Nitric Oxide Regulates Seed Germination by Integrating Multiple Signalling Pathways
Abstract
:1. Introduction
2. Main Text
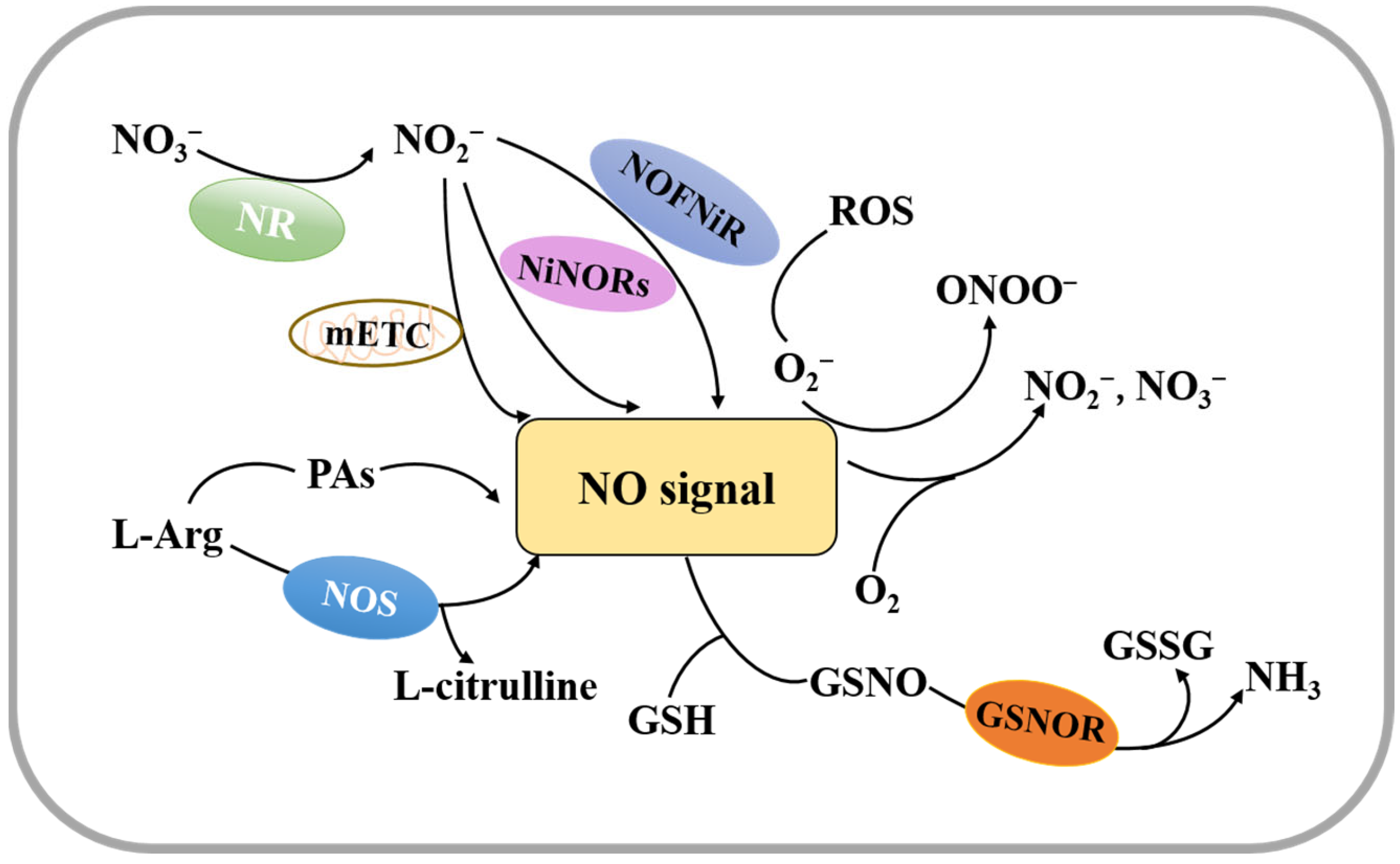
2.1. Synthesis and Decomposition of Nitric Oxide in Plants
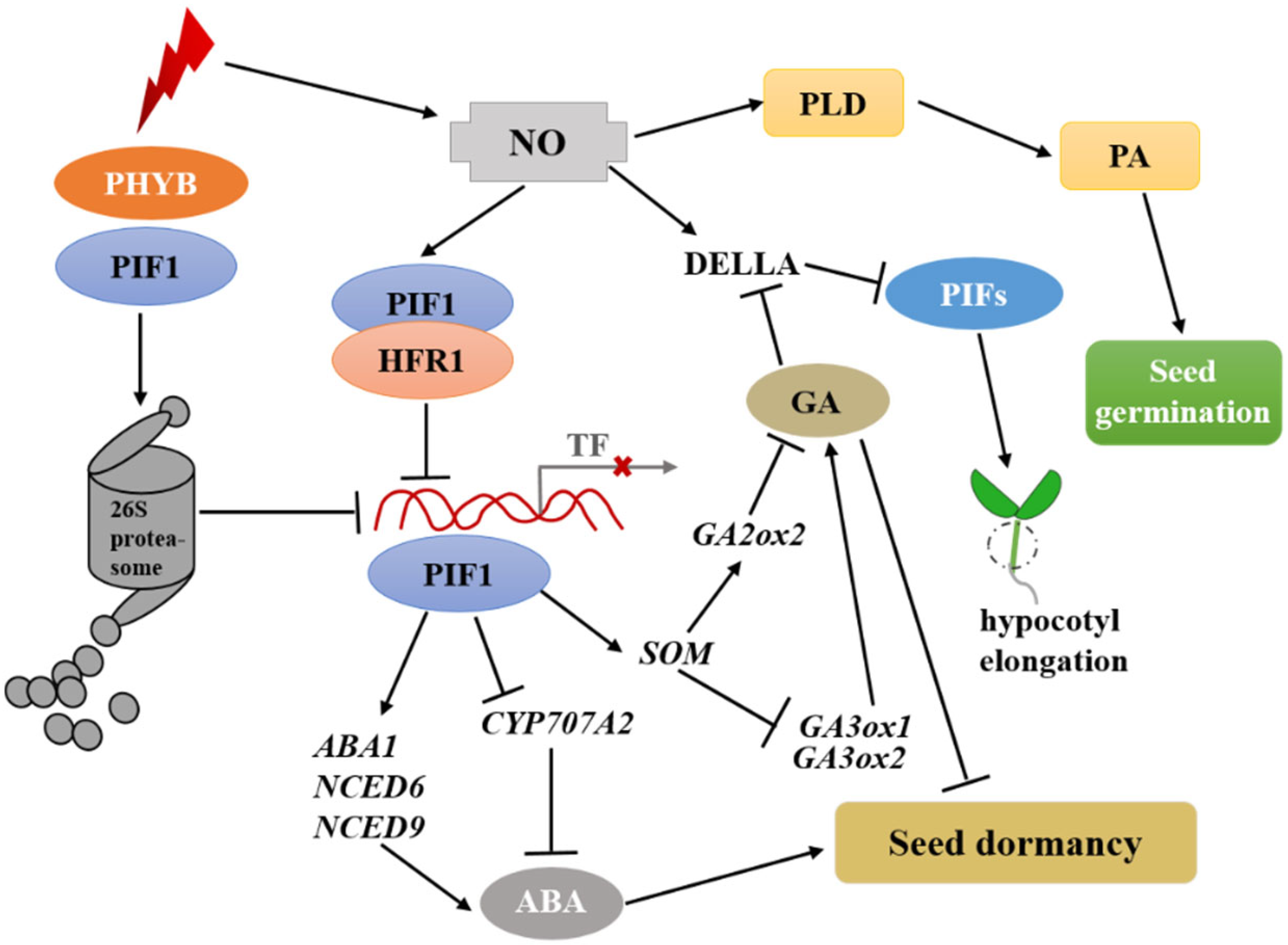
2.2. Nitric Oxide and Phytochrome Signaling Pathways Jointly Regulate Seed Germination under Light
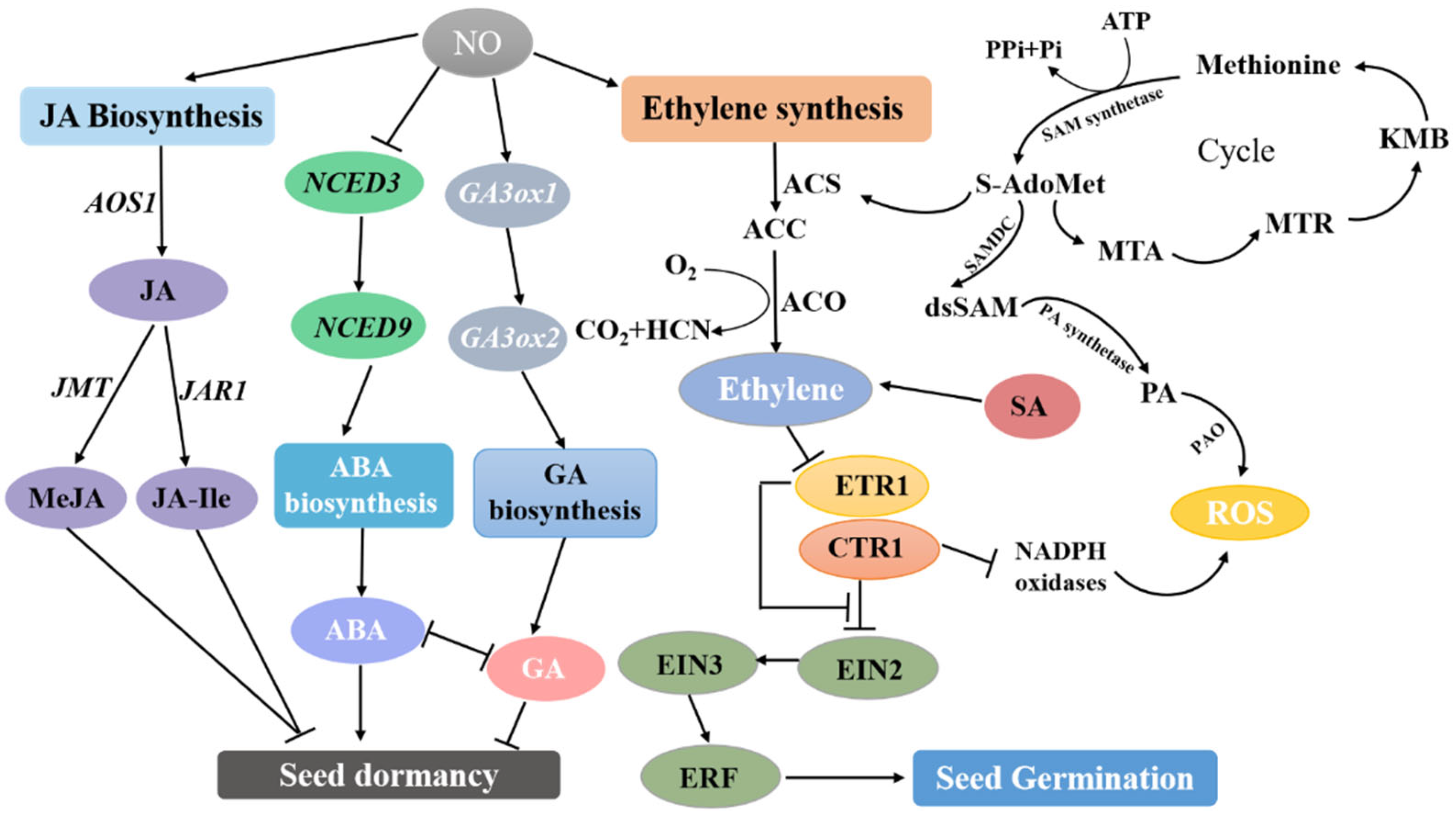
2.3. The Crosstalk between Nitric Oxide and Plant Hormone Signaling Pathways in Seed Dormancy and Germination
2.3.1. Nitric Oxide and Abscisic Acid Signaling Pathways Jointly Regulate Seed Germination
2.3.2. Nitric Oxide and Auxin Signaling Pathways Jointly Regulate Seed Germination
2.3.3. Nitric Oxide and Gibberellin Signaling Pathways Jointly Regulate Seed Germination
2.3.4. Nitric Oxide Regulates Seed Germination Together with Polyamines and Jasmonic Acid Signaling Pathways
2.4. Regulation of Nitric Oxide on Seed Germination under Abiotic Stress
2.4.1. Mechanism of Nitric Oxide Action on Seed Germination under Salt Stress
2.4.2. Mechanism of Nitric Oxide Action on Seed Germination under Drought Stress
2.4.3. Involvement of Nitric Oxide in the Regulation of Seed Germination by Ambient Temperature
| Stress Types | Gene | Species | Function Description | References |
|---|---|---|---|---|
| Salt Stress | OsNOA1 | Oryza sativa | Overexpression of OsNOA1 gene improved plant salt tolerance by reducing Na+/K+ ratio in the mutant. | [104] |
| GmNOS2 GmNR1 | Glycine max | The Na+/K+ ratio was decreased and the balance of ABA, GA and IAA hormones was maintained, improving the salt tolerance index of soybean. | [106] | |
| SlGR | Solanum lycopersicum | Overexpression of SlGR significantly increased the activity of antioxidant enzymes and reduced the oxidative damage of tobacco seeds. | [111] | |
| NRT1.1 | Arabidopsis thaliana | Na+ accumulation was promoted after NO3- treatment, and Cl- accumulation was promoted after NH4+ treatment. | [121,122] | |
| ANR1 | Arabidopsis thaliana | Overexpression of ANR1 produces a salt-sensitive phenotype and inhibited seed germination | [123] | |
| NLP8 | Arabidopsis thaliana | Activate ABA catabolic gene CYP707A2 to promote germination. | [127] | |
| NLP2 | Arabidopsis thaliana | The NLP2-NR pathway activates the expression of ABA catabolic genes ABA8ox1 and ABAox2. | [130] | |
| NLP7 | Arabidopsis thaliana | Increasing the transcription level of ABA biosynthesis gene NCED3 to inhibit seed germination. | [133] | |
| NIA1,2 | Arabidopsis thaliana | It can resist salt stress by promoting EIN3 expression and transcription of downstream ethylene response genes. | [135] | |
| Drought stress | OsPIP1;3 | Oryza sativa | Overexpression of OsPIP1;3 induced germination under water stress conditions. | [22] |
| Drought stress | OtNOS | Arabidopsis thaliana | Overexpression of OtNOS elevated NO accumulation and enhanced seed tolerance to salt stress. | [138] |
| GAP1 | Arabidopsis thaliana | Functionally deficient mutants showed insensitivity to ABA. | [139] | |
| SnRK2.6 | Arabidopsis thaliana | NO inhibits the activity of SNF1-associated protein kinase (SnRK2.6) through GSNO, thereby inhibiting ABA signaling in guard cells. | [75] | |
| PRT6-1 NLP7-1 | Arabidopsis thaliana | The interaction between NLP7 and PRT6 enhanced seed tolerance to ABA. | [146] | |
| Heat stress | GSNOR1 | Arabidopsis thaliana | Overexpression of GSNOR1 increased the nitrosation modification level of ABI5 and induced the degradation of downstream ABI5 protein increasing seed germination | [160] |
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reed, R.C.; Bradford, K.J.; Khanday, I. Seed germination and vigor: Ensuring crop sustainability in a changing climate. Heredity 2022, 128, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2017, 68, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J. Seed germination and dormancy. Plant Cell 1997, 9, 1055. [Google Scholar] [CrossRef]
