Cannabidiol Modulates Alterations in PFC microRNAs in a Rat Model of Depression
Abstract
:1. Introduction
2. Results
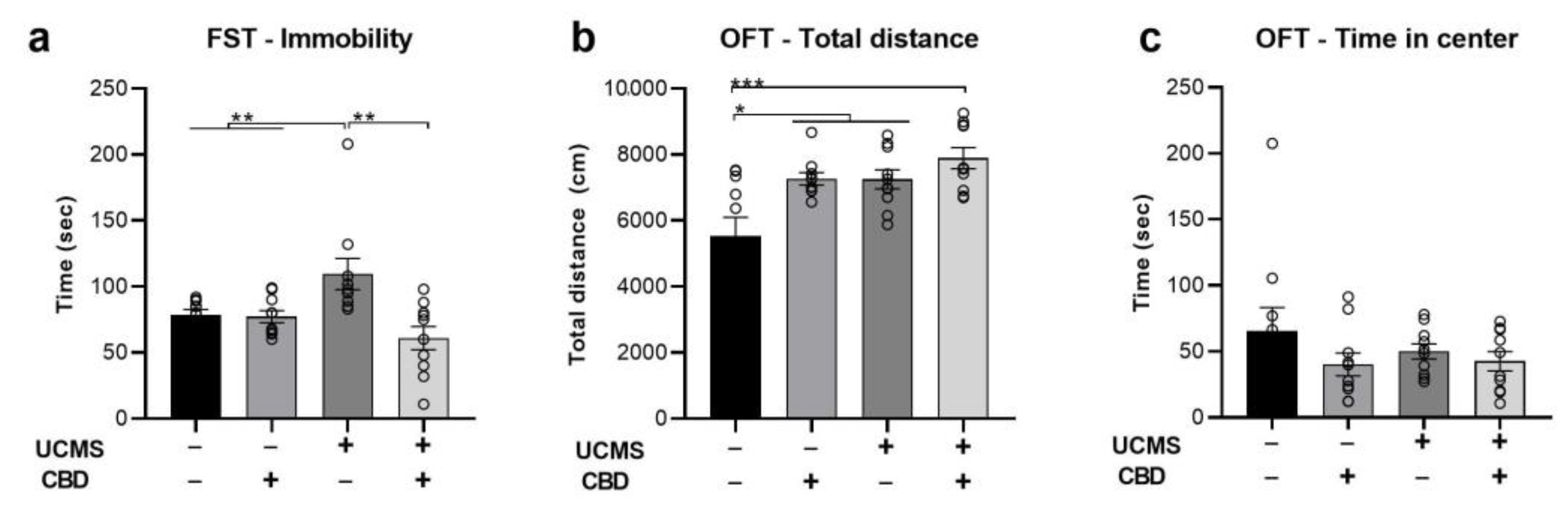
2.1. The Effects of Chronic CBD Administration during UCMS on Behavior
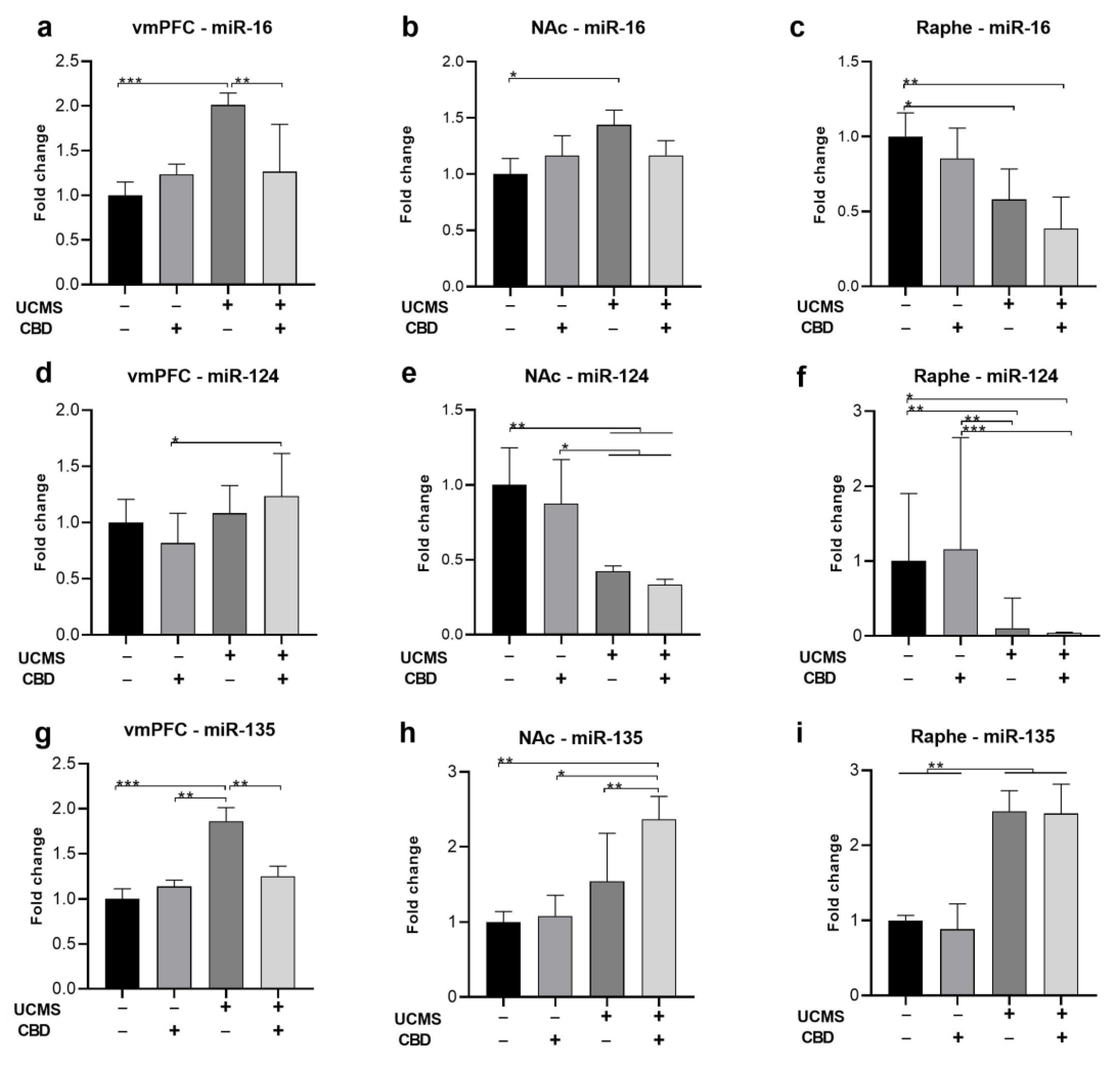
2.2. The Effects of Chronic CBD Administration during UCMS on miRNA Expression
2.2.1. miR-16
2.2.2. miR-124
2.2.3. miR-135
2.3. The Effects of Chronic CBD Administration during UCMS on Possible Target Genes
2.3.1. htr1a
2.3.2. slc6a4
2.3.3. ctnnb1
2.3.4. cnr1
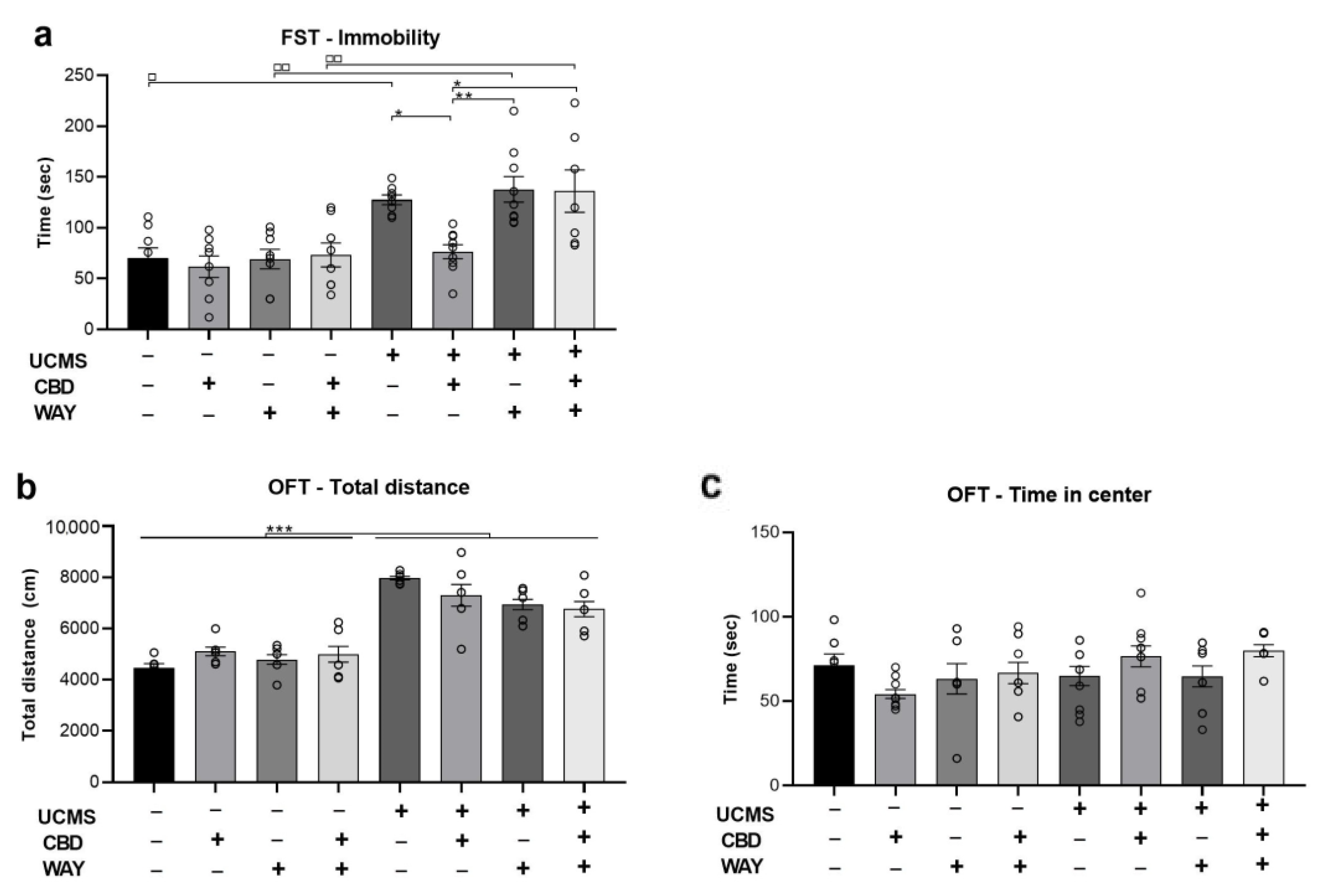
2.4. Does the 5-HT1a Antagonist WAY100635 Block the Effects of Chronic CBD Administration during UCMS on Behavior
3. Discussion
3.1. Alterations in miRNAs
3.1.1. miR-16
3.1.2. miR-124
3.1.3. miR-135
3.2. Alterations in Serotonergic Targets, β-catenin, and CB1
3.2.1. htr1a (5HT1a Gene)
3.2.2. slc6a4 (SERT Gene)
3.2.3. ctnnb1 (β-catenin Gene)
3.2.4. cnr1 (CB1 Gene)
4. Materials and Methods
4.1. Subjects
4.2. UCMS Protocol
4.3. Pharmacological Agents
4.4. Behavioral Tests
4.4.1. Locomotor Activity and Anxiety-like Behavior
4.4.2. Forced Swim Test (FST)
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maletic, V.; Robinson, M.; Oakes, T.; Iyengar, S.; Ball, S.G.; Russell, J. Neurobiology of depression: An integrated view of key findings. Int. J. Clin. Pract. 2007, 61, 2030–2040. [Google Scholar] [CrossRef] [Green Version]
- Pigott, H.E.; Leventhal, A.M.; Alter, G.S.; Boren, J.J. Efficacy and effectiveness of antidepressants: Current status of research. Psychother. Psychosom. 2010, 79, 267–279. [Google Scholar] [CrossRef]
- Shbiro, L.; Hen-Shoval, D.; Hazut, N.; Rapps, K.; Dar, S.; Zalsman, G.; Mechoulam, R.; Weller, A.; Shoval, G. Effects of cannabidiol in males and females in two different rat models of depression. Physiol. Behav. 2019, 201, 59–63. [Google Scholar] [CrossRef]
- Zanelati, T.V.; Biojone, C.; Moreira, F.A.; Guimarães, F.S.; Joca, S.R.L. Antidepressant-like effects of cannabidiol in mice: Possible involvement of 5-HT1A receptors. Br. J. Pharmacol. 2010, 159, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Réus, G.Z.; Stringari, R.B.; Ribeiro, K.F.; Luft, T.; Abelaira, H.M.; Fries, G.R.; Aguiar, B.W.; Kapczinski, F.; Hallak, J.E.; Zuardi, A.W.; et al. Administration of cannabidiol and imipramine induces antidepressant-like effects in the forced swimming test and increases brain-derived neurotrophic factor levels in the rat amygdala. Acta Neuropsychiatr. 2011, 23, 241–248. [Google Scholar] [CrossRef]
- Shoval, G.; Shbiro, L.; Hershkovitz, L.; Hazut, N.; Zalsman, G.; Mechoulam, R.; Weller, A. Prohedonic effect of cannabidiol in a rat model of depression. Neuropsychobiology 2016, 73, 123–129. [Google Scholar] [CrossRef]
