Short- and Long-Term Regulation of HuD: A Molecular Switch Mediated by Folic Acid?
Abstract
1. Introduction
2. Results
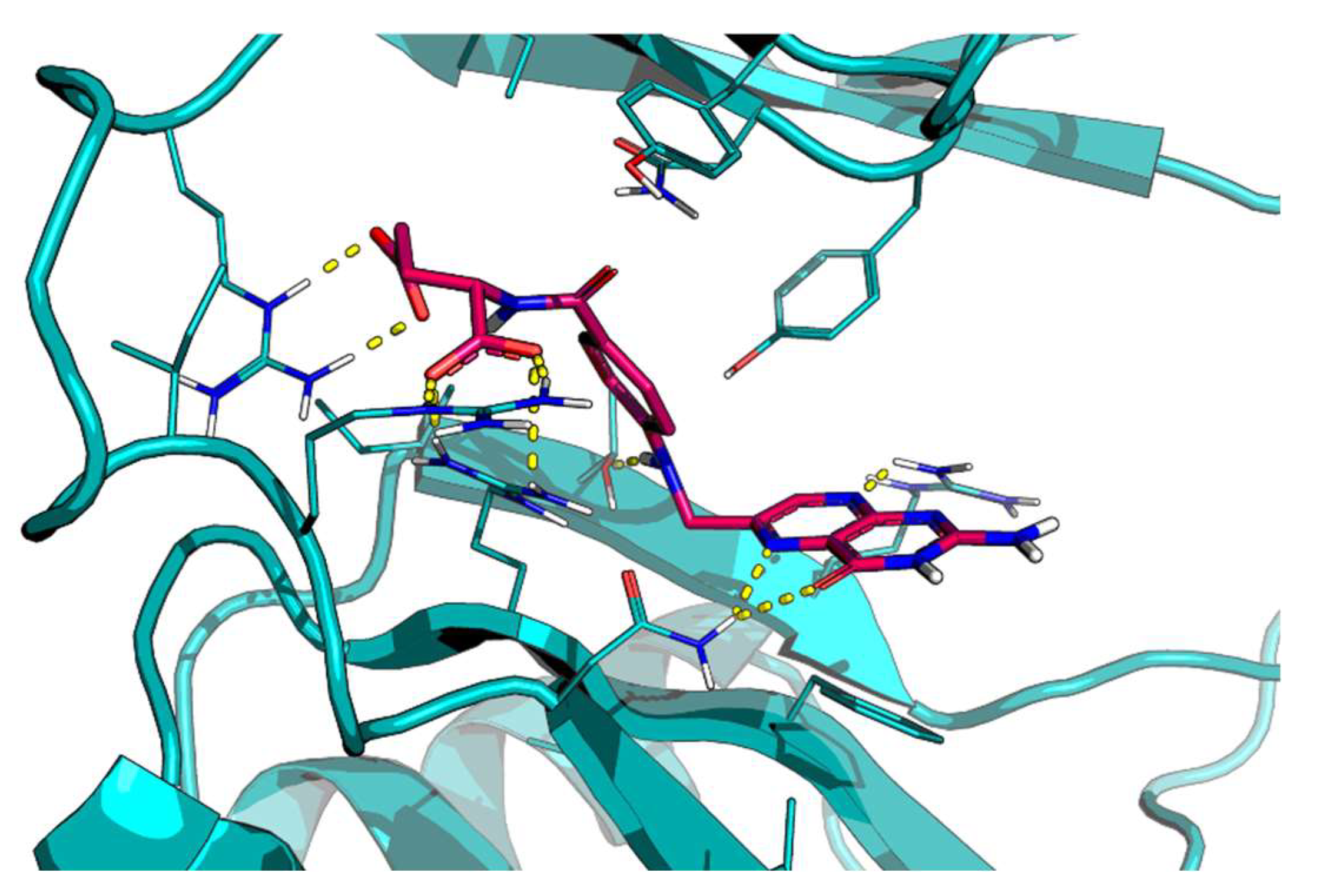
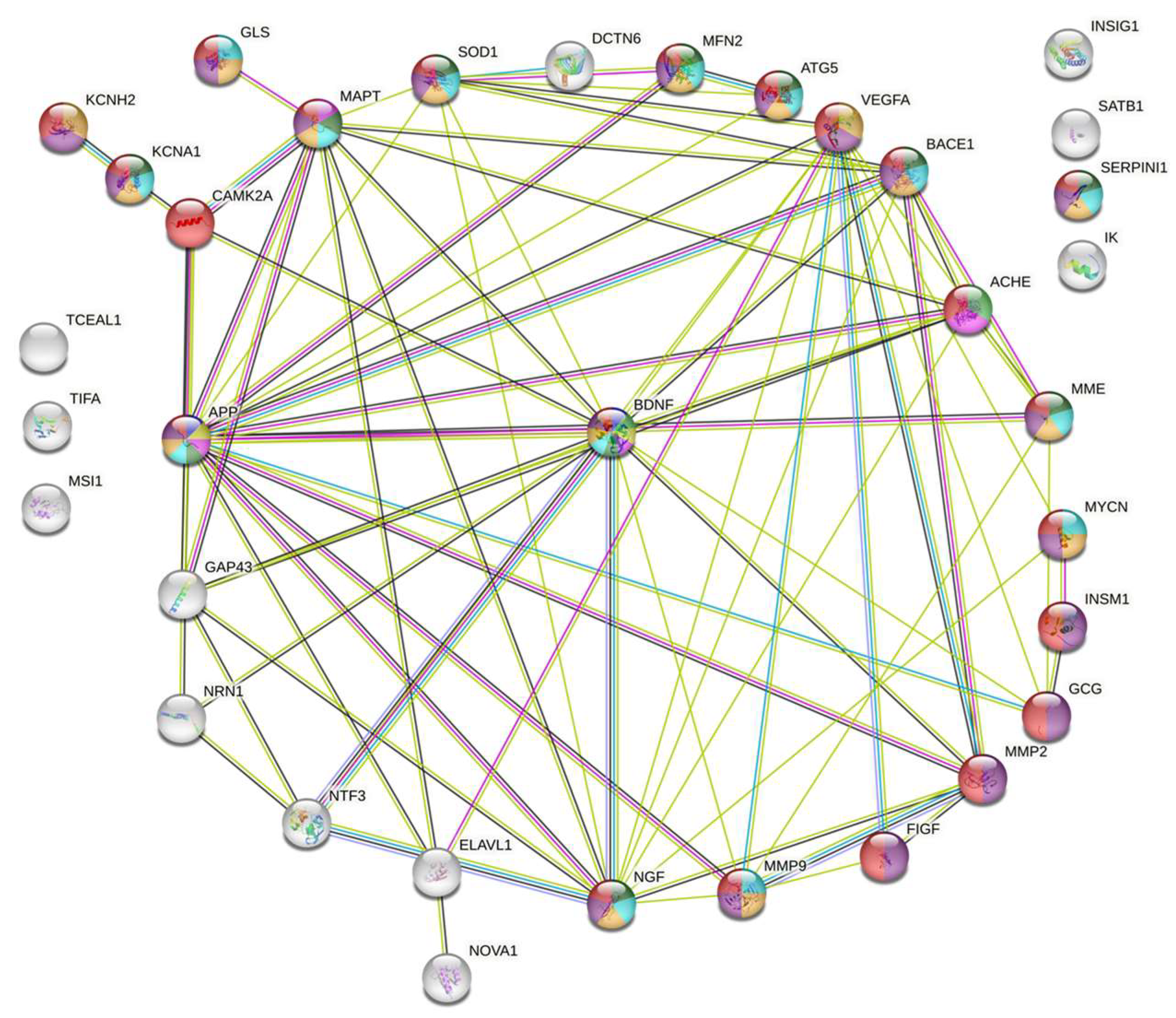
2.1. Protein–Protein Interaction Network of the Targets Controlled by HuD
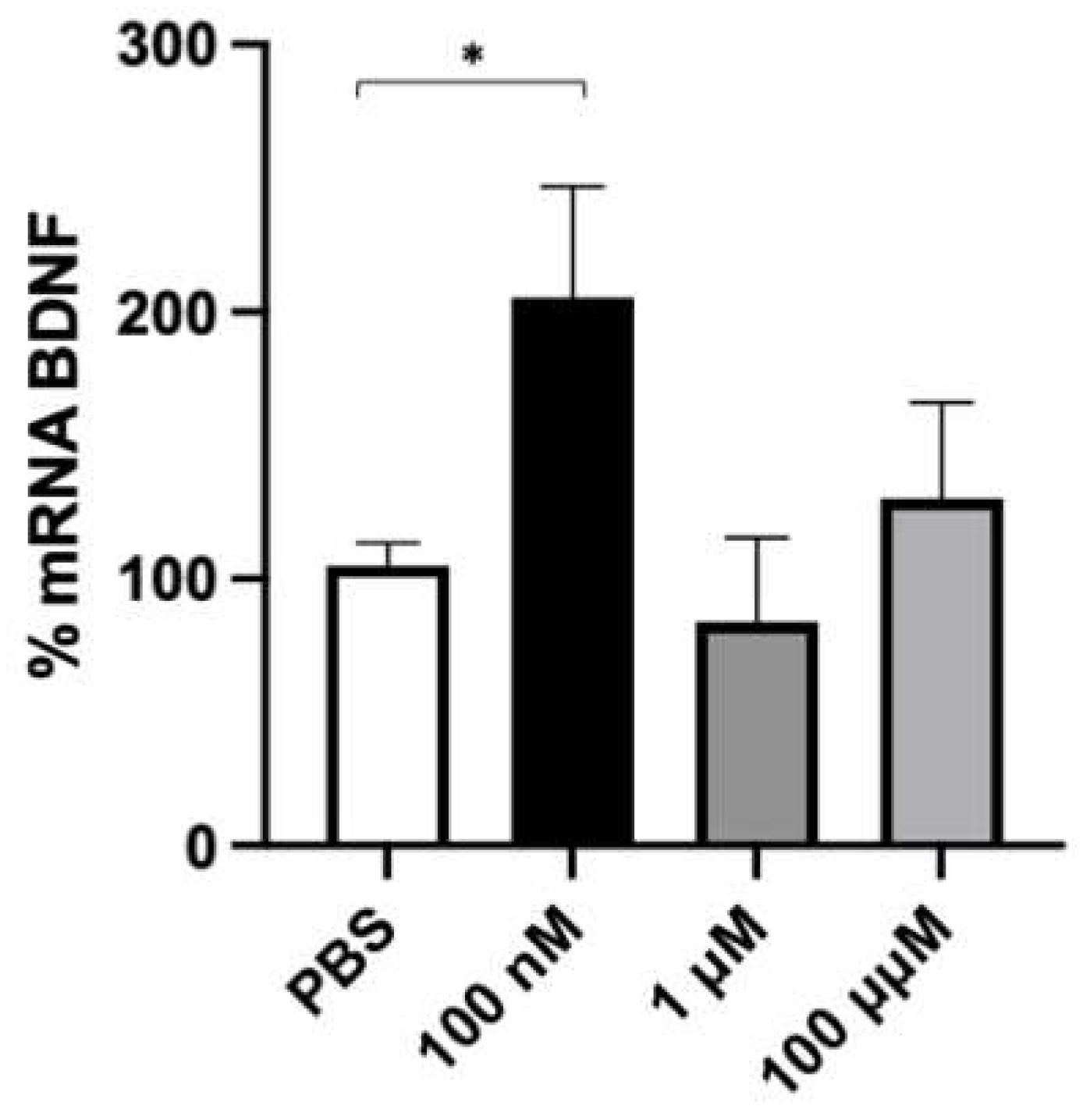
2.2. Effect of Folic Acid on Cell Viability and BDNF mRNA Content
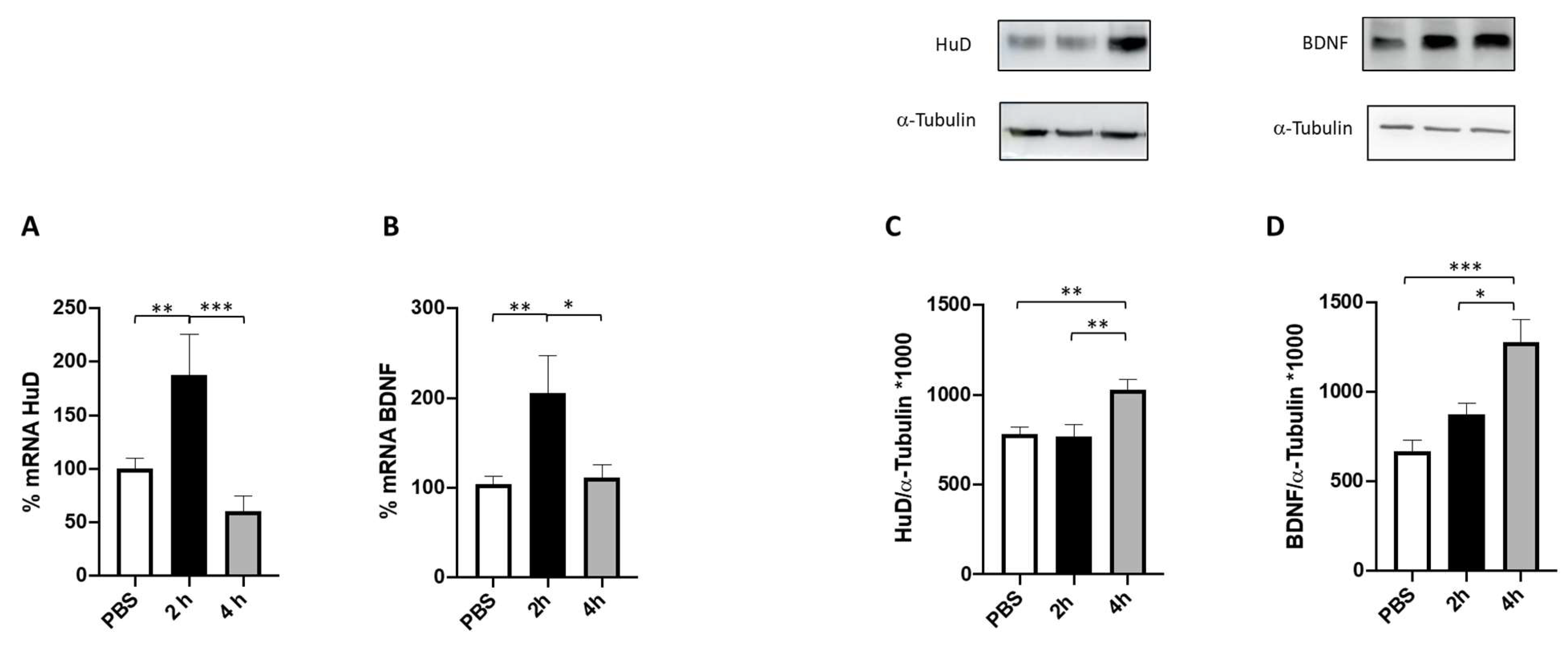
2.3. Effect of Folic Acid on HuD and BDNF Expression
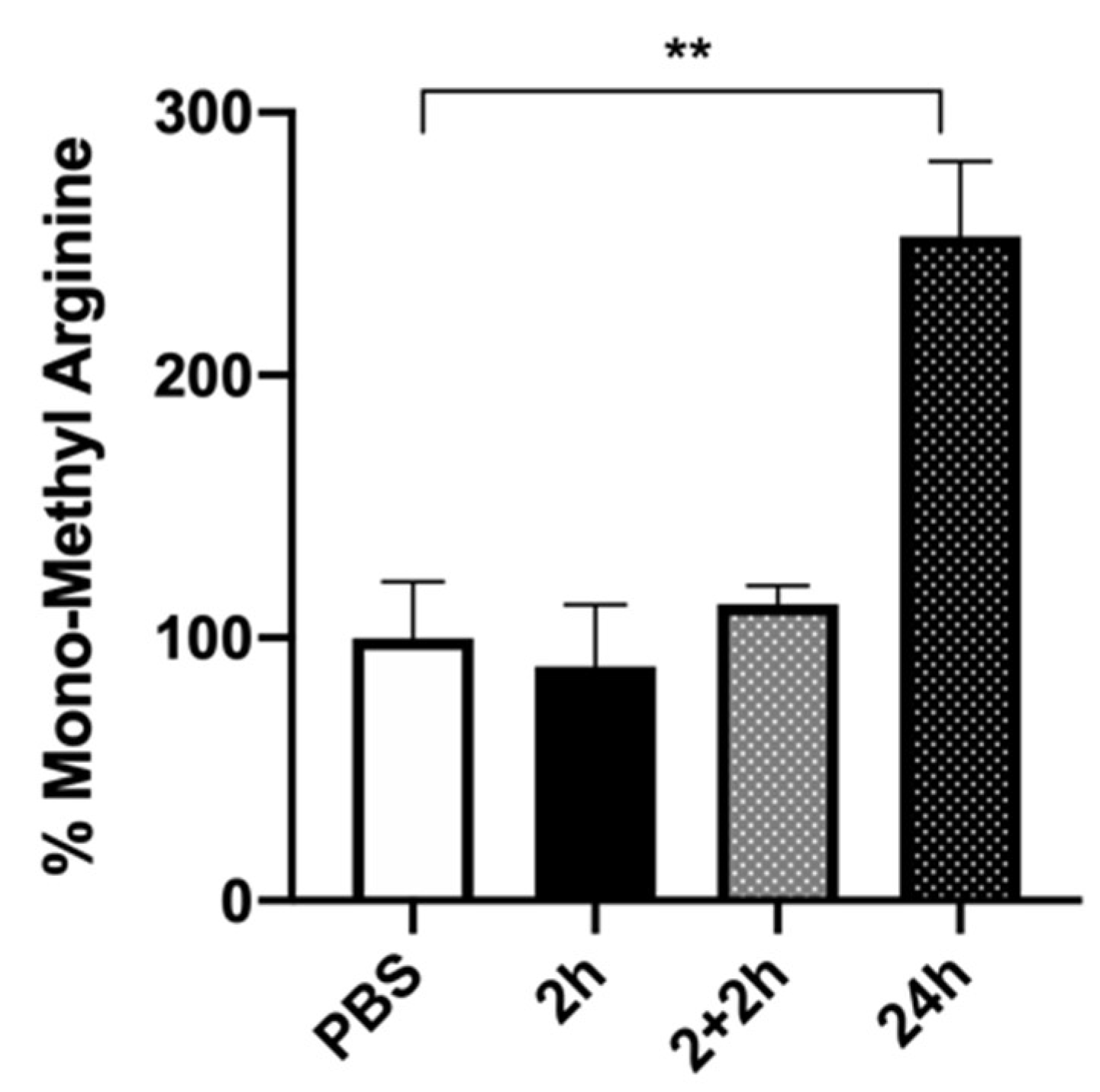
2.4. HuD Post-Translational Modification after Prolonged Folic Acid Treatment
3. Discussion
4. Material and Methods
4.1. Protein–Protein Interaction (PPI) Network
4.2. Cell Culture
4.3. MTT Assay
4.4. Western Blotting
4.5. Real-Time Quantitative RT-PCR
4.6. Immunoprecipitation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hussain, R.; Zubair, H.; Pursell, S.; Shahab, M. Neurodegenerative Diseases: Regenerative Mechanisms and Novel Therapeutic Approaches. Brain Sci. 2018, 8, E177. [Google Scholar] [CrossRef] [PubMed]
- Varesi, A.; Campagnoli, L.I.M.; Barbieri, A.; Rossi, L.; Ricevuti, G.; Esposito, C.; Chirumbolo, S.; Marchesi, N.; Pascale, A. RNA Binding Proteins in Senescence: A Potential Common Linker for Age-Related Diseases? Ageing Res. Rev. 2023, 88. [Google Scholar] [CrossRef] [PubMed]
- Lenzken, S.C.; Achsel, T.; Carrì, M.T.; Barabino, S.M.L. Neuronal RNA-Binding Proteins in Health, and Disease. Wiley Interdiscip. Rev. RNA 2014, 5, 565–576. [Google Scholar] [CrossRef]
- Pascale, A.; Govoni, S. The Complex World of Post-Transcriptional Mechanisms: Is Their Deregulation a Common Link for Diseases? Focus on ELAV-like RNA-Binding Proteins. Cell. Mol. Life Sci. 2012, 69, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Prashad, S.; Gopal, P.P. RNA-Binding Proteins in Neurological Development and Disease. RNA Biol. 2021, 18, 972–987. [Google Scholar] [CrossRef]
