Biochemical and Structural Insights into a Thiamine Diphosphate-Dependent α-Ketoglutarate Decarboxylase from Cyanobacterium Microcystis aeruginosa NIES-843
Abstract
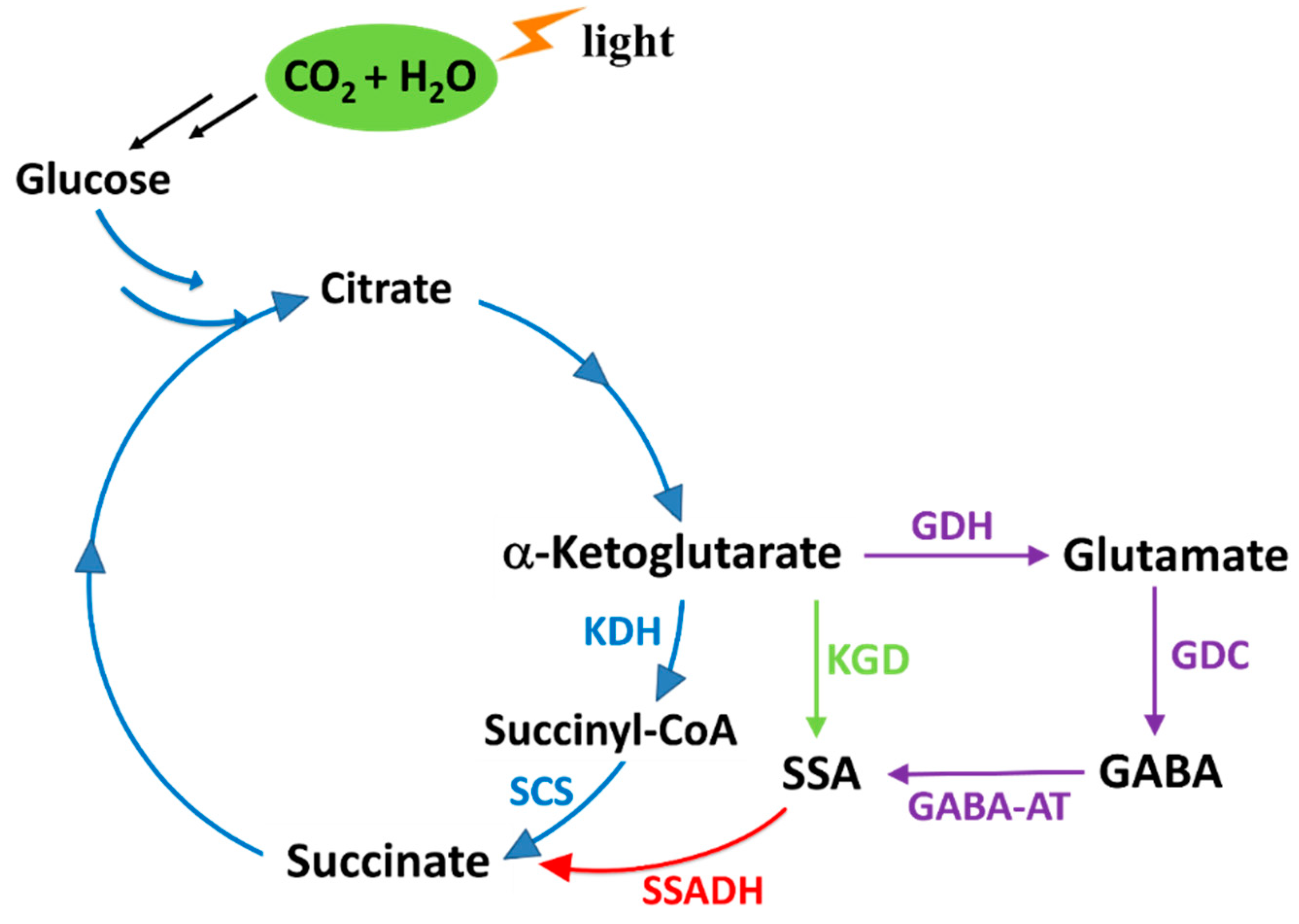
1. Introduction
2. Results and Discussion
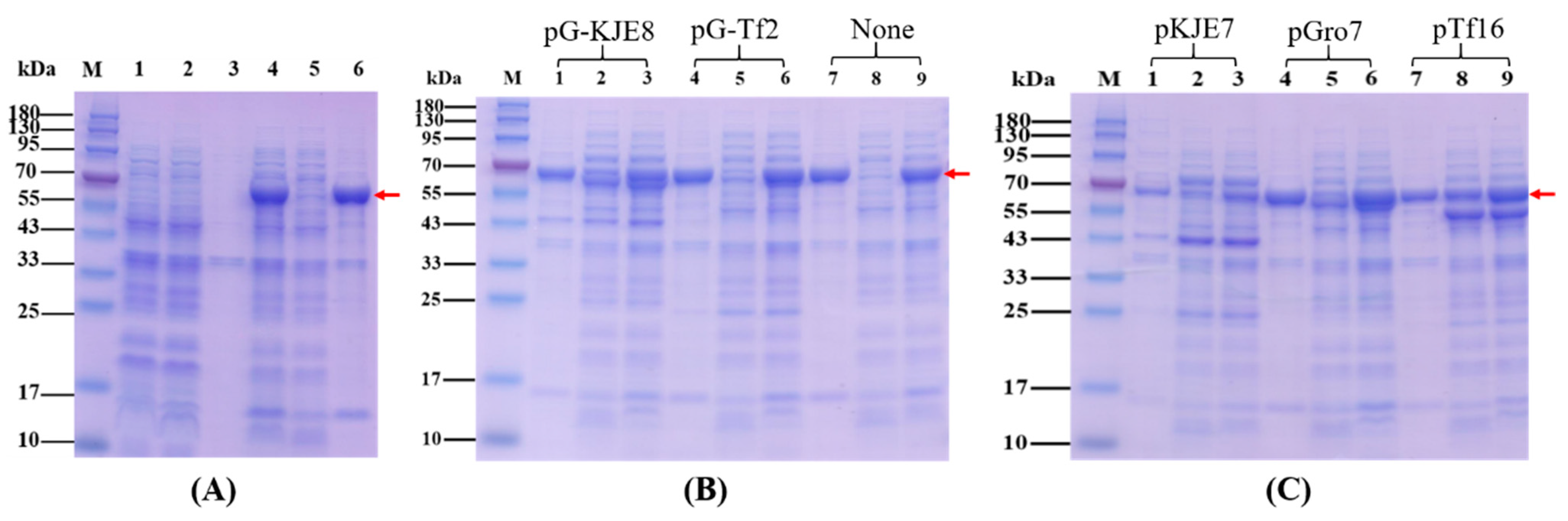
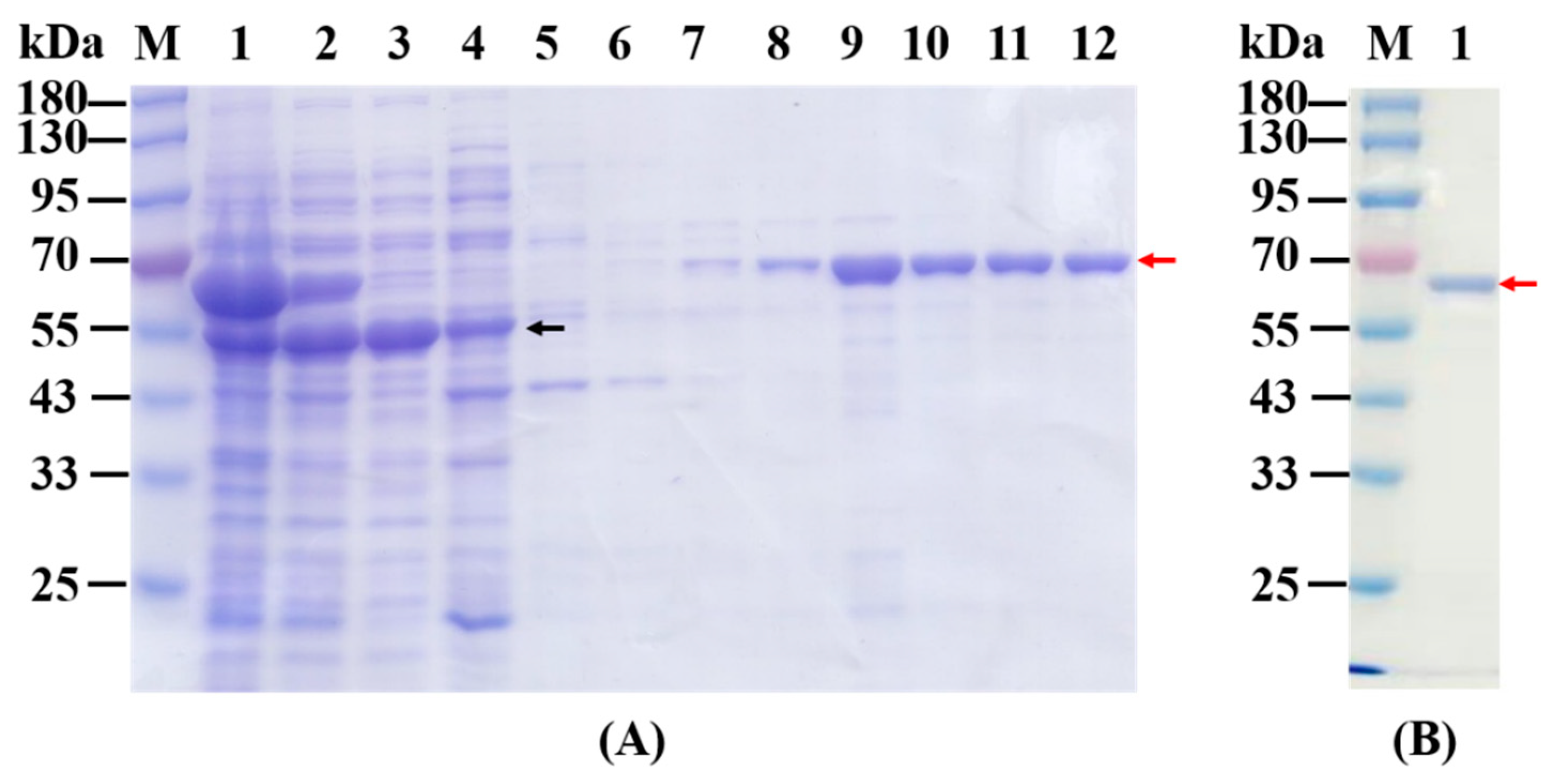
2.1. Overexpression, Purification and Native Mass Determination of Recombinant KGDs
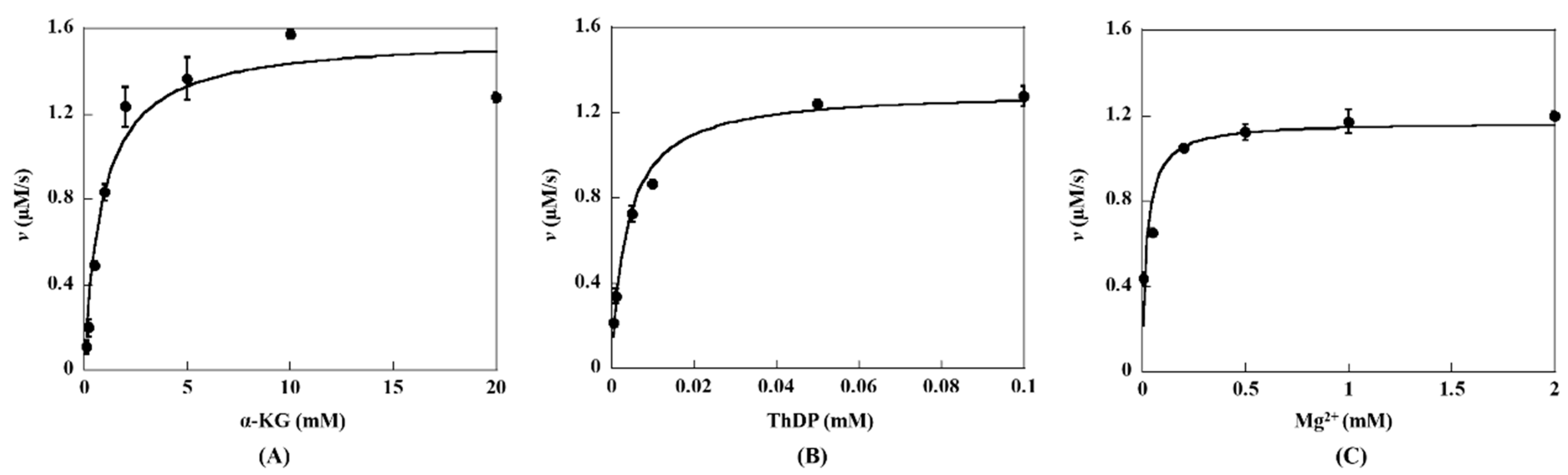
2.2. Steady-State Kinetic Assay of KGDs Wild-Type
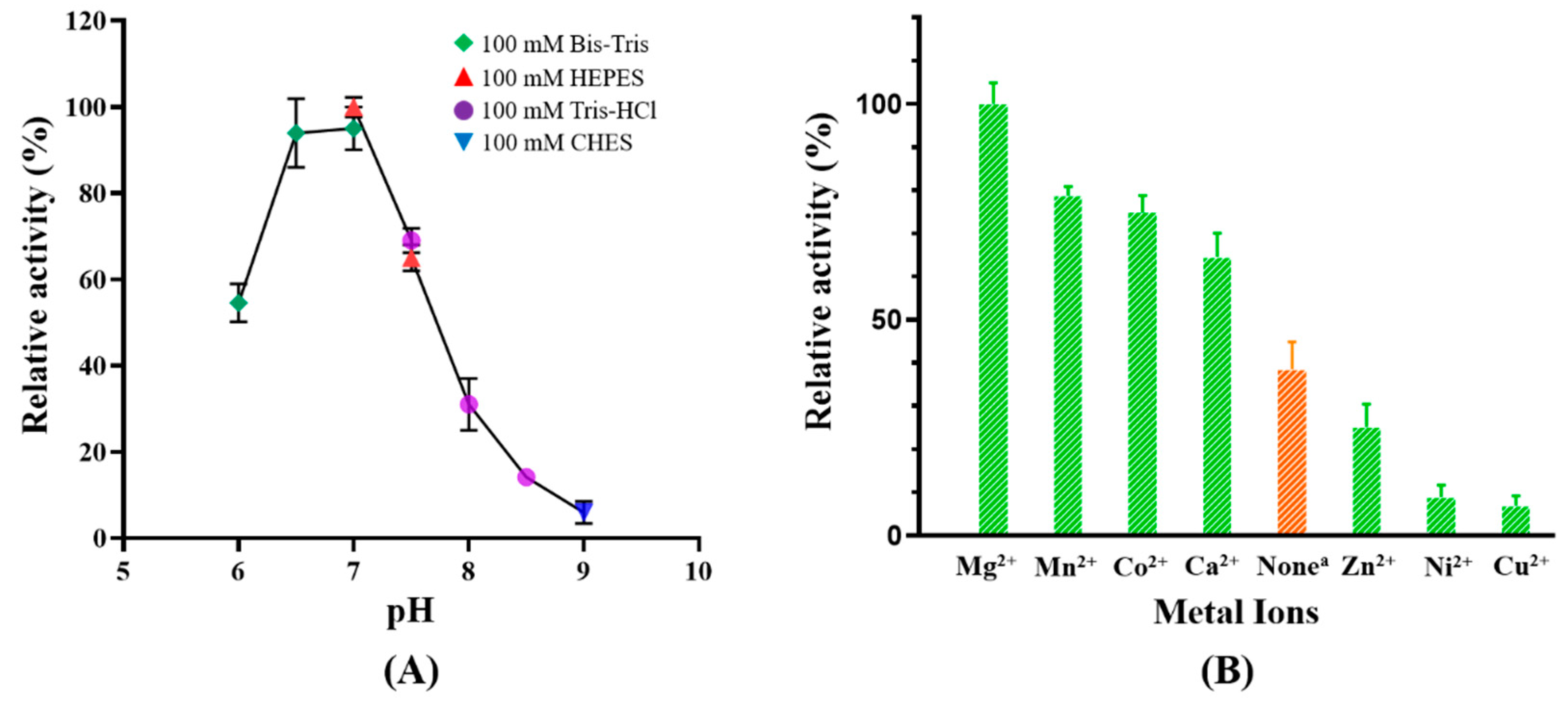
2.3. Effects of pH and Divalent Cations on Catalytic Activity of MaKGD
2.4. Determination of Substrate Specificity of MaKGD
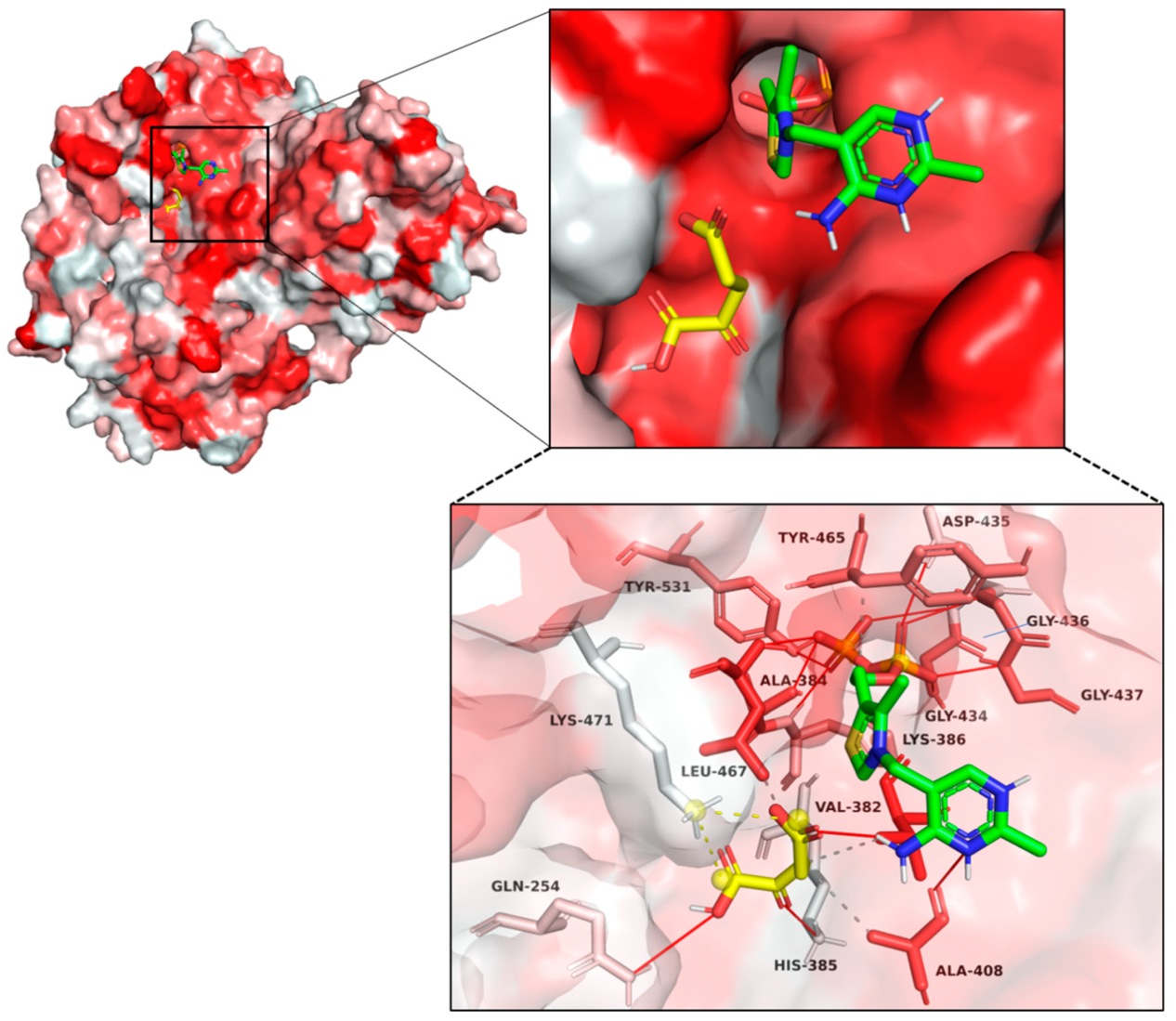
2.5. Structural Modeling and Autodocking of MaKGD with α-ketoacids
2.6. Kinetic Characterization of MaKGD Variants
3. Materials and Methods
3.1. Materials
3.2. Cloning, Overexpression and Purification of MaKGD and MiKGD
3.3. Catalytic Activity Determination of MaKGD and MiKGD
3.4. Effects of pH on the Activity of MaKGD
3.5. Effects of Divalent Cations on the Activity of MaKGD
3.6. Substrate Specificity Detection of MaKGD
3.7. Structural Modeling, Model Validation and Autodocking of MaKGD with α-ketoacids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehdizadeh Allaf, M.; Peerhossaini, H. Cyanobacteria: Model Microorganisms and Beyond. Microorganisms 2022, 10, 696. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakajima, N.; Okamoto, S.; Suzuki, I.; Tanabe, Y.; Tamaoki, M.; Nakamura, Y.; Kasai, F.; Watanabe, A.; Kawashima, K.; et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2007, 14, 247–256. [Google Scholar] [CrossRef]