- Linge, R.; Jiménez-Sánchez, L.; Campa, L.; Pilar-Cuéllar, F.; Vidal, R.; Pazos, A.; Adell, A.; Díaz, Á. Cannabidiol induces rapid-acting antidepressant-like effects and enhances cortical 5-HT/glutamate neurotransmission: Role of 5-HT1A receptors. Neuropharmacology 2016, 103, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Ferber, S.G.; Hazani, R.; Shoval, G.; Weller, A. Targeting the Endocannabinoid System in Borderline Personality Disorder: Corticolimbic and Hypothalamic Perspectives. Curr. Neuropharmacol. 2021, 19, 360–371. [Google Scholar]
- Zhornitsky, S.; Potvin, S. Cannabidiol in humans—The quest for therapeutic targets. Pharmaceuticals 2012, 5, 529–552. [Google Scholar] [CrossRef]
- McPartland, J.M.; Glass, M.; Pertwee, R.G. Meta-analysis of cannabinoid ligand binding affinity and receptor distribution: Interspecies differences. Br. J. Pharmacol. 2007, 152, 583–593. [Google Scholar] [CrossRef] [Green Version]
- Campos, A.C.; Moreira, F.A.; Gomes, F.V.; Del Bel, E.A.; Guimaraes, F.S. Multiple mechanisms involved in the large-spectrum therapeutic potential of cannabidiol in psychiatric disorders. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 3364–3378. [Google Scholar] [CrossRef]
- Resstel, L.B.; Tavares, R.F.; Lisboa, S.F.; Joca, S.R.; Corrêa, F.M.; Guimarães, F.S. 5-HT1A receptors are involved in the cannabidiol-induced attenuation of behavioural and cardiovascular responses to acute restraint stress in rats. Br. J. Pharmacol. 2009, 156, 181–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartim, A.G.; Guimarães, F.S.; Joca, S.R.L. Antidepressant-like effect of cannabidiol injection into the ventral medial prefrontal cortex—Possible involvement of 5-HT1A and CB1 receptors. Behav. Brain Res. 2016, 303, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Fogaça, M.V.; Campos, A.C.; Guimarães, F.S. Cannabidiol and 5-HT1A receptors. In Neuropathology of Drug Addictions and Substance Misuse; Academic Press: Cambridge, MA, USA, 2016; pp. 749–759. [Google Scholar]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1977, 16, 3797–3804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, C.H.; Soga, T.; Parhar, I.S. Brain beta-catenin signalling during stress and depression. Neurosignals 2018, 26, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Mizrachi Zer-Aviv, T.; Islami, L.; Hamilton, P.J.; Parise, E.M.; Nestler, E.J.; Sbarski, B.; Akirav, I. Enhancing Endocannabinoid Signaling via β-Catenin in the Nucleus Accumbens Attenuates PTSD-and Depression-like Behavior of Male Rats. Biomedicines 2022, 10, 1789. [Google Scholar] [CrossRef]
- Dias, C.; Feng, J.; Sun, H.; Mazei-Robison, M.S.; Damez-Werno, D.; Scobie, K.; Bagot, R.; LaBonté, B.; Ribeiro, E.; Liu, X.; et al. β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 2014, 516, 51–55. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.F.; Obrietan, K. MicroRNA as therapeutic targets for treatment of depression. Neuropsychiatr. Dis. Treat. 2013, 9, 1011. [Google Scholar]