- Schieweck, R.; Ninkovic, J.; Kiebler, M.A. RNA-Binding Proteins Balance Brain Function in Health and Disease. Physiol. Rev. 2021, 101, 1309–1370. [Google Scholar] [CrossRef]
- Colombrita, C.; Silani, V.; Ratti, A. ELAV Proteins along Evolution: Back to the Nucleus? Mol. Cell. Neurosci. 2013, 56, 447–455. [Google Scholar] [CrossRef]
- Hinman, M.N.; Lou, H. Diverse Molecular Functions of Hu Proteins. Cell Mol. Life Sci. 2008, 65, 3168–3181. [Google Scholar] [CrossRef]
- Bolognani, F.; Contente-Cuomo, T.; Perrone-Bizzozero, N.I. Novel Recognition Motifs and Biological Functions of the RNA-Binding Protein HuD Revealed by Genome-Wide Identification of Its Targets. Nucleic Acids Res. 2009, 38, 117–130. [Google Scholar] [CrossRef]
- Wang, F.; Tidei, J.J.; Polich, E.D.; Gao, Y.; Zhao, H.; Perrone-Bizzozero, N.I.; Guo, W.; Zhao, X. Positive Feedback between RNA-Binding Protein HuD and Transcription Factor SATB1 Promotes Neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E4995–E5004. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Hutchison, E.R.; Lee, E.K.; Kuwano, Y.; Kim, M.M.; Masuda, K.; Srikantan, S.; Subaran, S.S.; Marasa, B.S.; Mattson, M.P.; et al. MiR-375 Inhibits Differentiation of Neurites by Lowering HuD Levels. Mol. Cell Biol. 2010, 30, 4197–4210. [Google Scholar] [CrossRef] [PubMed]
- Bronicki, L.M.; Jasmin, B.J. Emerging Complexity of the HuD/ELAVl4 Gene; Implications for Neuronal Development, Function, and Dysfunction. RNA 2013, 19, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Good, P.J. A Conserved Family of Elav-like Genes in Vertebrates. Proc. Natl. Acad. Sci. USA 1995, 92, 4557–4561. [Google Scholar] [CrossRef]
