Type One Protein Phosphatase 4aD Negatively Regulates Cotton (Gossypium hirsutum) Salt Tolerance by Inhibiting the Phosphorylation of Kinases That Respond to Abscisic Acid
Abstract
1. Introduction
2. Results
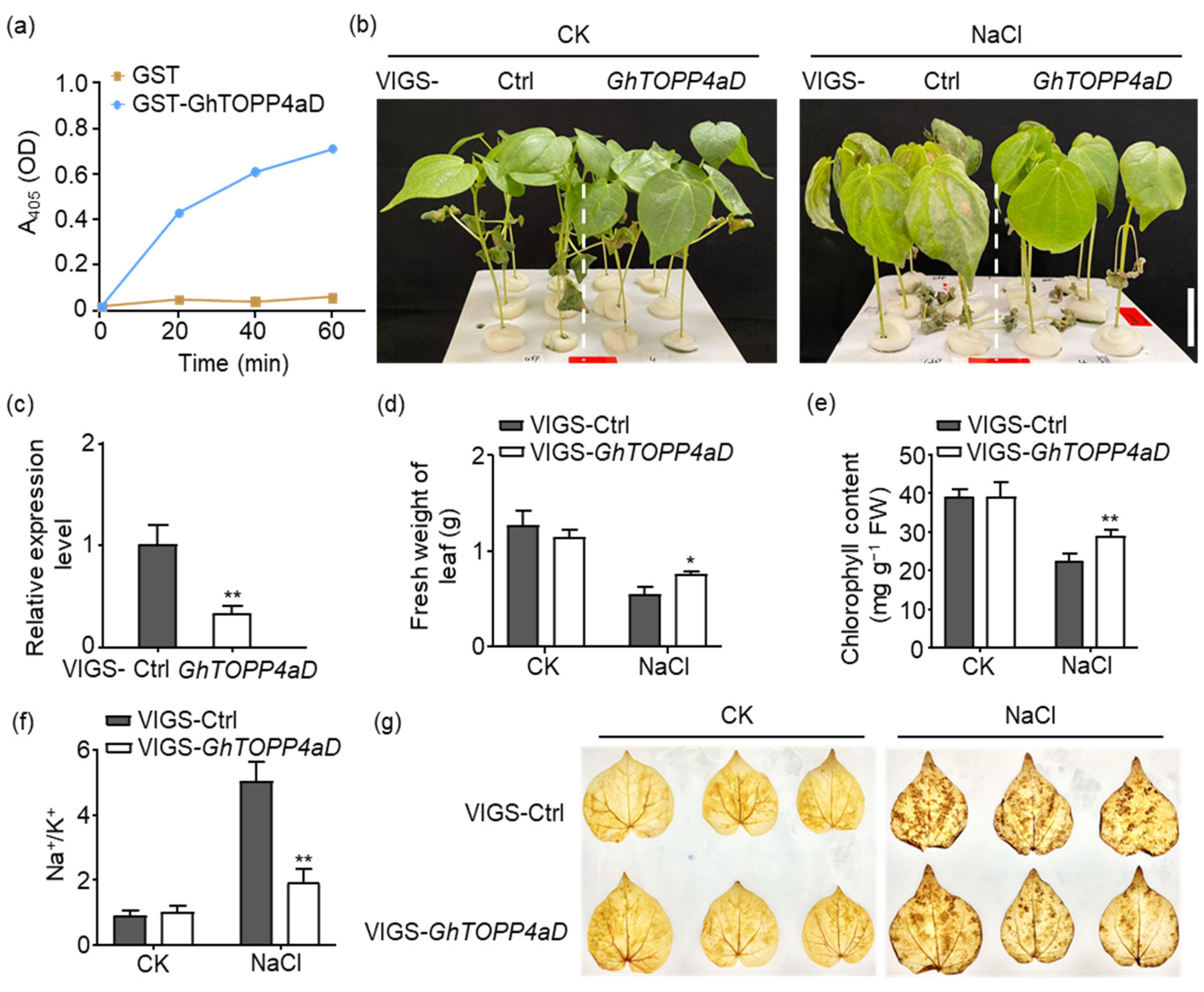
2.1. GhTOPP4aD Negatively Regulates Cotton Salt Response
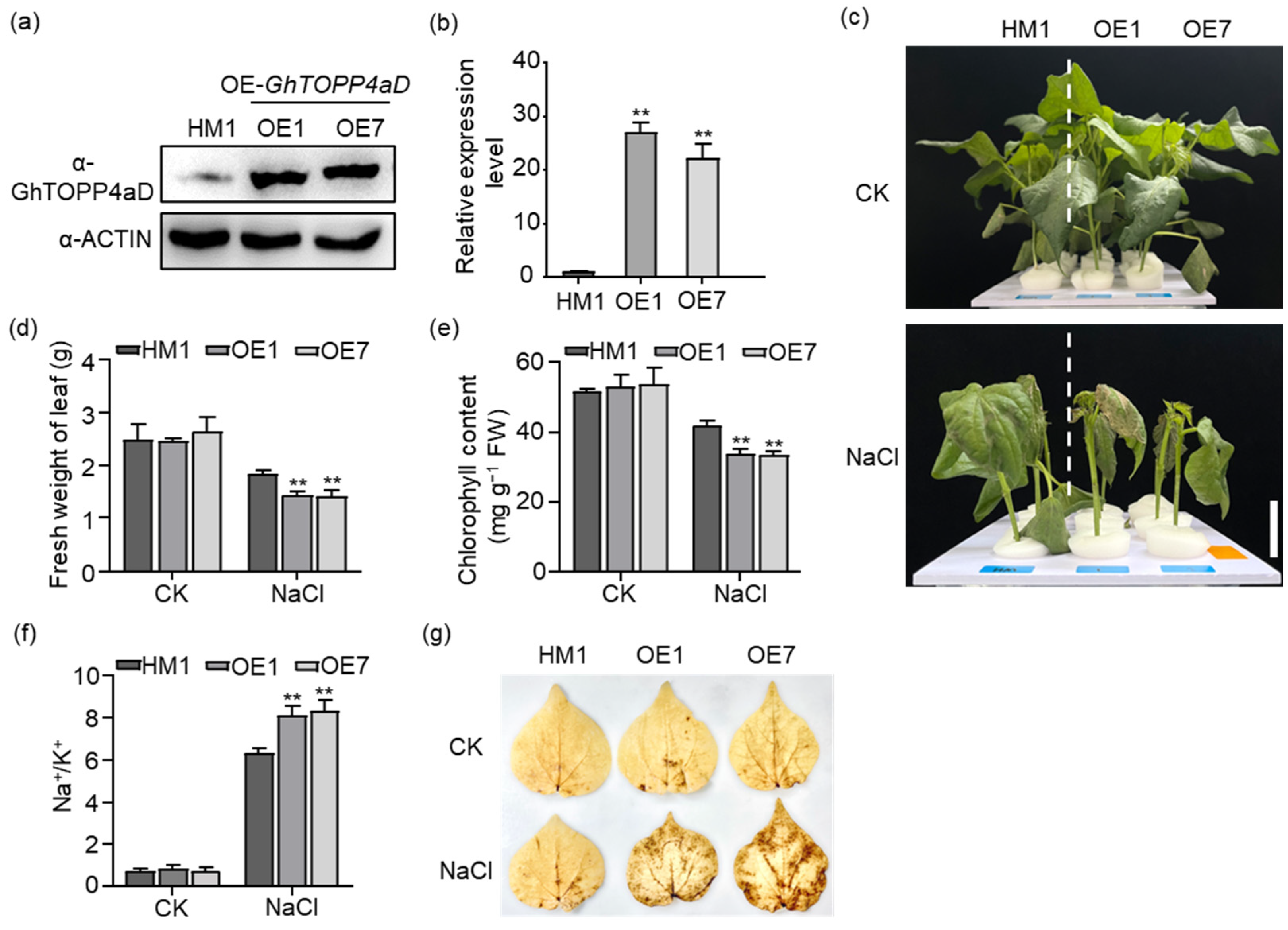
2.2. Overexpressing GhTOPP4aD Plants Are More Sensitive to Salt-Stress than the Recipient (HM1)
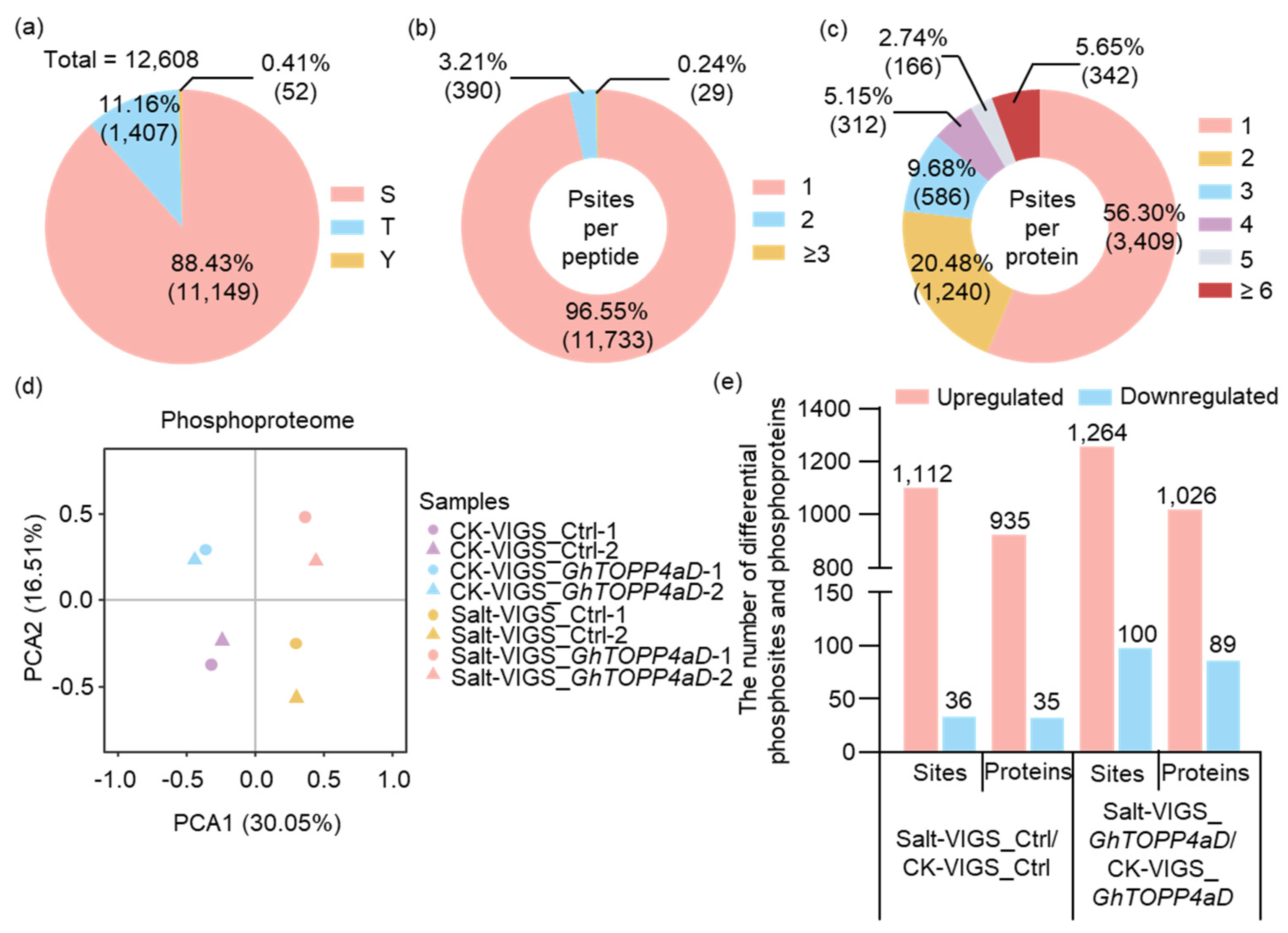
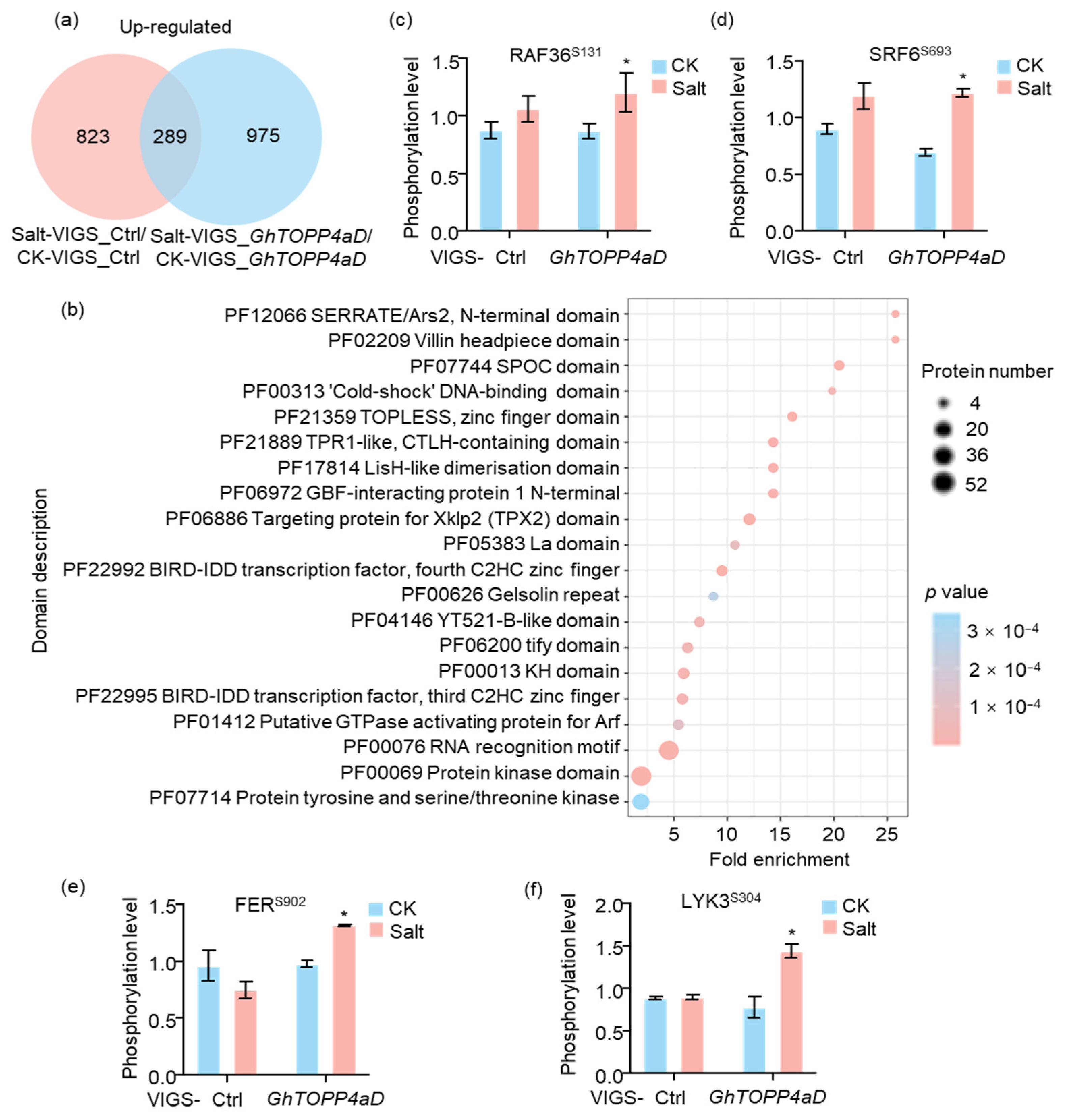
2.3. Salt-Responsive Phosphosites and Phosphoproteins Modulated by GhTOPP4aD
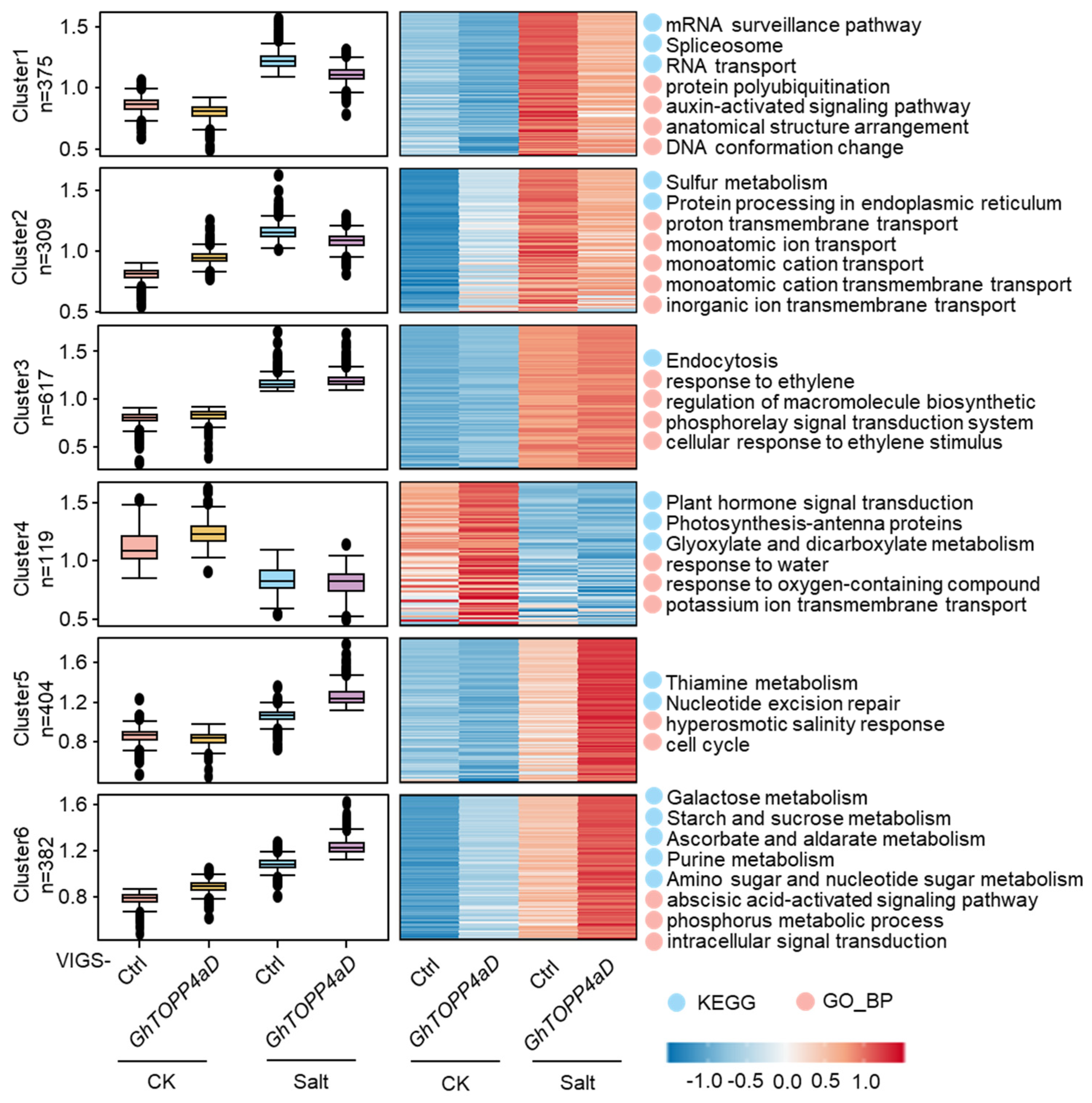
2.4. GhTOPP4aD Regulates Protein Phosphorylation Levels in Diverse Biological Processes and Pathways
2.5. GhTOPP4aD Regulates the Dephosphorylation of Numerous Kinases Under Salt Stress
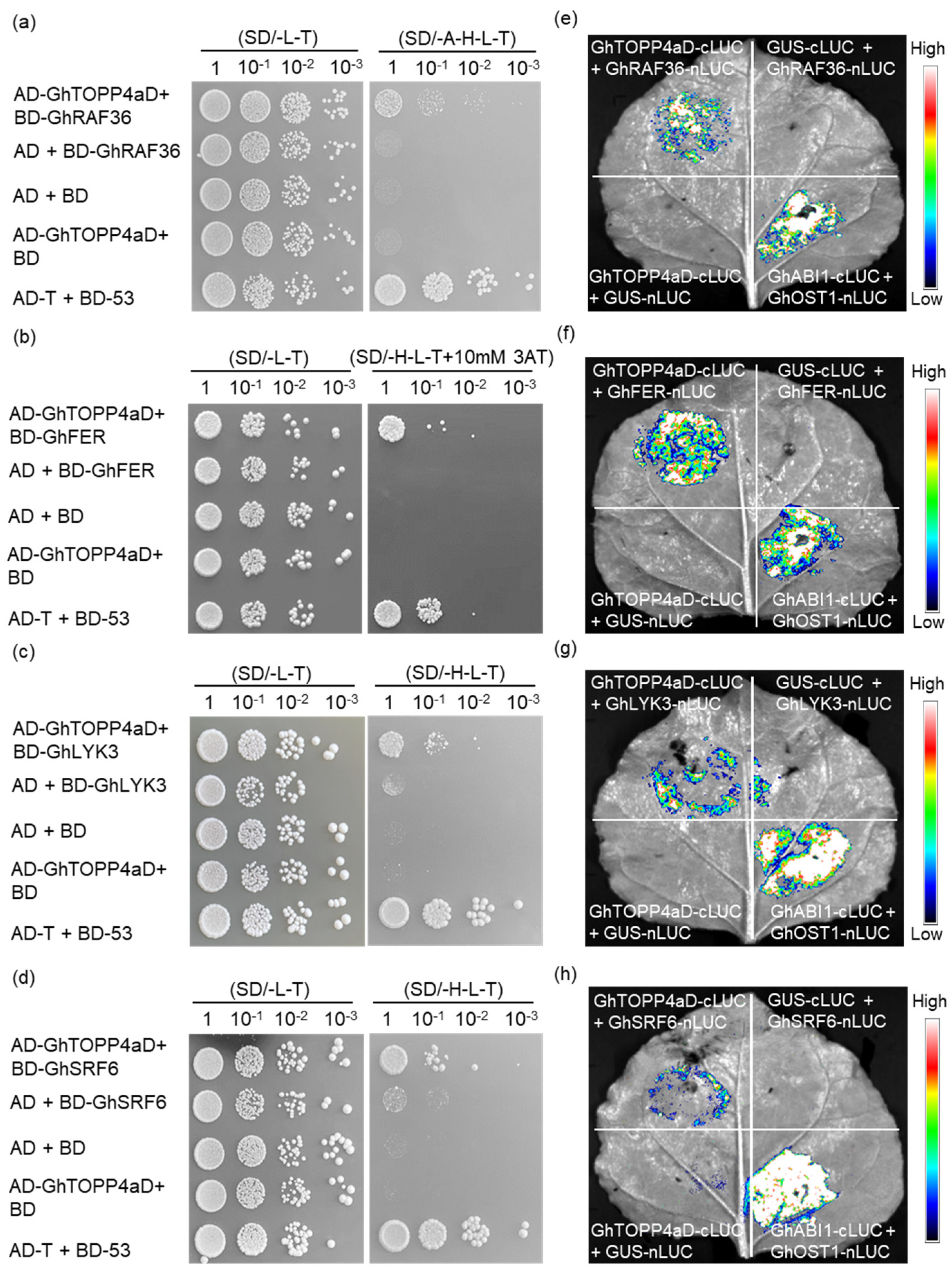
2.6. GhTOPP4aD and the Candidate Tyrosine Kinases Interact In Vivo
2.7. In Vitro Dephosphorylation Assay to Analyze the Effect of GhTOPP4aD on Tyrosine Kinase Activity
2.8. Silencing GhTOPP4aD Enhances NaCl-Induced Phosphorylation of GhRAF36, GhLYK3, and GhFER Kinases
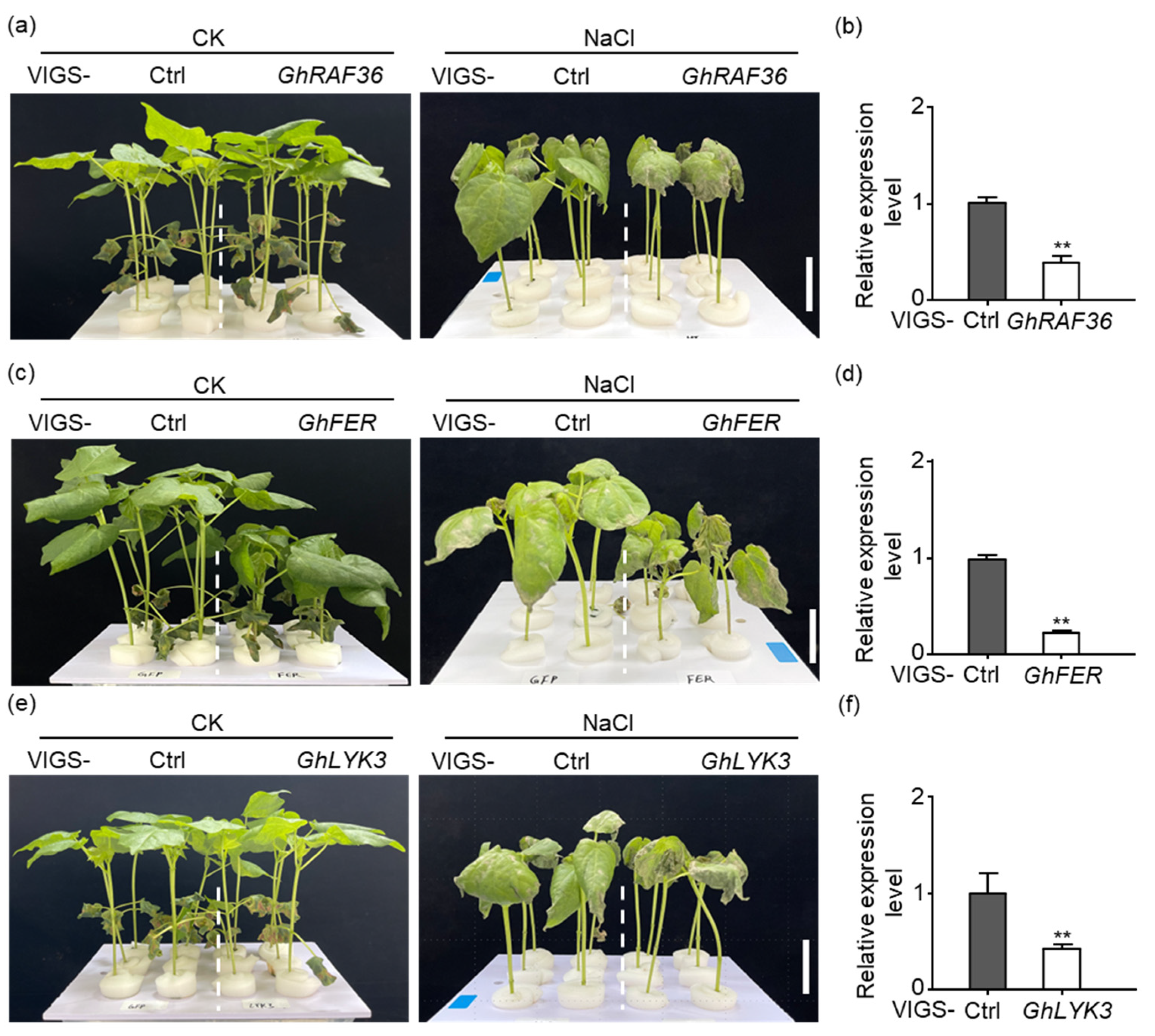
2.9. Silencing GhRAF36, GhFER, and GhLYK3 Enhance Cotton Salt Stress Sensitivity
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Virus-Induced Gene Silencing (VIGS) Assay
4.3. Real-Time Quantitative PCR (RT-qPCR)
4.4. Measurement of Sodium and Potassium Ion Content
4.5. Determination of Chlorophyll Content
4.6. Diaminobenzidine (DAB) Staining
4.7. Measurements of Phosphatase Activity
4.8. Recombinant Protein Expression and Purification
4.9. Detecting GhTOPP4aD Protein Abundance
4.10. Yeast Two-Hybrid Assay
4.11. Luciferase Complementation Imaging (LCI) Assay
4.12. In Vitro Dephosphorylation Assay
4.13. In Vitro Kinase Assay