- O’connor, R.M.; Dinan, T.G.; Cryan, J.F. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol. Psychiatry 2012, 17, 359–376. [Google Scholar] [CrossRef] [Green Version]
- Serafini, G.; Pompili, M.; Hansen, K.F.; Obrietan, K.; Dwivedi, Y.; Shomron, N.; Girardi, P. The involvement of microRNAs in major depression, suicidal behavior, and related disorders: A focus on miR-185 and miR-491-3p. Cell. Mol. Neurobiol. 2014, 34, 17–30. [Google Scholar] [CrossRef]
- Ferrúa, C.P.; Giorgi, R.; da Rosa, L.C.; do Amaral, C.C.; Ghisleni, G.C.; Pinheiro, R.T.; Nedel, F. MicroRNAs expressed in depression and their associated pathways: A systematic review and a bioinformatics analysis. J. Chem. Neuroanat. 2019, 100, 101650. [Google Scholar] [CrossRef]
- Allen, L.; Dwivedi, Y. MicroRNA mediators of early life stress vulnerability to depression and suicidal behavior. Mol. Psychiatry 2020, 25, 308–320. [Google Scholar] [CrossRef]
- Muiños-Gimeno, M.; Espinosa-Parrilla, Y.; Guidi, M.; Kagerbauer, B.; Sipilä, T.; Maron, E.; Pettai, K.; Kananen, L.; Navinés, R.; Martín-Santos, R.; et al. Human microRNAs miR-22, miR-138-2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol. Psychiatry 2011, 69, 526–533. [Google Scholar] [CrossRef]
- Haramati, S.; Navon, I.; Issler, O.; Ezra-Nevo, G.; Gil, S.; Zwang, R.; Hornstein, E.; Chen, A. MicroRNA as repressors of stress-induced anxiety: The case of amygdalar miR-34. J. Neurosci. 2011, 31, 14191–14203. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.; Zhu, X.; Zhang, Y.; Zhang, S.; Zhang, L.; Xue, L.; Yi, J.; Yao, S.; Zhang, X. Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS ONE 2012, e46921. [Google Scholar] [CrossRef]
- Issler, O.; Haramati, S.; Paul, E.D.; Maeno, H.; Navon, I.; Zwang, R.; Gil, S.; Mayberg, H.S.; Dunlop, B.W.; Menke, A.; et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 2014, 83, 344–360. [Google Scholar] [CrossRef] [Green Version]
- Higuchi, F.; Uchida, S.; Yamagata, H.; Abe-Higuchi, N.; Hobara, T.; Hara, K.; Kobayashi, A.; Shintaku, T.; Itoh, Y.; Suzuki, T.; et al. Hippocampal microRNA-124 enhances chronic stress resilience in mice. J. Neurosci. 2016, 36, 7253–7267. [Google Scholar] [CrossRef] [Green Version]
- Volk, N.; Pape, J.C.; Engel, M.; Zannas, A.S.; Cattane, N.; Cattaneo, A.; Binder, E.B.; Chen, A. Amygdalar microRNA-15a is essential for coping with chronic stress. Cell Rep. 2016, 17, 1882–1891. [Google Scholar] [CrossRef] [Green Version]
- Maurel, O.M.; Torrisi, S.A.; Barbagallo, C.; Purrello, M.; Salomone, S.; Drago, F.; Leggio, G.M. Dysregulation of miR-15a-5p, miR-497a-5p and miR-511-5p is associated with modulation of BDNF and FKBP5 in brain areas of PTSD-related susceptible and resilient mice. Int. J. Mol. Sci. 2021, 22, 5157. [Google Scholar] [CrossRef]
- Song, M.F.; Dong, J.Z.; Wang, Y.W.; He, J.; Ju, X.; Zhang, L.; Zhang, Y.H.; Shi, J.F.; Lv, Y.Y. CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J. Affect. Disord. 2015, 178, 25–31. [Google Scholar] [CrossRef]