- Hilhorst, H.W. A critical update on seed dormancy. I. Primary dormancy1. Seed Sci. Res. 1995, 5, 61–73. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New. Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Deng, G.; Sun, H.; Hu, Y.; Yang, Y.; Li, P.; Chen, Y.; Zhu, Y.; Zhou, Y.; Huang, J.; Neill, S.; et al. A Transcription Factor WRKY36 Interacts with AFP2 to Break Primary Seed Dormancy by Progressively Silencing DOG1 in Arabidopsis. New. Phytol. 2023, 238, 688–704. [Google Scholar] [CrossRef]
- Grainge, G.; Nakabayashi, K.; Iza, F.; Leubner-Metzger, G.; Steinbrecher, T. Gas-Plasma-Activated Water Impact on Photo-Dependent Dormancy Mechanisms in Nicotiana tabacum Seeds. Int. J. Mol. Sci. 2022, 23, 6709. [Google Scholar] [CrossRef]
- Née, G.; Xiang, Y.; Soppe, W.J. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 2017, 35, 8–14. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Chen, Z. The control of seed dormancy and germination by temperature, light and nitrate. Bot. Rev. 2020, 86, 39–75. [Google Scholar] [CrossRef]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Augusto, O.; Bonini, M.G.; Amanso, A.M.; Linares, E.; Santos, C.C.; De Menezes, S.L. Nitrogen dioxide and carbonate radical anion: Two emerging radicals in biology. Free Radic. Biol. Med. 2002, 32, 841–859. [Google Scholar] [CrossRef]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijo, J.A. Nitric oxide: A multitasked signaling gas in plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef]
- Saddhe, A.A.; Malvankar, M.R.; Karle, S.B.; Kumar, K. Reactive nitrogen species: Paradigms of cellular signaling and regulation of salt stress in plants. Environ. Exp. Bot. 2019, 161, 86–97. [Google Scholar] [CrossRef]
- Rather, B.A.; Mir, I.R.; Masood, A.; Anjum, N.A.; Khan, N.A. Nitric Oxide Pre-Treatment Advances Seed Germination and Alleviates Copper-Induced Photosynthetic Inhibition in Indian Mustard. Plants 2020, 9, 776. [Google Scholar] [CrossRef]
- Bethke, P.C.; Libourel, I.G.L.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2005, 57, 517–526. [Google Scholar] [CrossRef]
- Krasuska, U.; Ciacka, K.; Orzechowski, S.; Fettke, J.; Bogatek, R.; Gniazdowska, A. Modification of the endogenous NO level influences apple embryos dormancy by alterations of nitrated and biotinylated protein patterns. Planta 2016, 244, 877–891. [Google Scholar] [CrossRef]
- Pandey, S.; Kumari, A.; Shree, M.; Kumar, V.; Singh, P.; Bharadwaj, C.; Loake, G.J.; Parida, S.K.; Masakapalli, S.K.; Gupta, K.J. Nitric oxide accelerates germination via the regulation of respiration in chickpea. J. Exp. Bot. 2019, 70, 4539–4555. [Google Scholar] [CrossRef]
- Liu, H.Y.; Yu, X.; Cui, D.Y.; Sun, M.H.; Sun, W.N.; Tang, Z.C.; Kwak, S.S.; Su, W.A. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 2007, 17, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.X.; Li, X.; Li, C.; Zhao, L. The role of nitric oxide in plant responses to salt stress. Int. J. Mol. Sci. 2022, 23, 6167. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Oliveira, P.J.; Oliveira, H.C.; Kolbert, Z.; Freschi, L. The light and dark sides of nitric oxide: Multifaceted roles of nitric oxide in plant responses to light. J. Exp. Bot. 2021, 72, 885–903. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef]
- León, J.; Costa-Broseta, Á. Present knowledge and controversies, deficiencies, and misconceptions on nitric oxide synthesis, sensing, and signaling in plants. Plant Cell Environ. 2020, 43, 1–15. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New. Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 2008, 59, 21–39. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef]
- Yamasaki, H. Nitrite-dependent nitric oxide production pathway: Implications for involvement of active nitrogen species in photoinhibition in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, Y.; Liu, L.; Liu, X.; Li, B.; Jin, C.; Lin, X. Molecular functions of nitric oxide and its potential applications in horticultural crops. Hort. Res. 2021, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Zemojtel, T.; Fröhlich, A.; Palmieri, M.C.; Kolanczyk, M.; Mikula, I.; Wyrwicz, L.S.; Wanker, E.E.; Mundlos, S.; Vingron, M.; Martasek, P.; et al. Plant nitric oxide synthase: A never-ending story? Trends Plant Sci. 2006, 11, 524–525. [Google Scholar] [CrossRef] [PubMed]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Sandalio, L.M.; Valderrama, R.; Palma, J.; Lupiánez, J.A.; del Rıo, L.A. Localization of nitric-oxide synthase in plant peroxisomes. J. Biol. Chem. 1999, 274, 36729–36733. [Google Scholar] [CrossRef]