4.14. Phosphoproteome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flowers, T.J.; Galal, H.K.; Bromham, L. Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 2010, 37, 604–612. [Google Scholar]
- Rodriguez-Uribe, L.; Higbie, S.M.; Stewart, J.M.; Wilkins, T.; Lindemann, W.; Sengupta-Gopalan, C.; Zhang, J. Identification of salt responsive genes using comparative microarray analysis in Upland cotton (Gossypium hirsutum L.). Plant Sci. 2011, 180, 461–469. [Google Scholar] [PubMed]
- Khan, A.; Pan, X.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51, 47. [Google Scholar]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar]
- Bonhomme, L.; Valot, B.; Tardieu, F.; Zivy, M. Phosphoproteome dynamics upon changes in plant water status reveal early events associated with rapid growth adjustment in maize leaves. Mol. Cell. Proteom. 2012, 11, 957–972. [Google Scholar]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhao, X. Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS ONE 2014, 9, e112807. [Google Scholar]
- Sharif, I.; Aleem, S.; Farooq, J.; Rizwan, M.; Younas, A.; Sarwar, G.; Chohan, S.M. Salinity stress in cotton: Effects, mechanism of tolerance and its management strategies. Physiol. Mol. Biol. Plants 2019, 25, 807–820. [Google Scholar]
- Chitteti, B.R.; Peng, Z. Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 2007, 6, 1718–1727. [Google Scholar]
- Gupta, R.; Min, C.W.; Kim, Y.J.; Kim, S.T. Identification of Msp1-Induced Signaling Components in Rice Leaves by Integrated Proteomic and Phosphoproteomic Analysis. Int. J. Mol. Sci. 2019, 20, 4135. [Google Scholar] [CrossRef]
- Nohzadeh Malakshah, S.; Habibi Rezaei, M.; Heidari, M.; Salekdeh, G.H. Proteomics reveals new salt responsive proteins associated with rice plasma membrane. Biosci. Biotechnol. Biochem. 2007, 71, 2144–2154. [Google Scholar] [CrossRef] [PubMed]