- Pascale, A.; Gusev, P.A.; Amadio, M.; Dottorini, T.; Govoni, S.; Alkon, D.L.; Quattrone, A. Increase of the RNA-Binding Protein HuD and Posttranscriptional up-Regulation of the GAP-43 Gene during Spatial Memory. Proc. Natl. Acad. Sci. USA 2004, 101, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Perrone-Bizzozero, N.; Bird, C.W. Role of HuD in Nervous System Function and Pathology. Front. Biosci. 2013, 5, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Dell’Orco, M.; Sardone, V.; Gardiner, A.S.; Pansarasa, O.; Bordoni, M.; Perrone-Bizzozero, N.I.; Cereda, C. HuD Regulates SOD1 Expression during Oxidative Stress in Differentiated Neuroblastoma Cells and Sporadic ALS Motor Cortex. Neurobiol. Dis. 2021, 148, 105211. [Google Scholar] [CrossRef]
- Tebaldi, T.; Zuccotti, P.; Peroni, D.; Köhn, M.; Gasperini, L.; Potrich, V.; Bonazza, V.; Dudnakova, T.; Rossi, A.; Sanguinetti, G.; et al. HuD Is a Neural Translation Enhancer Acting on MTORC1-Responsive Genes and Counteracted by the Y3 Small Non-Coding RNA. Mol. Cell 2018, 71, 256–270. [Google Scholar] [CrossRef]
- Dell’Orco, M.; Oliver, R.J.; Perrone-Bizzozero, N. HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes. Front. Genet. 2020, 11, 790. [Google Scholar] [CrossRef]
- Nasti, R.; Rossi, D.; Amadio, M.; Pascale, A.; Unver, M.Y.; Hirsch, A.K.H.; Collina, S. Compounds Interfering with Embryonic Lethal Abnormal Vision (ELAV) Protein-RNA Complexes: An Avenue for Discovering New Drugs. J. Med. Chem. 2017, 60, 8257–8267. [Google Scholar] [CrossRef]
- Silvestri, B.; Mochi, M.; Garone, M.G.; Rosa, A. Emerging Roles for the RNA-Binding Protein HuD (ELAVL4) in Nervous System Diseases. Int. J. Mol. Sci. 2022, 23, 14606. [Google Scholar] [CrossRef]