- Tun, N.N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.; Scherer, G.F. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef]
- Gupta, K.J.; Stoimenova, M.; Kaiser, W.M. In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 2005, 56, 2601–2609. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; López-Jaramillo, J.; Padilla, M.N.; Carreras, A.; Corpas, F.J.; Barroso, J.B. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 2014, 65, 527–538. [Google Scholar] [CrossRef]
- Gaupels, F.; Spiazzi-Vandelle, E.; Yang, D.; Delledonne, M. Detection of peroxynitrite accumulation in Arabidopsis thaliana during the hypersensitive defense response. Nitric Oxide 2011, 25, 222–228. [Google Scholar] [CrossRef]
- Kwon, E.; Feechan, A.; Yun, B.W.; Hwang, B.H.; Pallas, J.A.; Kang, J.G.; Loake, G.J. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, Z.; Gomez, A.; Liu, B.; Lin, C.; Oka, Y. Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J. Plant Res. 2016, 129, 137–148. [Google Scholar] [CrossRef]
- Tripathi, S.; Hoang, Q.T.; Han, Y.J.; Kim, J.I. Regulation of photomorphogenic development by plant phytochromes. Int. J. Mol. Sci. 2019, 20, 6165. [Google Scholar] [CrossRef] [PubMed]
- Quail, P.H. Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 2002, 14, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Nambara, E.; Choi, G.; Yamaguchi, S. Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 2009, 69, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Lamattina, L. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 2000, 210, 215–221. [Google Scholar] [CrossRef]
- Liu, J.; Xue, T.; Shen, Y.J.H. Effect of Nitric Oxide on Seed Germination and Dormancy in Empress Trees. Hort Technol. 2019, 29, 271–275. [Google Scholar] [CrossRef]
- Batak, I.; Dević, M.; Gibal, Z.; Grubišić, D.; Poff, K.L.; Konjević, R. The effects of potassium nitrate and NO-donors on phytochrome A-and phytochrome B-specific induced germination of Arabidopsis thaliana seeds. Seed Sci. Res. 2002, 12, 253–259. [Google Scholar] [CrossRef]
- Shen, H.; Moon, J.; Huq, E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005, 44, 1023–1035. [Google Scholar] [CrossRef]
- Lozano-Juste, J.; León, J. Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis. Plant Physiol. 2011, 156, 1410–1423. [Google Scholar] [CrossRef]
- Bai, S.; Yao, T.; Li, M.; Guo, X.; Zhang, Y.; Zhu, S.; He, Y. PIF3 is involved in the primary root growth inhibition of Arabidopsis induced by nitric oxide in the light. Mol. Plant 2014, 7, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. Corrigendum to: SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent Seed Germination Downstream of PIL5. Plant Cell 2021, 33, 2093–2095. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhong, S.; Mo, X.; Liu, N.; Nezames, C.D.; Deng, X.W. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 2013, 25, 3770–3784. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jia, Y.; Yu, L.; Yang, W.; Chen, Z.; Chen, H.; Hu, X. Nitric oxide promotes light-initiated seed germination by repressing PIF1 expression and stabilizing HFR1. Plant Physiol. Biochem. 2018, 123, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.V.; Barrero, J.M.; Hughes, T.; Julkowska, M.; Taylor, J.M.; Xu, Q.; Gubler, F. Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 2013, 238, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Piterková, J.; Luhová, L.; Hofman, J.; Turecková, V.; Novák, O.; Petrivalsky, M.; Fellner, M. Nitric oxide is involved in light-specific responses of tomato during germination under normal and osmotic stress conditions. Ann. Bot. 2012, 110, 767–776. [Google Scholar] [CrossRef]
- Wang, X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 2005, 139, 566–573. [Google Scholar] [CrossRef]
- Berridge, M.J. Inositol trisphosphate and calcium signalling. Nature 1993, 361, 315–325. [Google Scholar] [CrossRef]