- Baudry, A.; Mouillet-Richard, S.; Schneider, B.; Launay, J.M.; Kellermann, O. miR-16 targets the serotonin transporter: A new facet for adaptive responses to antidepressants. Science 2010, 329, 1537–1541. [Google Scholar] [CrossRef] [Green Version]
- Portugalov, A.; Zaidan, H.; Gaisler-Salomon, I.; Hillard, C.J.; Akirav, I. FAAH inhibition restores early life stress-induced alterations in PFC microRNAs associated with depressive-like behavior in male and female rats. IJMS 2022, 23, 16101. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, D.; Xu, J.; Jiang, H.; Pan, F. Early adolescent stress-induced changes in prefrontal cortex miRNA-135a and hippocampal miRNA-16 in male rats. Dev. Psychobiol. 2017, 59, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Adlakha, Y.K.; Saini, N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol. Cancer 2014, 13, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Q.; Xia, W.; Liu, J.C.; Yang, J.Y.; Lee, D.F.; Xia, J.; Bartholomeusz, G.; Li, Y.; Pan, Y.; Li, Z.; et al. Erk associates with and primes GSK-3β for its inactivation resulting in upregulation of β-catenin. Mol. Cell 2005, 19, 159–170. [Google Scholar] [CrossRef]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Bower, K.A.; Chen, G.; Shi, X.; Ke, Z.J.; Luo, J. Interaction between ERK and GSK3β mediates basic fibroblast growth factor-induced apoptosis in SK-N-MC neuroblastoma cells. J. Biol. Chem. 2008, 283, 9248–9256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewernick, B.H.; Hurlemann, R.; Matusch, A.; Kayser, S.; Grubert, C.; Hadrysiewicz, B.; Axmacher, N.; Lemke, M.; Cooper-Mahkorn, D.; Cohen, M.X.; et al. Nucleus accumbens deep brain stimulation decreases ratings of depression and anxiety in treatment-resistant depression. Biol. Psychiatry 2010, 67, 110–116. [Google Scholar] [CrossRef]
- Campbell, S.; MacQueen, G. The role of the hippocampus in the pathophysiology of major depression. J. Psychiatry Neurosci. 2004, 29, 417–426. [Google Scholar]
- Koenigs, M.; Huey, E.D.; Calamia, M.; Raymont, V.; Tranel, D.; Grafman, J. Distinct regions of prefrontal cortex mediate resistance and vulnerability to depression. J. Neurosci. 2008, 28, 12341–12348. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Shi, R.; Wang, J.; Wang, J.F.; Li, X.M. Unpredictable chronic mild stress not chronic restraint stress induces depressive behaviours in mice. Neuroreport 2014, 25, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.D.; An, S.C.; Zhang, X. Involvement of hippocampal serotonin and neuropeptide Y in depression induced by chronic unpredicted mild stress. Brain Res. Bull. 2008, 77, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, M.K.; Bishnoi, M.; Kulkarni, S.K. Anti-depressant like effect of curcumin and its combination with piperine in unpredictable chronic stress-induced behavioral, biochemical and neurochemical changes. Pharmacol. Biochem. Behav. 2009, 92, 39–43. [Google Scholar] [CrossRef]