- Pascale, A.; Amadio, M.; Scapagnini, G.; Lanni, C.; Racchi, M.; Provenzani, A.; Govoni, S.; Alkon, D.L.; Quattrone, A. Neuronal ELAV Proteins Enhance MRNA Stability by a PKCalpha-Dependent Pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 12065–12070. [Google Scholar] [CrossRef] [PubMed]
- Talman, V.; Amadio, M.; Osera, C.; Sorvari, S.; Boije Af Gennäs, G.; Yli-Kauhaluoma, J.; Rossi, D.; Govoni, S.; Collina, S.; Ekokoski, E.; et al. The C1 Domain-Targeted Isophthalate Derivative HMI-1b11 Promotes Neurite Outgrowth and GAP-43 Expression through PKCα Activation in SH-SY5Y Cells. Pharmacol. Res. 2013, 73, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, N.; Amadio, M.; Colombrita, C.; Govoni, S.; Ratti, A.; Pascale, A. PKC Activation Counteracts ADAM10 Deficit in HuD-Silenced Neuroblastoma Cells. J. Alzheimer’s Dis. 2016, 54, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Mori, Y.; Chu, D.L.; Koyama, Y.; Miyata, S.; Tanaka, H.; Yachi, K.; Kubo, T.; Yoshikawa, H.; Tohyama, M. CARM1 Regulates Proliferation of PC12 Cells by Methylating HuD. Mol. Cell Biol. 2006, 26, 2273–2285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Hall, T.M. Structural Basis for Recognition of AU-Rich Element RNA by the HuD Protein. Nat. Struct. Biol. 2001, 8, 141–145. [Google Scholar] [CrossRef]
- Della Volpe, S.; Linciano, P.; Listro, R.; Tumminelli, E.; Amadio, M.; Bonomo, I.; Elgaher, W.A.M.; Adam, S.; Hirsch, A.K.H.; Boeckler, F.M.; et al. Identification of N,N-Arylalkyl-Picolinamide Derivatives Targeting the RNA-Binding Protein HuR, by Combining Biophysical Fragment-Screening and Molecular Hybridization. Bioorg Chem. 2021, 116, 105305. [Google Scholar] [CrossRef]