- An, Z.; Zhou, C.J. Light induces lettuce seed germination through promoting nitric oxide production and phospholipase D-derived phosphatidic acid formation. S. Afr. J. Bot. 2017, 108, 416–422. [Google Scholar] [CrossRef]
- Freschi, L. Nitric oxide and phytohormone interactions: Current status and perspectives. Front. Plant Sci. 2013, 4, 398. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Ye, N.; Liu, R.; Jia, W.; Zhang, J. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009, 183, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.G.; Chen, L.; Zhang, L.L.; Zhang, W.H. Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol. 2009, 151, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Sarath, G.; Hou, G.; Baird, L.M.; Mitchell, R.B. ABA, ROS and NO are Key Players During Switchgrass Seed Germination. Plant Signal. Behav. 2007, 2, 492–493. [Google Scholar] [CrossRef]
- Sarath, G.; Hou, G.; Baird, L.M.; Mitchell, R.B. Reactive oxygen species, ABA and nitric oxide interactions on the germination of warm-season C4-grasses. Planta 2007, 226, 697–708. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, A.; Vandewiele, M.; de Rycke, R.; van Damme, J.; van Montagu, M.; Krebbers, E.; Vandekerckhove, J. Expression and Processing of an Arabidopsis 2S Albumin in Transgenic Tobacco. Plant Physiol. 1990, 92, 899–907. [Google Scholar] [CrossRef]
- Penfield, S.; Rylott, E.L.; Gilday, A.D.; Graham, S.; Larson, T.R.; Graham, I.A. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 2004, 16, 2705–2718. [Google Scholar] [CrossRef]
- Bethke, P.C.; Libourel, I.G.; Aoyama, N.; Chung, Y.Y.; Still, D.W.; Jones, R.L. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007, 143, 1173–1188. [Google Scholar] [CrossRef]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J. Rapid accumulation of NO regulates ABA catabolism and seed dormancy during imbibition in Arabidopsis. Plant Signal. Behav. 2009, 4, 905–907. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, R.; Zhao, M.; Wang, F.; Zhang, N.; Si, H. NO and ABA Interaction Regulates Tuber Dormancy and Sprouting in Potato. Front. Plant Sci. 2020, 11, 311. [Google Scholar] [CrossRef]
- Andryka-Dudek, P.; Ciacka, K.; Wiśniewska, A.; Bogatek, R.; Gniazdowska, A. Nitric Oxide-Induced Dormancy Removal of Apple Embryos Is Linked to Alterations in Expression of Genes Encoding ABA and JA Biosynthetic or Transduction Pathways and RNA Nitration. Int. J. Mol. Sci. 2019, 20, 1007. [Google Scholar] [CrossRef]
- Sun, L.R.; Yue, C.M.; Hao, F.S. Update on roles of nitric oxide in regulating stomatal closure. Plant Signal. Behav. 2019, 14, e1649569. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Considine, M.J. Nitric Oxide Enables Germination by a Four-Pronged Attack on ABA-Induced Seed Dormancy. Front. Plant Sci. 2018, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhu, J.K.; Lang, Z. Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal. Behav. 2015, 10, e1031939. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marín-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Biol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [CrossRef]
- Azhar, B.J.; Zulfiqar, A.; Shakeel, S.N.; Schaller, G.E. Amplification and adaptation in the ethylene signaling pathway. Small Methods 2020, 4, 1900452. [Google Scholar] [CrossRef]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2014, 5, 539. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Ortiz, S.; Garnica-Vergara, A.; Esparza-Reynoso, S.; García-Cárdenas, E.; Raya-González, J.; Francisco Ruiz-Herrera, L.; López-Bucio, J. Jasmonic acid-ethylene crosstalk via ETHYLENE INSENSITIVE 2 reprograms Arabidopsis root system architecture through nitric oxide accumulation. J. Plant Growth Regul. 2018, 37, 438–451. [Google Scholar] [CrossRef]
- Wang, H.Q.; Sun, L.P.; Wang, L.X.; Fang, X.W.; Li, Z.Q.; Zhang, F.F.; Hu, X.; Qi, C.; He, J.M. Ethylene mediates salicylic-acid-induced stomatal closure by controlling reactive oxygen species and nitric oxide production in Arabidopsis. Plant Sci. 2020, 294, 110464. [Google Scholar] [CrossRef] [PubMed]