- Bangasser, D.A.; Cuarenta, A. Sex differences in anxiety and depression: Circuits and mechanisms. Nat. Rev. Neurosci. 2021, 22, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.S.; Gobinath, A.R.; Galea, L.A. Sex differences in depression: Insights from clinical and preclinical studies. Prog. Neurobiol. 2019, 176, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Zurawek, D.; Kusmider, M.; Faron-Gorecka, A.; Gruca, P.; Pabian, P.; Kolasa, M.; Solich, J.; Szafran-Pilch, K.; Papp, M.; Dziedzicka-Wasylewska, M. Time-dependent miR-16 serum fluctuations together with reciprocal changes in the expression level of miR-16 in mesocortical circuit contribute to stress resilient phenotype in chronic mild stress–an animal model of depression. Eur. Neuropsychopharmacol. 2016, 26, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Gheysarzadeh, A.; Sadeghifard, N.; Afraidooni, L.; Pooyan, F.; Mofid, M.R.; Valadbeigi, H.; Bakhtiari, H.; Keikhavani, S. Serum-based microRNA biomarkers for major depression: MiR-16, miR-135a, and miR-1202. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2018, 23, 39. [Google Scholar] [CrossRef]
- Dwivedi, Y.; Roy, B.; Lugli, G.; Rizavi, H.; Zhang, H.; Smalheiser, N.R. Chronic corticosterone-mediated dysregulation of microRNA network in prefrontal cortex of rats: Relevance to depression pathophysiology. Transl. Psychiatry 2015, 5, e682. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Pan, J.; Chen, L. MiR-124 suppression in the prefrontal cortex reduces depression-like behavior in mice. Biosci. Rep. 2019, 39, BSR20190186. [Google Scholar] [CrossRef] [Green Version]
- Lou, D.; Wang, J.; Wang, X. miR-124 ameliorates depressive-like behavior by targeting STAT3 to regulate microglial activation. Mol. Cell. Probes 2019, 48, 101470. [Google Scholar] [CrossRef]
- Periyasamy, P.; Liao, K.; Kook, Y.H.; Niu, F.; Callen, S.E.; Guo, M.L.; Buch, S. Cocaine-mediated downregulation of miR-124 activates microglia by targeting KLF4 and TLR4 signaling. Mol. Neurobiol. 2018, 55, 3196–3210. [Google Scholar] [CrossRef]
- Cabana-Domínguez, J.; Arenas, C.; Cormand, B.; Fernàndez-Castillo, N. MiR-9, miR-153 and miR-124 are down-regulated by acute exposure to cocaine in a dopaminergic cell model and may contribute to cocaine dependence. Transl. Psychiatry 2018, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.; Balasubramaniam, M.; Martínez-Rivera, F.J.; Godino, A.; Peck, E.G.; Patnaik, S.; Suar, M.; Calipari, E.S.; Nestler, E.J.; Villalta, F.; et al. Cocaine-regulated microRNA miR-124 controls poly (ADP-ribose) polymerase-1 expression in neuronal cells. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Yang, W.; Liu, M.; Zhang, Q.; Zhang, J.; Chen, J.; Chen, Q.; Suo, L. Knockdown of miR-124 reduces depression-like behavior by targeting CREB1 and BDNF. Curr. Neurovascular Res. 2020, 17, 196–203. [Google Scholar] [CrossRef]