- Ajadi, A.A.; Cisse, A.; Ahmad, S.; Yifeng, W.; Yazhou, S.; Shufan, L.; Xixi, L.; Bello, B.K.; Tajo, S.M.; Xiaohong, T.; et al. Protein Phosphorylation and Phosphoproteome: An Overview of Rice. Rice Sci. 2020, 27, 184–200. [Google Scholar]
- Farkas, I.; Dombradi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ceulemans, H.; Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004, 84, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, S.; Santos, M.; Martins, F.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A. Protein phosphatase 1 is a key player in nuclear events. Cell Signal. 2015, 27, 2589–2598. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Guo, X.; Qin, Q.; Yan, J.; Niu, Y.; Huang, B.; Guan, L.; Li, Y.; Ren, D.; Li, J.; Hou, S. TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol. 2015, 167, 1058–1075. [Google Scholar] [CrossRef]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 Regulates the Stability of PHYTOCHROME INTERACTING FACTOR5 during Photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef]
- Hou, Y.J.; Zhu, Y.; Wang, P.; Zhao, Y.; Xie, S.; Batelli, G.; Wang, B.; Duan, C.G.; Wang, X.; Xing, L.; et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet. 2016, 12, e1005835. [Google Scholar] [CrossRef]
- Reinders, J.; Sickmann, A. State-of-the-art in phosphoproteomics. Proteomics 2005, 5, 4052–4061. [Google Scholar] [CrossRef]
- Miyata, K.; Hayafune, M.; Kobae, Y.; Kaku, H.; Nishizawa, Y.; Masuda, Y.; Shibuya, N.; Nakagawa, T. Evaluation of the Role of the LysM Receptor-Like Kinase, OsNFR5/OsRLK2 for AM Symbiosis in Rice. Plant Cell Physiol. 2016, 57, 2283–2290. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Barrena, M.J.; Fujii, H.; Angulo, I.; Martinez-Ripoll, M.; Zhu, J.K.; Albert, A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 2007, 26, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Barbier-Brygoo, H.; Lauriere, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar]