- Della Volpe, S.; Nasti, R.; Queirolo, M.; Unver, M.Y.; Jumde, V.K.; Dömling, A.; Vasile, F.; Potenza, D.; Ambrosio, F.A.; Costa, G.; et al. Novel Compounds Targeting the RNA-Binding Protein HuR. Structure-Based Design, Synthesis, and Interaction Studies. ACS Med. Chem. Lett. 2019, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Šponer, J.; Bussi, G.; Krepl, M.; Banáš, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurečka, P.; et al. RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef] [PubMed]
- Vasile, F.; Della Volpe, S.; Ambrosio, F.A.; Costa, G.; Unver, M.Y.; Zucal, C.; Rossi, D.; Martino, E.; Provenzani, A.; Hirsch, A.K.H.; et al. Exploration of Ligand Binding Modes towards the Identification of Compounds Targeting HuR: A Combined STD-NMR and Molecular Modelling Approach. Sci. Rep. 2018, 8, 13780. [Google Scholar] [CrossRef]
- Uusitalo, J.J.; Ingólfsson, H.I.; Marrink, S.J.; Faustino, I. Martini Coarse-Grained Force Field: Extension to RNA. Biophys. J. 2017, 113, 246–256. [Google Scholar] [CrossRef]
- Ambrosio, F.A.; Coricello, A.; Costa, G.; Lupia, A.; Micaelli, M.; Marchesi, N.; Sala, F.; Pascale, A.; Rossi, D.; Vasile, F.; et al. Identification of Compounds Targeting HuD. Another Brick in the Wall of Neurodegenerative Disease Treatment. J. Med. Chem. 2021, 64, 9989–10000. [Google Scholar] [CrossRef] [PubMed]
- Gliwińska, A.; Czubilińska-Łada, J.; Więckiewicz, G.; Świętochowska, E.; Badeński, A.; Dworak, M.; Szczepańska, M. The Role of Brain-Derived Neurotrophic Factor (BDNF) in Diagnosis and Treatment of Epilepsy, Depression, Schizophrenia, Anorexia Nervosa and Alzheimer’s Disease as Highly Drug-Resistant Diseases: A Narrative Review. Brain Sci. 2023, 13, 163. [Google Scholar] [CrossRef]