- Fakhoury, M. Revisiting the serotonin hypothesis: Implications for major depressive disorders. Mol. Neurobiol. 2016, 53, 2778–2786. [Google Scholar] [CrossRef]
- Kalueff, A.V.; Olivier, J.D.A.; Nonkes, L.J.P.; Homberg, J.R. Conserved role for the serotonin transporter gene in rat and mouse neurobehavioral endophenotypes. Neurosci. Biobehav. Rev. 2010, 34, 373–386. [Google Scholar] [CrossRef]
- Moya, P.R.; Wendland, J.R.; Salemme, J.; Fried, R.L.; Murphy, D.L. miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int. J. Neuropsychopharmacol. 2013, 16, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Dahlhoff, M.; Siegmund, A.; Golub, Y.; Wolf, E.; Holsboer, F.; Wotjak, C.T. AKT/GSK-3β/β-catenin signalling within hippocampus and amygdala reflects genetically determined differences in posttraumatic stress disorder like symptoms. Neuroscience 2010, 169, 1216–1226. [Google Scholar] [CrossRef]
- Chen, Y.C.; Tan, Q.R.; Dang, W.; Wang, H.N.; Zhang, R.B.; Li, Z.Y.; Lin, H.; Liu, R. The effect of citalopram on chronic stress-induced depressive-like behavior in rats through GSK3β/β-catenin activation in the medial prefrontal cortex. Brain Res. Bull. 2012, 88, 338–344. [Google Scholar] [CrossRef]
- Pilar-Cuéllar, F.; Vidal, R.; Pazos, A. Subchronic treatment with fluoxetine and ketanserin increases hippocampal brain-derived neurotrophic factor, β-catenin and antidepressant-like effects. Br. J. Pharmacol. 2012, 165, 1046–1057. [Google Scholar] [CrossRef] [Green Version]
- Vallée, A.; Lecarpentier, Y.; Vallée, J.N. Possible actions of cannabidiol in obsessive-compulsive disorder by targeting the WNT/β-catenin pathway. Mol. Psychiatry 2022, 27, 230–248. [Google Scholar] [CrossRef] [PubMed]
- Sbarski, B.; Akirav, I. Cannabinoids as therapeutics for PTSD. Pharmacol. Ther. 2020, 211, 107551. [Google Scholar] [CrossRef] [PubMed]
- Hillard, C.J.; Beatka, M.; Sarvaideo, J. Endocannabinoid signaling and the hypothalamic-pituitary-adrenal axis. Compr. Physiol. 2016, 7, 1. [Google Scholar] [PubMed]
- Hill, M.N.; McLaughlin, R.J.; Bingham, B.; Shrestha, L.; Lee, T.T.; Gray, J.M.; Hillard, C.J.; Gorzalka, B.B.; Viau, V. Endogenous cannabinoid signaling is essential for stress adaptation. Proc. Natl. Acad. Sci. USA 2010, 107, 9406–9411. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.N.; Karatsoreos, I.N.; Hillard, C.J.; McEwen, B.S. Rapid elevations in limbic endocannabinoid content by glucocorticoid hormones in vivo. Psychoneuroendocrinology 2010, 35, 1333–1338. [Google Scholar] [CrossRef] [Green Version]
- Alteba, S.; Zer-Aviv, T.M.; Tenenhaus, A.; David, G.B.; Adelman, J.; Hillard, C.J.; Doron, R.; Akirav, I. Antidepressant-like effects of URB597 and JZL184 in male and female rats exposed to early life stress. Eur. Neuropsychopharmacol. 2020, 39, 70–86. [Google Scholar] [CrossRef]