- Zhang, S.; Qian, X.; Chang, S.; Dismukes, G.C.; Bryant, D.A. Natural and Synthetic Variants of the Tricarboxylic Acid Cycle in Cyanobacteria: Introduction of the GABA Shunt into Synechococcus sp. PCC 7002. Front. Microbiol. 2016, 7, 1972. [Google Scholar] [CrossRef]
- Shigeoka, S.; Onishi, T.; Maeda, K.; Nakano, Y.; Kitaoka, S. Occurrence of thiamin pyrophosphate-dependent 2-oxoglutarate decarboxylase in mitochondria of Euglena gracilis. FEBS Lett. 1986, 195, 43–47. [Google Scholar] [CrossRef]
- Jordan, F.; Patel, H. Catalysis in Enzymatic Decarboxylations: Comparison of Selected Cofactor-dependent and Cofactor-independent Examples. ACS Catal. 2013, 3, 1601–1617. [Google Scholar] [CrossRef]
- Atsumi, S.; Li, Z.; Liao, J.C. Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 6306–6311. [Google Scholar] [CrossRef]
- Yep, A.; Kenyon, G.L.; McLeish, M.J. Determinants of substrate specificity in KdcA, a thiamin diphosphate-dependent decarboxylase. Bioorg. Chem. 2006, 34, 325–336. [Google Scholar] [CrossRef]
- De la Plaza, M.; Fernandez de Palencia, P.; Pelaez, C.; Requena, T. Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett. 2004, 238, 367–374. [Google Scholar] [CrossRef]
- Schutz, A.; Golbik, R.; Tittmann, K.; Svergun, D.I.; Koch, M.H.; Hubner, G.; Konig, S. Studies on structure-function relationships of indolepyruvate decarboxylase from Enterobacter cloacae, a key enzyme of the indole acetic acid pathway. Eur. J. Biochem. FEBS 2003, 270, 2322–2331. [Google Scholar] [CrossRef]