- Gniazdowska, A.; Dobrzyńska, U.; Babańczyk, T.; Bogatek, R. Breaking the apple embryo dormancy by nitric oxide involves the stimulation of ethylene production. Planta 2007, 225, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Gniazdowska, A.; Krasuska, U.; Czajkowska, K.; Bogatek, R. Nitric oxide, hydrogen cyanide and ethylene are required in the control of germination and undisturbed development of young apple seedlings. Plant Growth Regul. 2010, 61, 75–84. [Google Scholar] [CrossRef]
- Gniazdowska, A.; Krasuska, U.; Bogatek, R. Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 2010, 232, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, S.; Wang, J.; Shen, H.; Yang, L. Interaction between reactive oxygen species and hormones during the breaking of embryo dormancy in Sorbus pohuashanensis by exogenous nitric oxide. J. For. Res. 2022, 33, 435–444. [Google Scholar] [CrossRef]
- El-Maarouf-Bouteau, H.; Sajjad, Y.; Bazin, J.; Langlade, N.; Cristescu, S.M.; Balzergue, S.; Baudouin, E.; Bailly, C. Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ. 2015, 38, 364–374. [Google Scholar] [CrossRef]
- Dixon, K.; Merritt, D.; Flematti, G.; Ghisalberti, E.L. Karrikinolide—A phytoreactive compound derived from smoke with applications in horticulture, ecological restoration and agriculture. Acta Hortic. 2009, 813, 155–170. [Google Scholar] [CrossRef]
- Sami, A.; Riaz, M.W.; Zhou, X.; Zhu, Z.; Zhou, K. Alleviating dormancy in Brassica oleracea seeds using NO and KAR1 with ethylene biosynthetic pathway, ROS and antioxidant enzymes modifications. BMC Plant Biol. 2019, 19, 577. [Google Scholar] [CrossRef]
- Sami, A.; Rehman, S.; Tanvir, M.A.; Zhou, X.Y.; Zhu, Z.H.; Zhou, K. Assessment of the germination potential of Brassica oleracea seeds treated with karrikin 1 and cyanide, which modify the ethylene biosynthetic pathway. J. Plant Growth Regul. 2021, 40, 1257–1269. [Google Scholar] [CrossRef]
- Kępczyński, J. Gas-priming as a novel simple method of seed treatment with ethylene, hydrogen cyanide or nitric oxide. Acta Physiol. Plant. 2021, 43, 117. [Google Scholar] [CrossRef]
- Kara, Z.; Doğan, O.; Vergili, E. Sodium Nitroprusside and Gibberellin Effects on Seed Germination and Seedling Development of Grapevine (Vitis vinifera L.) Cvs. Ekşi Kara and Gök Üzüm. Erwerbs-Obstbau 2020, 62, 61–68. [Google Scholar] [CrossRef]
- Kępczyński, J.; Sznigir, P. Response of Amaranthus retroflexus L. seeds to gibberellic acid, ethylene and abscisic acid depending on duration of stratification and burial. Plant Growth Regul. 2013, 70, 15–26. [Google Scholar] [CrossRef]
- Kępczyński, J.; Sznigir, P. Participation of GA3, ethylene, NO and HCN in germination of Amaranthus retroflexus L. seeds with various dormancy levels. Acta Physiol. Plant. 2014, 36, 1463–1472. [Google Scholar] [CrossRef]
- Kępczyński, J.; Cembrowska-Lech, D.; Sznigir, P. Interplay between nitric oxide, ethylene, and gibberellic acid regulating the release of Amaranthus retroflexus seed dormancy. Acta Physiol. Plant. 2017, 39, 1–13. [Google Scholar] [CrossRef]
- Matilla, A.J. Polyamines and seed germination. Seed Sci. Res. 1996, 6, 81–93. [Google Scholar] [CrossRef]
- Silveira, V.; Santa-Catarina, C.; Tun, N.N.; Scherer, G.F.; Handro, W.; Guerra, M.P.; Floh, E.I. Polyamine effects on the endogenous polyamine contents, nitric oxide release, growth and differentiation of embryogenic suspension cultures of Araucaria angustifolia (Bert.) O. Ktze. Plant Sci. 2006, 171, 91–98. [Google Scholar] [CrossRef]
- Pal Bais, H.; Ravishankar, G.A. Role of polyamines in the ontogeny of plants and their biotechnological applications. Plant Cell, Tissue Organ Cult. 2002, 69, 1–34. [Google Scholar] [CrossRef]
- Cona, A.; Cenci, F.; Cervelli, M.; Federico, R.; Mariottini, P.; Moreno, S.; Angelini, R. Polyamine oxidase, a hydrogen peroxide-producing enzyme, is up-regulated by light and down-regulated by auxin in the outer tissues of the maize mesocotyl. Plant Physiol. 2003, 131, 803–813. [Google Scholar] [CrossRef]
