xu, G. Molecular Insights into TT2-Type MYB Regulators Illuminate the Complexity of Floral Flavonoids Biosynthesis in Freesia hybrida. Hortic. Res. 2024, 12, uhae352. [Google Scholar] [CrossRef] [PubMed]
- Imayama, T.; Yoshihara, N.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ino, I.; Yabuya, T. Isolation and characterization of a cDNA clone of UDP-glucose: Anthocyanin 5-O-glucosyltransferase in Iris hollandica. Plant Sci. 2004, 167, 1243–1248. [Google Scholar] [CrossRef]
- Poustka, F.; Irani, N.G.; Feller, A.; Lu, Y.; Pourcel, L.; Frame, K.; Grotewold, E. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 2007, 145, 1323–1335. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Han, T.; Zhang, J.; Wang, Y.; Kimani, S.; Wang, L.; Gao, X. MYB repressors and MBW activation complex collaborate to fine-tune flower coloration in Freesia hybrida. Commun. Biol. 2020, 3, 396. [Google Scholar] [CrossRef]
- Sui, X.; Gao, X.; Ao, M.; Wang, Q.; Yang, D.; Wang, M.; Fu, Y.; Wang, L. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Freesia hybrida. Plant Cell Rep. 2011, 30, 1209–1218. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Wu, Y.; Han, T.; Lyu, L.; Li, W.; Wu, W. Research progress in understanding the biosynthesis and regulation of plant anthocyanins. Sci. Hortic. 2023, 321, 112374. [Google Scholar] [CrossRef]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef]
- Shen, Y.; Rao, Y.; Ma, M.; Li, Y.; He, Y.; Wang, Z.; Liang, M.; Ning, G. Coordination among flower pigments, scents and pollinators in ornamental plants. Hortic. Adv. 2024, 2, 6. [Google Scholar] [CrossRef]
- Kubo, A.; Arai, Y.; Nagashima, S.; Yoshikawa, T. Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 2004, 429, 198–203. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- He, F.; Chen, W.-K.; Yu, K.-J.; Ji, X.-N.; Duan, C.-Q.; Reeves, M.J.; Wang, J. Molecular and biochemical characterization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry 2015, 117, 363–372. [Google Scholar] [CrossRef]
- Jonsson, L.; Aarsman, M.; Van Diepen, J.; De Vlaming, P.; Smit, N.; Schram, A. Properties and genetic control of anthocyanin 5-O-glucosyltransferase in flowers of Petunia hybrida. Planta 1984, 160, 341–347. [Google Scholar] [CrossRef]
- Griesser, M.; Hoffmann, T.; Bellido, M.L.; Rosati, C.; Fink, B.; Kurtzer, R.; Aharoni, A.; Munoz-Blanco, J.; Schwab, W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008, 146, 1528–1539. [Google Scholar] [CrossRef]
- Yabuya, T.; Yamaguchi, M.-a.; Imayama, T.; Katoh, K.; Ino, I. Anthocyanin 5-O-glucosyltransferase in flowers of Iris ensata. Plant Sci. 2002, 162, 779–784. [Google Scholar] [CrossRef]
- Li, Y.; Bao, T.; Zhang, J.; Li, H.; Shan, X.; Yan, H.; Kimani, S.; Zhang, L.; Gao, X. The coordinated interaction or regulation between floral pigments and volatile organic compounds. Hortic. Plant J. 2024, 11, 463–485. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Gao, R.; Li, Y.; Shan, X.; Wang, Y.; Yang, S.; Ma, S.; Xia, Z.; Zheng, H.; Wei, C.; Tong, L. A single nucleotide polymorphism affects protein translation and leads to post-anthesis color change variation in closely related Lotus species. Plant J. 2025, 121, e17188. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Kimani, S.; Li, Y.; Li, H.; Yang, S.; Zhang, J.; Wang, Q.; Wang, Z.; Ning, G.; Wang, L. Allelic variation of terpene synthases drives terpene diversity in the wild species of the Freesia genus. Plant Physiol. 2023, 192, 2419–2435. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, N.; Kimani, S.; Li, Y.; Bao, T.; Ning, G.; Li, L.; Liu, B.; Wang, L.; Gao, X. Characterization of Terpene synthase variation in flowers of wild Aquilegia species from Northeastern Asia. Hortic. Res. 2022, 9, uhab020. [Google Scholar] [CrossRef]
- Yamazaki, M.; Yamagishi, E.; Gong, Z.; Fukuchi-Mizutani, M.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Yamaguchi, M.; Saito, K. Two flavonoid glucosyltransferases from Petunia hybrida: Molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol. Biol. 2002, 48, 401–411. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan; Bao, T.; Zheng, X.; Pang, Y.; Gao, R.; Shan, X.; Zhu, S.; Kimani, S.K.; Gao, X.; Li, Y. Functional Characterization of 5-O-Glycosyltranferase Transforming 3-O Anthocyanins into 3,5-O Anthocyanins in Freesia hybrida. Int. J. Mol. Sci. 2025, 26, 4542. https://doi.org/10.3390/ijms26104542
Adnan, Bao T, Zheng X, Pang Y, Gao R, Shan X, Zhu S, Kimani SK, Gao X, Li Y. Functional Characterization of 5-O-Glycosyltranferase Transforming 3-O Anthocyanins into 3,5-O Anthocyanins in Freesia hybrida. International Journal of Molecular Sciences. 2025; 26(10):4542. https://doi.org/10.3390/ijms26104542
Chicago/Turabian StyleAdnan, Tingting Bao, Xiang Zheng, Yicong Pang, Ruifang Gao, Xiaotong Shan, Shirui Zhu, Shadrack Kanyonji Kimani, Xiang Gao, and Yueqing Li. 2025. "Functional Characterization of 5-O-Glycosyltranferase Transforming 3-O Anthocyanins into 3,5-O Anthocyanins in Freesia hybrida" International Journal of Molecular Sciences 26, no. 10: 4542. https://doi.org/10.3390/ijms26104542
APA StyleAdnan, Bao, T., Zheng, X., Pang, Y., Gao, R., Shan, X., Zhu, S., Kimani, S. K., Gao, X., & Li, Y. (2025). Functional Characterization of 5-O-Glycosyltranferase Transforming 3-O Anthocyanins into 3,5-O Anthocyanins in Freesia hybrida. International Journal of Molecular Sciences, 26(10), 4542. https://doi.org/10.3390/ijms26104542