- Polovnikova, E.S.; McLeish, M.J.; Sergienko, E.A.; Burgner, J.T.; Anderson, N.L.; Bera, A.K.; Jordan, F.; Kenyon, G.L.; Hasson, M.S. Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent. Biochemistry 2003, 42, 1820–1830. [Google Scholar] [CrossRef]
- Shigeoka, S.; Nakano, Y. Characterization and molecular properties of 2-oxoglutarate decarboxylase from Euglena gracilis. Arch. Biochem. Biophys. 1991, 288, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Kapol, R.; Radler, F. Purification and characterization of 2-oxoglutarate decarboxylase of Leuconostoc oenos. J. Gen. Microbiol. 1990, 136, 1497–1499. [Google Scholar] [CrossRef]
- Green, L.S.; Li, Y.; Emerich, D.W.; Bergersen, F.J.; Day, D.A. Catabolism of alpha-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: Evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 2000, 182, 2838–2844. [Google Scholar] [CrossRef]
- Palaniappan, C.; Sharma, V.; Hudspeth, M.E.; Meganathan, R. Menaquinone (vitamin K2) biosynthesis: Evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J. Bacteriol. 1992, 174, 8111–8118. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, L.P.; Zhao, H.; Dickinson, C.E.; Arango, N.M.; Lima, C.D.; Fischer, S.M.; Ouerfelli, O.; Nathan, C.; Rhee, K.Y. Activity-based metabolomic profiling of enzymatic function: Identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase. Chem. Biol. 2010, 17, 323–332. [Google Scholar] [CrossRef]