- Beckel-Mitchener, A.C.; Miera, A.; Keller, R.; Perrone-Bizzozero, N.I. Poly(A) Tail Length-Dependent Stabilization of GAP-43 MRNA by the RNA-Binding Protein HuD. J. Biol. Chem. 2002, 277, 27996–28002. [Google Scholar] [CrossRef]
- Bolognani, F.; Tanner, D.C.; Nixon, S.; Okano, H.J.; Okano, H.; Perrone-Bizzozero, N.I. Coordinated Expression of HuD and GAP-43 in Hippocampal Dentate Granule Cells during Developmental and Adult Plasticity. Neurochem. Res. 2007, 32, 2142–2151. [Google Scholar] [CrossRef]
- Lim, C.S.; Alkon, D.L. Protein Kinase C Stimulates HuD-Mediated MRNA Stability and Protein Expression of Neurotrophic Factors and Enhances Dendritic Maturation of Hippocampal Neurons in Culture. Hippocampus 2012, 22, 2303–2319. [Google Scholar] [CrossRef]
- Kang, M.-J.; Abdelmohsen, K.; Hutchison, E.R.; Mitchell, S.J.; Grammatikakis, I.; Guo, R.; Noh, J.H.; Martindale, J.L.; Yang, X.; Lee, E.K.; et al. HuD Regulates Coding and Noncoding RNA to Induce APP→Aβ Processing. Cell Rep. 2014, 7, 1401–1409. [Google Scholar] [CrossRef]
- King, P.H. RNA-Binding Analyses of HuC and HuD with the VEGF and c-Myc 3’-Untranslated Regions Using a Novel ELISA-Based Assay. Nucleic Acids Res. 2000, 28, E20. [Google Scholar] [CrossRef] [PubMed]
- Castrén, E.; Berninger, B.; Leingärtner, A.; Lindholm, D. Regulation of Brain-Derived Neurotrophic Factor MRNA Levels in Hippocampus by Neuronal Activity. Prog. Brain Res. 1998, 117, 57–64. [Google Scholar] [CrossRef]
- Higashimoto, K.; Kuhn, P.; Desai, D.; Cheng, X.; Xu, W. Phosphorylation-Mediated Inactivation of Coactivator-Associated Arginine Methyltransferase 1. Proc. Natl. Acad. Sci. USA 2007, 104, 12318–12323. [Google Scholar] [CrossRef] [PubMed]