- Liu, X.; Jiang, W.; Li, Y.; Nie, H.; Cui, L.; Li, R.; Tan, L.; Peng, L.; Li, C.; Luo, J.; et al. FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat. Plants 2023, 9, 645–660. [Google Scholar] [PubMed]
- Kamiyama, Y.; Hirotani, M.; Ishikawa, S.; Minegishi, F.; Katagiri, S.; Rogan, C.J.; Takahashi, F.; Nomoto, M.; Ishikawa, K.; Kodama, Y.; et al. Arabidopsis group C Raf-like protein kinases negatively regulate abscisic acid signaling and are direct substrates of SnRK2. Proc. Natl. Acad. Sci. USA 2021, 118, e2100073118. [Google Scholar] [PubMed]
- Paparella, C.; Savatin, D.V.; Marti, L.; De Lorenzo, G.; Ferrari, S. The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 2014, 165, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ji, J.; Ma, X.; Xu, Q.; Qi, X.; Chen, X. Comparative Proteomic Analysis Provides Insight into the Key Proteins Involved in Cucumber (Cucumis sativus L.) Adventitious Root Emergence under Waterlogging Stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar]
- Xu, D.; Yuan, H.; Tong, Y.; Zhao, L.; Qiu, L.; Guo, W.; Shen, C.; Liu, H.; Yan, D.; Zheng, B. Comparative Proteomic Analysis of the Graft Unions in Hickory (Carya cathayensis) Provides Insights into Response Mechanisms to Grafting Process. Front. Plant Sci. 2017, 8, 676. [Google Scholar]
- Hao, J.; Guo, H.; Shi, X.; Wang, Y.; Wan, Q.; Song, Y.B.; Zhang, L.; Dong, M.; Shen, C. Comparative proteomic analyses of two Taxus species (Taxus x media and Taxus mairei) reveals variations in the metabolisms associated with paclitaxel and other metabolites. Plant Cell Physiol. 2017, 58, 1878–1890. [Google Scholar]