- Lei, G.; Wang, X.; Lai, C.; Li, Z.M.; Zhang, W.; Xie, C.; Wang, F.; Wu, X.; Li, Z. Expression and biochemical characterization of alpha-ketoglutarate decarboxylase from cyanobacterium Synechococcus sp. PCC7002. Int. J. Biol. Macromol. 2018, 114, 188–193. [Google Scholar] [CrossRef]
- Wang, X.; Lei, G.; Wu, X.; Wang, F.; Lai, C.; Li, Z. Expression, purification and characterization of sll1981 protein from cyanobacterium Synechocystis sp. PCC6803. Protein Expr. Purif. 2017, 139, 21–28. [Google Scholar] [CrossRef]
- Li, Z.-M.; Bai, F.; Wang, X.; Xie, C.; Wan, Y.; Li, Y.; Liu, J.; Li, Z. Kinetic Characterization and Catalytic Mechanism of N-Acetylornithine Aminotransferase Encoded by slr1022 Gene from Synechocystis sp. PCC6803. Int. J. Mol. Sci. 2023, 24, 5853. [Google Scholar] [CrossRef]
- Hasson, M.S.; Muscate, A.; McLeish, M.J.; Polovnikova, L.S.; Gerlt, J.A.; Kenyon, G.L.; Petsko, G.A.; Ringe, D. The crystal structure of benzoylformate decarboxylase at 1.6 Å Resolution: Diversity of catalytic residues in thiamin diphosphate-dependent enzymes. Biochemistry 1998, 37, 9918–9930. [Google Scholar] [CrossRef]
- Candy, J.M.; Duggleby, R.G. Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme. Biochim. Biophys. Acta—Protein Struct. Mol. Enzymol. 1998, 1385, 323–338. [Google Scholar] [CrossRef]
- Xiong, W.; Brune, D.; Vermaas, W.F. The gamma-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014, 93, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, L.; Liu, H.; Li, C.; Li, Z.; Wang, J.; Tan, X. CyanoOmicsDB: An integrated omics database for functional genomic analysis of cyanobacteria. Nucleic Acids Res. 2022, 50, D758–D764. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, M.; Nakano, Y.; Kitaoka, S. Subcellular localization of the GABA-shunt enzymes in Euglena gracilis strain Z. J. Protozool. 1979, 26, 471–473. [Google Scholar] [CrossRef]
- Kneen, M.M.; Stan, R.; Yep, A.; Tyler, R.P.; Saehuan, C.; McLeish, M.J. Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J. 2011, 278, 1842–1853. [Google Scholar] [CrossRef]
- Wang, B.; Bai, Y.; Fan, T.; Zheng, X.; Cai, Y. Characterisation of a thiamine diphosphate-dependent alpha-keto acid decarboxylase from Proteus mirabilis JN458. Food Chem. 2017, 232, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Neumann, P.; Parthier, C.; Friedemann, R.; Nemeria, N.; Jordan, F.; Tittmann, K. Double duty for a conserved glutamate in pyruvate decarboxylase: Evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate. Biochemistry 2010, 49, 8197–8212. [Google Scholar] [CrossRef]
- Nemeria, N.; Korotchkina, L.; McLeish, M.J.; Kenyon, G.L.; Patel, M.S.; Jordan, F. Elucidation of the chemistry of enzyme-bound thiamin diphosphate prior to substrate binding: Defining internal equilibria among tautomeric and ionization states. Biochemistry 2007, 46, 10739–10744. [Google Scholar] [CrossRef]
- Bao, W.; Li, X.; Liu, J.; Zheng, R.; Liu, L.; Zhang, H. The Characterization of an Efficient Phenylpyruvate Decarboxylase KDC4427, Involved in 2-Phenylethanol and IAA Production from Bacterial Enterobacter sp. CGMCC 5087. Microbiol. Spectr. 2022, 10, e0266021. [Google Scholar] [CrossRef]