- Alteba, S.; Portugalov, A.; Hillard, C.J.; Akirav, I. Inhibition of fatty acid amide hydrolase (FAAH) during adolescence and exposure to early life stress may exacerbate depression-like behaviors in male and female rats. Neuroscience 2021, 455, 89–106. [Google Scholar] [CrossRef]
- Burstein, O.; Shoshan, N.; Doron, R.; Akirav, I. Cannabinoids prevent depressive-like symptoms and alterations in BDNF expression in a rat model of PTSD. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 84, 129–139. [Google Scholar] [CrossRef]
- Laprairie, R.B.; Bagher, A.M.; Kelly, M.E.M.; Denovan-Wright, E. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.; Fierro, A.; Pessoa-Mahana, C.D. Cannabidiol binding and negative allosteric modulation at the cannabinoid type 1 receptor in the presence of delta-9-tetrahydrocannabinol: An In Silico study. PLoS ONE 2019, 14, e0220025. [Google Scholar] [CrossRef]
- Willner, P.; Muscat, R.; Papp, M. Chronic mild stress-induced anhedonia: A realistic animal model of depression. Neurosci. Biobehav. Rev. 1992, 16, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Grønli, J.; Murison, R.; Bjorvatn, B.; Sørensen, E.; Portas, C.M.; Ursin, R. Chronic mild stress affects sucrose intake and sleep in rats. Behav. Brain Res. 2004, 150, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Sales, A.J.; Fogaça, M.V.; Sartim, A.G.; Pereira, V.S.; Wegener, G.; Guimarães, F.S.; Joca, S.R. Cannabidiol induces rapid and sustained antidepressant-like effects through increased BDNF signaling and synaptogenesis in the prefrontal cortex. Mol. Neurobiol. 2019, 56, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Rogóż, Z.; Kabziński, M.; Sadaj, W.; Rachwalska, P.; Gądek-Michalska, A. Effect of co-treatment with fluoxetine or mirtazapine and risperidone on the active behaviors and plasma corticosterone concentration in rats subjected to the forced swim test. Pharmacol. Rep. 2012, 64, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Zaidan, H.; Ramaswami, G.; Barak, M.; Li, J.B.; Gaisler-Salomon, I. Pre-reproductive stress and fluoxetine treatment in rats affect offspring A-to-I RNA editing, gene expression and social behavior. Environ. Epigenetics 2018, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Iffland, K.; Grotenhermen, F. An update on safety and side effects of cannabidiol: A review of clinical data and relevant animal studies. Cannabis Cannabinoid Res. 2017, 2, 139–154. [Google Scholar] [CrossRef]

| miR-16 vmPFC | miR-16 NAC | miR-16 Raphe | miR-124 vmPFC | miR-124 NAC | miR-124 Raphe | miR-135 vmPFC | miR-135 NAC | miR-135 Raphe | |
|---|---|---|---|---|---|---|---|---|---|
| FST—immobility | r = 0.277 p = 0.102 | r = 0.250 p = 0.147 | r = 0.069 p = 716 | r = 0.087 p = 0.613 | r = −0.040 p = 0.816 | r = 0.054 p = 0.749 | r = 0.428 * p = 0.012 | r = −0.339 * p = 0.046 | r = 0.187 p = 0.340 |