- Krasuska, U.; Ciacka, K.; Bogatek, R.; Gniazdowska, A.J. Polyamines and Nitric Oxide Link in Regulation of Dormancy Removal and Germination of Apple (Malus domestica Borkh.) Embryos. J. Plant Growth Regul. 2014, 33, 590–601. [Google Scholar] [CrossRef]
- Krasuska, U.; Ciacka, K.; Gniazdowska, A. Nitric oxide-polyamines cross-talk during dormancy release and germination of apple embryos. Nitric Oxide 2017, 68, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.Q.; Okamoto, M.; Crawford, N.M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 2003, 302, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Xiao, S.; Yu, L.; Fan, L.M. Expression of a rice gene OsNOA1 re-establishes nitric oxide synthesis and stress-related gene expression for salt tolerance in Arabidopsis nitric oxide-associated 1 mutant Atnoa1. Environ. Exp. Bot. 2009, 65, 90–98. [Google Scholar] [CrossRef]
- Patel, P.; Kadur Narayanaswamy, G.; Kataria, S.; Baghel, L. Involvement of nitric oxide in enhanced germination and seedling growth of magnetoprimed maize seeds. Plant Signal. Behav. 2017, 12, e1293217. [Google Scholar] [CrossRef] [PubMed]
- Kataria, S.; Anand, A.; Raipuria, R.K.; Kumar, S.; Jain, M.; Watts, A.; Brestic, M. Magnetopriming Actuates Nitric Oxide Synthesis to Regulate Phytohormones for Improving Germination of Soybean Seeds under Salt Stress. Cells 2022, 11, 2174. [Google Scholar] [CrossRef]
- Shi, Q.; Ding, F.; Wang, X.; Wei, M. Exogenous nitric oxide protect cucumber roots against oxidative stress induced by salt stress. Plant Physiol. Biochem. 2007, 45, 542–550. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Al-Whaibi, M.H.; Basalah, M.O. Role of nitric oxide in tolerance of plants to abiotic stress. Protoplasma 2011, 248, 447–455. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, W.; He, J.; Zhang, L.; Wei, Y.; Yang, M. Nitric oxide alleviates salt stress in seed germination and early seedling growth of pakchoi (Brassica chinensis L.) by enhancing physiological and biochemical parameters. Ecotoxicol. Environ. Saf. 2020, 187, 109785. [Google Scholar] [CrossRef]
- Zhai, J.; Liang, Y.; Zeng, S.; Yan, J.; Li, K.; Xu, H. Overexpression of tomato glutathione reductase (SlGR) in transgenic tobacco enhances salt tolerance involving the S-nitrosylation of GR. Plant Physiol. Biochem. 2023, 196, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.L.D.; Pinheiro, D.T.; Borges, E.E.D.L.; Silva, L.J.D.; Dias, D.C.F.D.S. Effect of cyanide by sodium nitroprusside (SNP) application on germination, antioxidative system and lipid peroxidation of Senna macranthera seeds under saline stress. J. Seed Sci. 2019, 41, 086–096. [Google Scholar] [CrossRef]
- Hajihashemi, S.; Skalicky, M.; Brestic, M.; Pavla, V. Cross-talk between nitric oxide, hydrogen peroxide and calcium in salt-stressed Chenopodium quinoa Willd. At seed germination stage. Plant Physiol. Biochem. 2020, 154, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Ikeya, S.; Aoyanagi, T.; Ishizuka, I.; Takeuchi, A.; Kozaki, A. Nitrate Promotes Germination Under Inhibition by NaCl or High Concentration of Glucose. Plants 2020, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, F.; Naeem, M.; Akram, A.; Ashraf, M.Y.; Ahmad, K.S.; Zulfiqar, B.; Sardar, H.; Shabbir, R.N.; Majeed, S.; Shehzad, M.A.; et al. Seed priming with KNO3 mediates biochemical processes to inhibit lead toxicity in maize (Zea mays L.). J. Sci. Food Agric. 2017, 97, 4780–4789. [Google Scholar] [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef]
- Fang, X.Z.; Fang, S.Q.; Ye, Z.Q.; Liu, D.; Zhao, K.L.; Jin, C.W. NRT1.1 Dual-Affinity Nitrate Transport/Signalling and its Roles in Plant Abiotic Stress Resistance. Front. Plant Sci. 2021, 12, 715694. [Google Scholar] [CrossRef]
- Álvarez-Aragón, R.; Haro, R.; Benito, B.; Rodríguez-Navarro, A. Salt intolerance in Arabidopsis: Shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 2016, 243, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.X.; Zhu, Y.X.; Fang, X.Z.; Ye, J.Y.; Du, W.X.; Zhu, Q.Y.; Lin, X.Y.; Jin, C.W. Ammonium aggravates salt stress in plants by entrapping them in a chloride over-accumulation state in an NRT1.1-dependent manner. Sci. Total Environ. 2020, 746, 141244. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Yu, L.H.; Xiang, C.B. ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression. New Phytol. 2020, 225, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Konishi, M.; Okitsu, T.; Yanagisawa, S. Nitrate-responsive NIN-like protein transcription factors perform unique and redundant roles in Arabidopsis. J. Exp. Bot. 2021, 72, 5735–5750. [Google Scholar] [CrossRef]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef]