- Zhang, S.; Bryant, D.A. The tricarboxylic acid cycle in cyanobacteria. Science 2011, 334, 1551–1553. [Google Scholar] [CrossRef]
- Stribny, J.; Romagnoli, G.; Perez-Torrado, R.; Daran, J.M.; Querol, A. Characterisation of the broad substrate specificity 2-keto acid decarboxylase Aro10p of Saccharomyces kudriavzevii and its implication in aroma development. Microb. Cell Fact. 2016, 15, 51. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, D.; Kahyaoglu, A.; Farid, R.S.; Jordan, F. Is a hydrophobic amino acid required to maintain the reactive V conformation of thiamin at the active center of thiamin diphosphate-requiring enzymes? Experimental and computational studies of isoleucine 415 of yeast pyruvate decarboxylase. Biochemistry 1998, 37, 13379–13391. [Google Scholar] [CrossRef] [PubMed]
- Candy, J.M.; Koga, J.; Nixon, P.F.; Duggleby, R.G. The role of residues glutamate-50 and phenylalanine-496 in Zymomonas mobilis pyruvate decarboxylase. Biochem. J. 1996, 315 Pt 3, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Hübner, G.; Tittmann, K.; Killenberg-Jabs, M.; Schäffner, J.; Spinka, M.; Neef, H.; Kern, D.; Kern, G.; Schneider, G.; Wikner, C.; et al. Activation of thiamin diphosphate in enzymes. Biochim. Biophys. Acta 1998, 1385, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.; Kern, G.; Neef, H.; Tittmann, K.; Killenberg-Jabs, M.; Wikner, C.; Schneider, G.; Hübner, G. How thiamine diphosphate is activated in enzymes. Science 1997, 275, 67–70. [Google Scholar] [CrossRef]