| OFT—total distance | r = 0.176 p = 0.305 | r = 0.322 p = 0.060 | r = −0.473 ** p = 0.008 | r = −0.159 p = 0.355 | r = 0.455 ** p = 0.005 | r = 0.060 p = 0.718 | r = 0.012 p = 0.944 | r = 0.192 p = 0.268 | r = 0.474 * p = 0.011 |
| OFT—time in center | r = 0.0.14 p = 0.933 | r = −0.2.33 p = 0.198 | r = 0.276 p = 0.140 | r = −0.184 p = 0.282 | r = −0.142 p = 0.403 | r = 0.157 p = 0.347 | r = −0.75 p = 0.675 | r = −0.229 p = 0.186 | r = −0.70 p = 0.723 |
| hrt1a | slc6a4 | ctnnb1 | cnr1 | |
|---|---|---|---|---|
| miR-16 | r = 0.591 *** p = 0.000 | r = −0.432 * p = 0.031 | r = 0.140 p = 0.436 | r = 0.250 p = 0.160 |
| miR-124 | r = −0.131 p = 0.469 | r = 0.281 p = 0.183 | r = 0.027 p = 0.880 | r = −0.075 p = 0.682 |
| miR-135 | r = 0.478 ** p = 0.008 | r = −0.346 p = 0.091 | r = 0.001 p = 0.997 | r = 0.258 p = 0.154 |
| hrt1a | slc6a4 | ctnnb1 | cnr1 | |
|---|---|---|---|---|
| miR-16 | r = −0.404 p = 0.051 | r = −0.33 p = 0.878 | r = 0.479 ** p = 0.006 | r = 0.229 p = 0.215 |
| miR-124 | r = −0.272 p = 0.198 | r = −0.56 p = 0.794 | r = 0.463 ** p = 0.008 | r = 0.224 p = 0.234 |
| miR-135 | r = −0.324 p = 0.114 | r = −0.080 p = 0.724 | r = 0.222 p = 0.222 | r = 0.226 p = 0.231 |
| hrt1a | slc6a4 | ctnnb1 | cnr1 | |
|---|---|---|---|---|
| miR-16 | r = 0.125 p = 0.579 | r = 0.064 p = 0.795 | r = −0.205 p = 0.325 | r = 0.012 p = 0.956 |
| miR-124 | r = −0.358 p = 0.056 | r = 0.408 * p = 0.035 | r = −0.175 p = 0.330 | r = 0.147 p = 0.438 |
| miR-135 | r = 0.191 p = 0.382 | r = 0.232 p = 0.298 | r = 0.050 p = 0.815 | r = 0.324 p = 0.142 |
| Name | Description | GeneBankID (NM) | Protein Name | Primer Sequence | Efficacy | Description |
|---|---|---|---|---|---|---|
| Hprt | Housekeeping gene; used as a reference gene | NM_012583.2 | HPRT | F: 5′CGCCAGCTTCCTCCTCAG3′ | NM_012583.2 | HPRT |
| Htr1a | Serotonergic auto-receptor | R: 5′ATAACCTGGTTCATCATCACTAATCAC3′ | 99.83 | R: 5′ATAACCTGGTTCATCATCACTAATCAC3′ | 99.83 | |
| Slc6a4 | The serotonergic transporter | NM_012585.1 | 5HT1a | F: 5′CCACGGCTACACCATCTACTC3′ | NM_012585.1 | 5HT1a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bright, U.; Akirav, I. Cannabidiol Modulates Alterations in PFC microRNAs in a Rat Model of Depression. Int. J. Mol. Sci. 2023, 24, 2052. https://doi.org/10.3390/ijms24032052
Bright U, Akirav I. Cannabidiol Modulates Alterations in PFC microRNAs in a Rat Model of Depression. International Journal of Molecular Sciences. 2023; 24(3):2052. https://doi.org/10.3390/ijms24032052
Chicago/Turabian StyleBright, Uri, and Irit Akirav. 2023. "Cannabidiol Modulates Alterations in PFC microRNAs in a Rat Model of Depression" International Journal of Molecular Sciences 24, no. 3: 2052. https://doi.org/10.3390/ijms24032052
APA StyleBright, U., & Akirav, I. (2023). Cannabidiol Modulates Alterations in PFC microRNAs in a Rat Model of Depression. International Journal of Molecular Sciences, 24(3), 2052. https://doi.org/10.3390/ijms24032052