- Xue, L.; Wang, P.; Wang, L.; Renzi, E.; Radivojac, P.; Tang, H.; Arnold, R.; Zhu, J.K.; Tao, W.A. Quantitative measurement of phosphoproteome response to osmotic stress in arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol. Cell Proteom. 2013, 12, 2354–2369. [Google Scholar]
- Zhang, M.; Lv, D.; Ge, P.; Bian, Y.; Chen, G.; Zhu, G.; Li, X.; Yan, Y. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteom. 2014, 109, 290–308. [Google Scholar]
- Yu, B.; Li, J.; Koh, J.; Dufresne, C.; Yang, N.; Qi, S.; Zhang, Y.; Ma, C.; Duong, B.V.; Chen, S.; et al. Quantitative proteomics and phosphoproteomics of sugar beet monosomic addition line M14 in response to salt stress. J. Proteom. 2016, 143, 286–297. [Google Scholar]
- Yu, C.; Wu, Q.; Sun, C.; Tang, M.; Sun, J.; Zhan, Y. The Phosphoproteomic Response of Okra (Abelmoschus esculentus L.) Seedlings to Salt Stress. Int. J. Mol. Sci. 2019, 20, 1262. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chevalier, D.; Larue, C.; Ki Cho, S.; Walker, J.C. The Protein Phosphatases and Protein Kinases of Arabidopsis thaliana. Arab. Book. 2007, 5, e0106. [Google Scholar]
- Liu, Y.; Yan, J.; Qin, Q.; Zhang, J.; Chen, Y.; Zhao, L.; He, K.; Hou, S. Type one protein phosphatases (TOPPs) contribute to the plant defense response in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 360–377. [Google Scholar] [PubMed]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [PubMed]
- Meng, Y.C.; Zhang, H.F.; Pan, X.X.; Chen, N.; Hu, H.F.; Haq, S.U.; Khan, A.; Chen, R.G. CaDHN3, a Pepper (Capsicum annuum L.) Dehydrin Gene Enhances the Tolerance against Salt and Drought Stresses by Reducing ROS Accumulation. Int. J. Mol. Sci. 2021, 22, 3205. [Google Scholar] [CrossRef]