- Doxakis, E. RNA Binding Proteins: A Common Denominator of Neuronal Function and Dysfunction. Neurosci. Bull. 2014, 30, 610–626. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Perrone-Bizzozero, N.I. RNA-Protein Interactions and Control of MRNA Stability in Neurons. J. Neurosci. Res. 2008, 86, 481–489. [Google Scholar] [CrossRef]
- Hilgers, V. Regulation of Neuronal RNA Signatures by ELAV/Hu Proteins. WIREs RNA 2023, 14, e1733. [Google Scholar] [CrossRef]
- Jung, M.; Lee, E.K. RNA–Binding Protein HuD as a Versatile Factor in Neuronal and Non–Neuronal Systems. Biology 2021, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Li, C.; Zhao, J. Identification of Key Pathways and Genes in Colorectal Cancer Using Bioinformatics Analysis. Med. Oncol. 2016, 33, 111. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Wang, J.; Li, G.; Sun, Y.; Deng, Q.; Li, M.; Song, K.; Zhao, Z. Preliminary Therapeutic and Mechanistic Evaluation of S-Allylmercapto-N-Acetylcysteine in the Treatment of Pulmonary Emphysema. Int. Immunopharmacol. 2021, 98, 107913. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, N.; Barbieri, A.; Fahmideh, F.; Govoni, S.; Ghidoni, A.; Parati, G.; Vanoli, E.; Pascale, A.; Calvillo, L. Use of Dual-Flow Bioreactor to Develop a Simplified Model of Nervous-Cardiovascular Systems Crosstalk: A Preliminary Assessment. PLoS ONE 2020, 15, e0242627. [Google Scholar] [CrossRef] [PubMed]
- Fahmideh, F.; Marchesi, N.; Campagnoli, L.I.M.; Landini, L.; Caramella, C.; Barbieri, A.; Govoni, S.; Pascale, A. Effect of Troxerutin in Counteracting Hyperglycemia-Induced VEGF Upregulation in Endothelial Cells: A New Option to Target Early Stages of Diabetic Retinopathy? Front. Pharmacol. 2022, 13, 951833. [Google Scholar] [CrossRef]