- Xie, C.; Li, Z.M.; Bai, F.; Hu, Z.; Zhang, W.; Li, Z. Kinetic and structural insights into enzymatic mechanism of succinic semialdehyde dehydrogenase from Cyanothece sp. ATCC51142. PLoS ONE 2020, 15, e0239372. [Google Scholar] [CrossRef]
- Wang, X.; Lai, C.; Lei, G.; Wang, F.; Long, H.; Wu, X.; Chen, J.; Huo, G.; Li, Z. Kinetic characterization and structural modeling of an NADP(+)-dependent succinic semialdehyde dehydrogenase from Anabaena sp. PCC7120. Int. J. Biol. Macromol. 2018, 108, 615–624. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]

| Proteins | α-KG | ThDP | Mg2+ | ||
|---|---|---|---|---|---|
| KM (mM) | kcat (s−1) | kcat/KM (M−1s−1) | K0.5 (mM) a | K0.5 (mM) b | |
| StKGD [17] | 21 ± 4 | 0.26 ± 0.03 | 12.38 | 0.078 ± 0.028 | Not available |
| StKGD c | 3.83 ± 0.75 | 3.07 ± 0.42 | 0.80 × 103 | 0.063 ± 0.019 | 0.080 ± 0.007 |
| ScKGD [16] | 0.19 ± 0.02 | 1.20 ± 0.32 | 6.32 × 103 | 0.031 ± 0.004 | 0.059 ± 0.005 |
| MaKGD | 0.87 ± 0.23 | 7.80 ± 0.50 | 8.97 × 103 | 0.0056 ± 0.0007 | 0.022 ± 0.003 |
| MiKGD | 4.47 ± 0.63 | 4.33 ± 0.37 | 0.97 × 103 | 0.026 ± 0.006 | 0.037 ± 0.004 |
| Proteins | α-KG | Relative Catalytic Rate | ThDP | |
|---|---|---|---|---|
| KM (mM) | kcat (s−1) | (% of Wild-Type) | K0.5 (mM) | |
| wild-type | 0.87 ± 0.23 | 7.80 ± 0.50 | 100 | 0.0056 ± 0.0007 |
| L467A | 1.22 ± 0.51 | 2.70 ± 0.32 | 35 | 0.026 ± 0.004 |
| Y465A | 1.48 ± 0.27 | 1.23 ± 0.25 | 15 | 0.090 ± 0.021 |
| D435E | 1.74 ± 0.60 | 9.73 ± 0.63 | 120 | 0.041 ± 0.003 |
| D435A | NA | NA | NA | NA |
| D435N | NA | NA | NA | NA |
| D462E | 2.38 ± 0.63 | 9.00 ± 0.47 | 115 | 0.761 ± 0.143 |