- Fu, Z.W.; Feng, Y.R.; Gao, X.; Ding, F.; Li, J.H.; Yuan, T.T.; Lu, Y.T. Salt stress-induced chloroplastic hydrogen peroxide stimulates pdTPI sulfenylation and methylglyoxal accumulation. Plant Cell 2023, 35, 1593–1616. [Google Scholar]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca(2+) Signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar]
- Liu, X.; Wang, L.; Liu, L.; Li, Y.; Ogden, M.; Somssich, M.; Liu, Y.; Zhang, Y.; Ran, M.; Persson, S.; et al. FERONIA adjusts CC1 phosphorylation to control microtubule array behavior in response to salt stress. Sci. Adv. 2024, 10, eadq8717. [Google Scholar] [PubMed]
- Barjaktarovic, Z.; Schutz, W.; Madlung, J.; Fladerer, C.; Nordheim, A.; Hampp, R. Changes in the effective gravitational field strength affect the state of phosphorylation of stress-related proteins in callus cultures of Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 779–789. [Google Scholar] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [PubMed]
- Jia, W.; Wang, Y.; Zhang, S.; Zhang, J. Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 2002, 53, 2201–2206. [Google Scholar]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar]
- Johnson, R.R.; Wagner, R.L.; Verhey, S.D.; Walker-Simmons, M.K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002, 130, 837–846. [Google Scholar] [PubMed]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.; et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [PubMed]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar]
- Lopez-Molina, L.; Chua, N.H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000, 41, 541–547. [Google Scholar]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [PubMed]
- Finkelstein, R.; Gampala, S.S.; Lynch, T.J.; Thomas, T.L.; Rock, C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005, 59, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Xue, Q.; McCray, T.; Margavage, K.; Chen, F.; Lee, J.H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 2013, 25, 517–534. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Y.; Zhang, Z.; Liu, X.; Hsu, C.C.; Du, Y.; Sang, T.; Zhu, C.; Wang, Y.; Satheesh, V.; et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 2020, 11, 613. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Sang, T.; Lin, Z.; Li, R.; Ren, W.; Shen, X.; Zhao, B.; Wang, X.; Zhang, X.; et al. Cell type-specific proteomics uncovers a RAF15-SnRK2.6/OST1 kinase cascade in guard cells. J. Integr. Plant Biol. 2023, 65, 2122–2137. [Google Scholar] [CrossRef]
- Peng, Z.; Jiang, X.; Wang, Z.; Wang, X.; Li, H.; He, S.; Pan, Z.; Qayyum, A.; Rehman, A.; Du, X. Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton. Int. J. Mol. Sci. 2021, 22, 12649. [Google Scholar] [CrossRef]
- Mu, C.; Zhou, L.; Shan, L.; Li, F.; Li, Z. Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 2019, 223, 1856–1872. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Iliuk, A.; Martinez, J.S.; Hall, M.C.; Tao, W.A. Phosphorylation assay based on multifunctionalized soluble nanopolymer. Anal. Chem. 2011, 83, 2767–2774. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Tao, W.A. Universal non-antibody detection of protein phosphorylation using pIMAGO. Curr. Protoc. Chem. Biol. 2015, 7, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. Imeta 2023, 2, e85. [Google Scholar] [CrossRef]


| Title | Number |
|---|---|
| Total spectrums | 158,227 |
| Matched spectrums | 44,865 |
| Peptides | 14,991 |
| Modified peptides | 13,599 |
| Identified proteins | 6810 (6055) |
| Comparable proteins | 6048 (5597) |
| Identified sites | 17,919 (12,608) |
| Comparable sites | 13,126 (10,939) |
| Accession | Gene | ID | Description | Sequence | Phos-Site |
|---|---|---|---|---|---|
| A0A1U8IDN3 | GhABI5 | Gh_D09G1317 | protein ABSCISIC ACID-INSENSITIVE 5-like | TIGGVAPVSPVSSER | S309 |