- Mallucci, G.; Marchesi, N.; Campagnoli, L.I.M.; Boschi, F.; Fahmideh, F.; Fusco, S.; Tavazzi, E.; Govoni, S.; Bergamaschi, R.; Pascale, A. Evidence for Novel Cell Defense Mechanisms Sustained by Dimethyl Fumarate in Multiple Sclerosis Patients: The HuR/SOD2 Cascade. Mult. Scler. Relat. Disord. 2022, 68, 104197. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchesi, N.; Linciano, P.; Campagnoli, L.I.M.; Fahmideh, F.; Rossi, D.; Costa, G.; Ambrosio, F.A.; Barbieri, A.; Collina, S.; Pascale, A. Short- and Long-Term Regulation of HuD: A Molecular Switch Mediated by Folic Acid? Int. J. Mol. Sci. 2023, 24, 12201. https://doi.org/10.3390/ijms241512201
Marchesi N, Linciano P, Campagnoli LIM, Fahmideh F, Rossi D, Costa G, Ambrosio FA, Barbieri A, Collina S, Pascale A. Short- and Long-Term Regulation of HuD: A Molecular Switch Mediated by Folic Acid? International Journal of Molecular Sciences. 2023; 24(15):12201. https://doi.org/10.3390/ijms241512201
Chicago/Turabian StyleMarchesi, Nicoletta, Pasquale Linciano, Lucrezia Irene Maria Campagnoli, Foroogh Fahmideh, Daniela Rossi, Giosuè Costa, Francesca Alessandra Ambrosio, Annalisa Barbieri, Simona Collina, and Alessia Pascale. 2023. "Short- and Long-Term Regulation of HuD: A Molecular Switch Mediated by Folic Acid?" International Journal of Molecular Sciences 24, no. 15: 12201. https://doi.org/10.3390/ijms241512201
APA StyleMarchesi, N., Linciano, P., Campagnoli, L. I. M., Fahmideh, F., Rossi, D., Costa, G., Ambrosio, F. A., Barbieri, A., Collina, S., & Pascale, A. (2023). Short- and Long-Term Regulation of HuD: A Molecular Switch Mediated by Folic Acid? International Journal of Molecular Sciences, 24(15), 12201. https://doi.org/10.3390/ijms241512201