| D462N | 3.92 ± 0.45 | 1.50 ± 0.36 | 19 | 0.008 ± 0.001 |
| D462A | NA | NA | NA | NA |
| M410A | 0.53 ± 0.15 | 1.33 ± 0.22 | 17 | 0.046 ± 0.010 |
| E50A | NA | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.-M.; Hu, Z.; Wang, X.; Chen, S.; Yu, W.; Liu, J.; Li, Z. Biochemical and Structural Insights into a Thiamine Diphosphate-Dependent α-Ketoglutarate Decarboxylase from Cyanobacterium Microcystis aeruginosa NIES-843. Int. J. Mol. Sci. 2023, 24, 12198. https://doi.org/10.3390/ijms241512198
Li Z-M, Hu Z, Wang X, Chen S, Yu W, Liu J, Li Z. Biochemical and Structural Insights into a Thiamine Diphosphate-Dependent α-Ketoglutarate Decarboxylase from Cyanobacterium Microcystis aeruginosa NIES-843. International Journal of Molecular Sciences. 2023; 24(15):12198. https://doi.org/10.3390/ijms241512198
Chicago/Turabian StyleLi, Zhi-Min, Ziwei Hu, Xiaoqin Wang, Suhang Chen, Weiyan Yu, Jianping Liu, and Zhimin Li. 2023. "Biochemical and Structural Insights into a Thiamine Diphosphate-Dependent α-Ketoglutarate Decarboxylase from Cyanobacterium Microcystis aeruginosa NIES-843" International Journal of Molecular Sciences 24, no. 15: 12198. https://doi.org/10.3390/ijms241512198
APA StyleLi, Z.-M., Hu, Z., Wang, X., Chen, S., Yu, W., Liu, J., & Li, Z. (2023). Biochemical and Structural Insights into a Thiamine Diphosphate-Dependent α-Ketoglutarate Decarboxylase from Cyanobacterium Microcystis aeruginosa NIES-843. International Journal of Molecular Sciences, 24(15), 12198. https://doi.org/10.3390/ijms241512198