| A0A1U8JCI3 | GhRAF36 | Gh_D05G1535 | Group C Raf-like protein kinase RAF36 | SGSPAPSSISSPLR | S131 |
| A0A1U8K1V6 | GhFER | Gh_D11G1618 | Receptor-like protein kinase FERONIA | AVFSEIR | S902 |
| A0A1U8NWI1 | GhLYK3 | Gh_A02G1137 | LysM domain receptor-like kinase 3 | KPSFCCGSGR | S304 |
| A0A1U8JI82 | GhSRF6 | Gh_A06G1240 | Protein STRUBBELIG-RECEPTOR FAMILY 6 | TIGTDQGASPR | S693 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, P.; Zhao, M.; Liu, J.; Du, M.; Tian, X.; Li, F.; Li, Z. Type One Protein Phosphatase 4aD Negatively Regulates Cotton (Gossypium hirsutum) Salt Tolerance by Inhibiting the Phosphorylation of Kinases That Respond to Abscisic Acid. Int. J. Mol. Sci. 2025, 26, 3471. https://doi.org/10.3390/ijms26083471
Cao P, Zhao M, Liu J, Du M, Tian X, Li F, Li Z. Type One Protein Phosphatase 4aD Negatively Regulates Cotton (Gossypium hirsutum) Salt Tolerance by Inhibiting the Phosphorylation of Kinases That Respond to Abscisic Acid. International Journal of Molecular Sciences. 2025; 26(8):3471. https://doi.org/10.3390/ijms26083471
Chicago/Turabian StyleCao, Pengfei, Miao Zhao, Jinxin Liu, Mingwei Du, Xiaoli Tian, Fangjun Li, and Zhaohu Li. 2025. "Type One Protein Phosphatase 4aD Negatively Regulates Cotton (Gossypium hirsutum) Salt Tolerance by Inhibiting the Phosphorylation of Kinases That Respond to Abscisic Acid" International Journal of Molecular Sciences 26, no. 8: 3471. https://doi.org/10.3390/ijms26083471
APA StyleCao, P., Zhao, M., Liu, J., Du, M., Tian, X., Li, F., & Li, Z. (2025). Type One Protein Phosphatase 4aD Negatively Regulates Cotton (Gossypium hirsutum) Salt Tolerance by Inhibiting the Phosphorylation of Kinases That Respond to Abscisic Acid. International Journal of Molecular Sciences, 26(8), 3471. https://doi.org/10.3390/ijms26083471