- Gupta, K.J.; Fernie, A.R.; Kaiser, W.M.; van Dongen, J.T. On the origins of nitric oxide. Trends Plant Sci. 2011, 16, 160–168. [Google Scholar] [CrossRef]
- Reda, M.; Golicka, A.; Kabała, K.; Janicka, M. Involvement of NR and PM-NR in NO biosynthesis in cucumber plants subjected to salt stress. Plant Sci. 2018, 267, 55–64. [Google Scholar] [CrossRef]
- Yi, Y.; Peng, Y.; Song, T.; Lu, S.; Teng, Z.; Zheng, Q.; Zhao, F.; Meng, S.; Liu, B.; Peng, Y.; et al. NLP2-NR Module Associated NO Is Involved in Regulating Seed Germination in Rice under Salt Stress. Plants 2022, 11, 795. [Google Scholar] [CrossRef]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Ripoll, J.J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.T.; Lee, W.J.; Choi, J.H.; Nguyen, D.T.; Truong, H.A.; Lee, S.A.; Hong, S.W.; Lee, H. The Loss of Function of the NODULE INCEPTION-Like PROTEIN 7 Enhances Salt Stress Tolerance in Arabidopsis Seedlings. Front. Plant Sci. 2021, 12, 743832. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yang, L.; Paul, M.; Zu, Y.; Tang, Z. Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: Evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol. Biochem. 2013, 73, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Y.; Chang, B.; Wang, Y.; Tang, Z. NO Promotes Seed Germination and Seedling Growth Under High Salt May Depend on EIN3 Protein in Arabidopsis. Front. Plant Sci. 2015, 6, 1203. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, C.; Wang, X.; Shen, H.; Yang, L. Exogenous Ethylene Alleviates the Inhibition of Sorbus pohuashanensis Embryo Germination in a Saline-Alkali Environment (NaHCO3). Int. J. Mol. Sci. 2023, 24, 4244. [Google Scholar] [CrossRef]
- Garcia-Mata, C.; Lamattina, L. Abscisic acid (ABA) inhibits light-induced stomatal opening through calcium- and nitric oxide-mediated signaling pathways. Nitric Oxide 2007, 17, 143–151. [Google Scholar] [CrossRef]
- Foresi, N.; Mayta, M.L.; Lodeyro, A.F.; Scuffi, D.; Correa-Aragunde, N.; García-Mata, C.; Casalongué, C.; Carrillo, N.; Lamattina, L. Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis. Plant J. 2015, 82, 891–903. [Google Scholar] [CrossRef]
- Albertos, P.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Fernández-Arbaizar, A.; Nakabayashi, K.; Nambara, E.; Godoy, M.; Franco, J.M.; Solano, R.; et al. Redox feedback regulation of ANAC089 signaling alters seed germination and stress response. Cell Rep. 2021, 35, 109263. [Google Scholar] [CrossRef]
- Malcheska, F.; Ahmad, A.; Batool, S.; Müller, H.M.; Ludwig-Müller, J.; Kreuzwieser, J.; Randewig, D.; Hänsch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef]
- Lozano-Juste, J.; León, J. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 2010, 152, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Griffiths, R.; Hancock, J.; Neill, S. A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2002, 99, 16314–16318. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, D.M.; Desikan, R.; Bright, J.; Confraria, A.; Harrison, J.; Hancock, J.T.; Barros, R.S.; Neill, S.J.; Wilson, I.D. Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ. 2009, 32, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lau, E.; Lam, M.P.Y.; Chu, H.; Li, S.; Huang, G.; Guo, P.; Wang, J.; Jiang, L.; Chu, I.K.; et al. OsNOA1/RIF1 is a functional homolog of AtNOA1/RIF1: Implication for a highly conserved plant cGTPase essential for chloroplast function. New Phytol. 2010, 187, 83–105. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef]
- Castillo, M.C.; Costa-Broseta, Á.; Gayubas, B.; León, J. NIN-like protein7 and PROTEOLYSIS6 functional interaction enhances tolerance to sucrose, ABA, and submergence. Plant Physiol. 2021, 187, 2731–2748. [Google Scholar] [CrossRef]
- Krasuska, U.; Gniazdowska, A. Nitric oxide and hydrogen cyanide as regulating factors of enzymatic antioxidant system in germinating apple embryos. Acta Physiol. Plant. 2012, 34, 683–692. [Google Scholar] [CrossRef]
- Oracz, K.; El-Maarouf-Bouteau, H.; Kranner, I.; Bogatek, R.; Corbineau, F.; Bailly, C. The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiol. 2009, 150, 494–505. [Google Scholar] [CrossRef]
- Dębska, K.; Krasuska, U.; Budnicka, K.; Bogatek, R.; Gniazdowska, A. Dormancy removal of apple seeds by cold stratification is associated with fluctuation in H2O2, NO production and protein carbonylation level. J. Plant Physiol. 2013, 170, 480–488. [Google Scholar] [CrossRef]
- Ciacka, K.; Tyminski, M.; Gniazdowska, A.; Krasuska, U. Cold stratification-induced dormancy removal in apple (Malus domestica Borkh.) seeds is accompanied by an increased glutathione pool in embryonic axes. J. Plant Physiol. 2022, 274, 153736. [Google Scholar] [CrossRef]
- Broniowska, K.A.; Diers, A.R.; Hogg, N. S-nitrosoglutathione. Biochim. Biophys. Acta 2013, 1830, 3173–3181. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Klessig, D.F. Nitric oxide as a signal in plants. Curr. Opin. Plant Biol. 1999, 2, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Jensen, D.E.; Belka, G.K.; Du Bois, G.C. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem. J. 1998, 331, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Ciacka, K.; Krasuska, U.; Otulak-Kozieł, K.; Gniazdowska, A. Dormancy removal by cold stratification increases glutathione and S-nitrosoglutathione content in apple seeds. Plant Physiol. Biochem. 2019, 138, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.H.; Sami, A.; Xu, Q.Q.; Wu, L.L.; Zheng, W.Y.; Chen, Z.P.; Jin, X.Z.; Zhang, H.; Li, Y.; Yu, Y.; et al. Effects of seed priming treatments on the germination and development of two rapeseed (Brassica napus L.) varieties under the co-influence of low temperature and drought. PLoS ONE 2021, 16, e0257236. [Google Scholar] [CrossRef]
- Amooaghaie, R.; Nikzad, K. The role of nitric oxide in priming-induced low-temperature tolerance in two genotypes of tomato. Seed Sci. Res. 2013, 23, 123–131. [Google Scholar] [CrossRef]
- Bian, L.; Yang, L.; Wang, J.A.; Shen, H.L. Effects of KNO3 pretreatment and temperature on seed germination of Sorbus pohuashanensis. J. For. Res. 2013, 24, 309–316. [Google Scholar] [CrossRef]
- Majláth, I.; Éva, C.; Hamow, K.Á.; Kun, J.; Pál, M.; Rahman, A.; Palla, B.; Nagy, Z.; Gyenesei, A.; Szalai, G.; et al. Methylglyoxal induces stress signaling and promotes the germination of maize at low temperature. Physiol. Plant. 2022, 174, e13609. [Google Scholar] [CrossRef]
- Sita, K.; Sehgal, A.; Bhardwaj, A.; Bhandari, K.; Kumar, S.; Prasad, P.V.; Jha, U.; Siddique, K.H.M.; Nayyar, H. Nitric oxide secures reproductive efficiency in heat-stressed lentil (Lens culinaris Medik.) plants by enhancing the photosynthetic ability to improve yield traits. Physiol. Mol. Biol. Plants 2021, 27, 2549–2566. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Yang, W.; Li, X.; Wei, J.; Hu, X.; Li, P. S-Nitrosoglutathion reductase activity modulates the thermotolerance of seeds germination by controlling ABI5 stability under high temperature. Phyton-Int. J. Exp. Bot. 2021, 90, 1075. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, R.; Wang, X.; Zhao, C.; Shen, H.; Yang, L. Nitric Oxide Regulates Seed Germination by Integrating Multiple Signalling Pathways. Int. J. Mol. Sci. 2023, 24, 9052. https://doi.org/10.3390/ijms24109052
Zhang Y, Wang R, Wang X, Zhao C, Shen H, Yang L. Nitric Oxide Regulates Seed Germination by Integrating Multiple Signalling Pathways. International Journal of Molecular Sciences. 2023; 24(10):9052. https://doi.org/10.3390/ijms24109052
Chicago/Turabian StyleZhang, Yue, Ruirui Wang, Xiaodong Wang, Caihong Zhao, Hailong Shen, and Ling Yang. 2023. "Nitric Oxide Regulates Seed Germination by Integrating Multiple Signalling Pathways" International Journal of Molecular Sciences 24, no. 10: 9052. https://doi.org/10.3390/ijms24109052





