Plant Dehydrins: Expression, Regulatory Networks, and Protective Roles in Plants Challenged by Abiotic Stress
Abstract
:1. Introduction
2. Dehydrin Structure, Classification, and Localization
3. Dehydrins Participate in Plant Response to Abiotic Stress
3.1. Dehydrin Expression under Drought Condition
3.2. Dehydrin Accumulation during Cold Stress
3.3. Dehydrin Expression Is Induced by Salt Stress
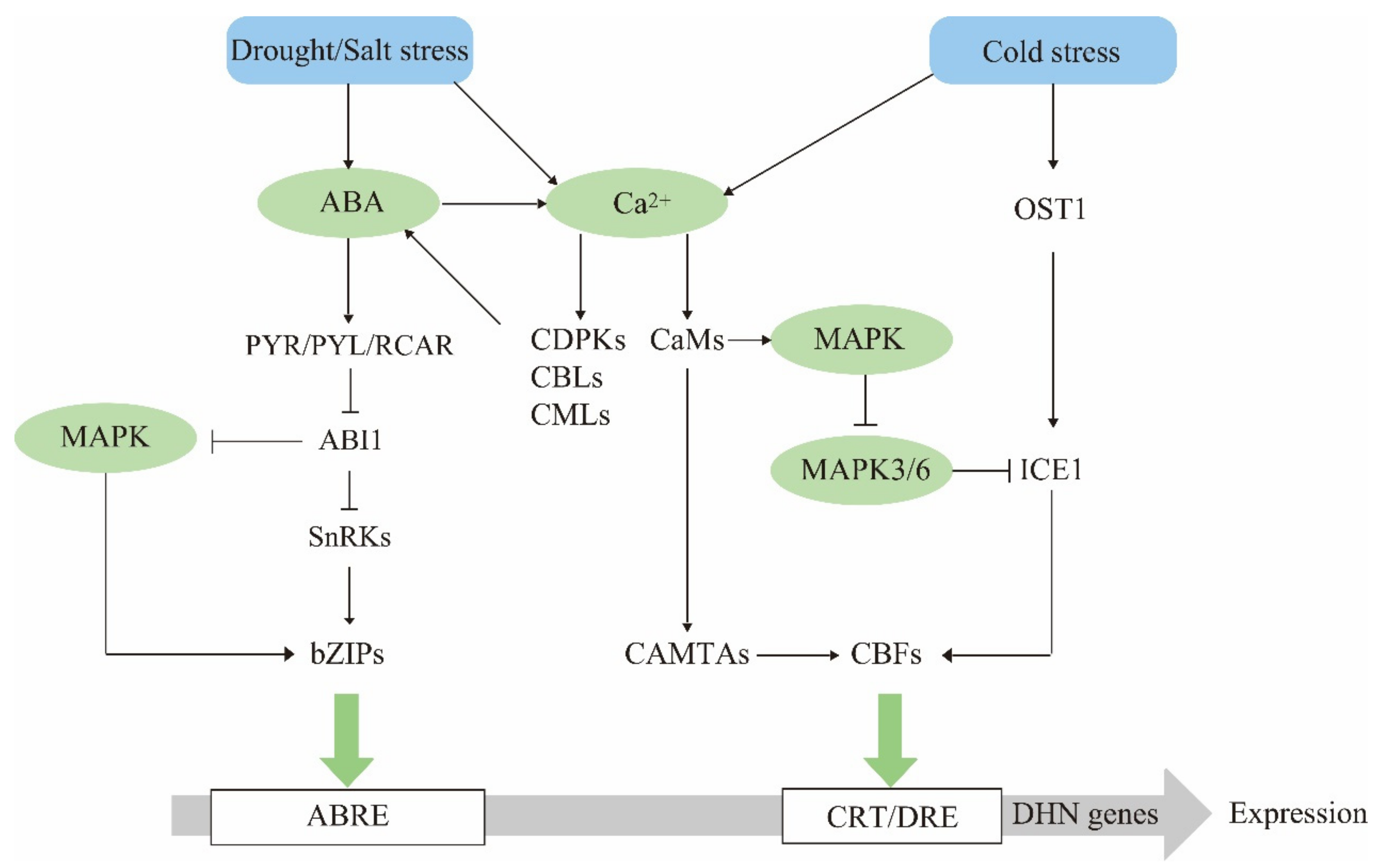
4. Dehydrin Gene Expression Regulatory Networks
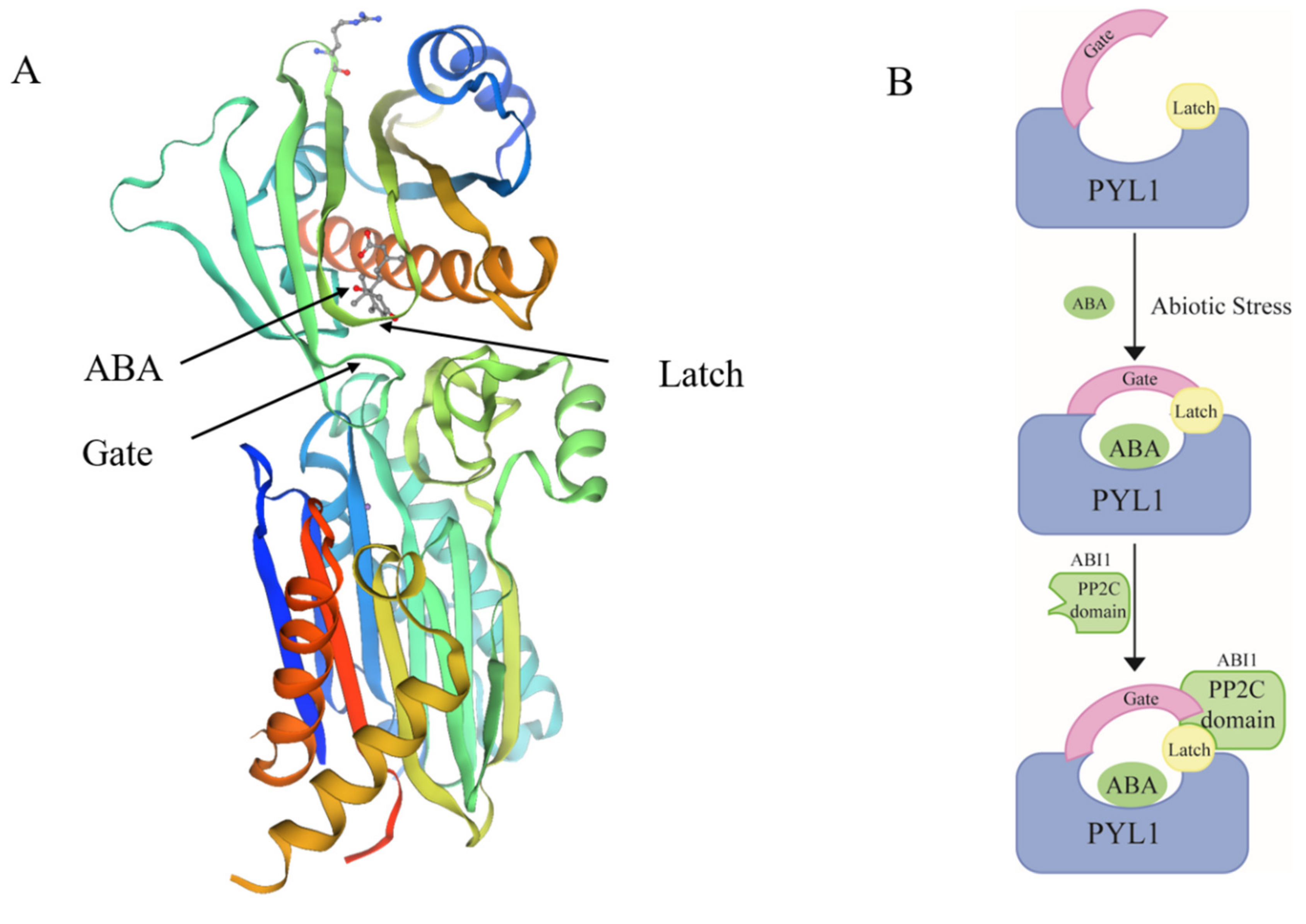
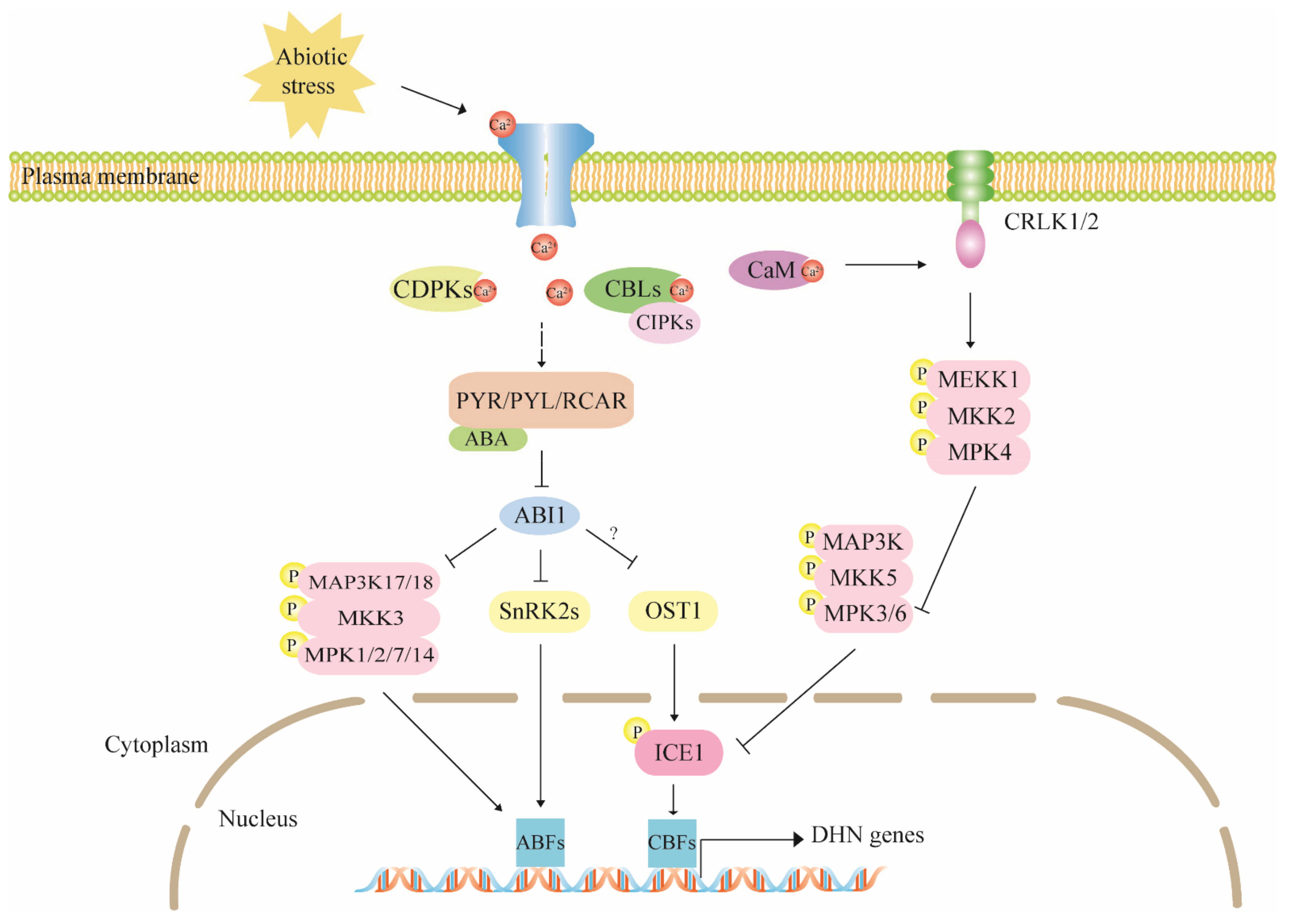
4.1. ABA Is a Dehydrin Mediator in Stress Response
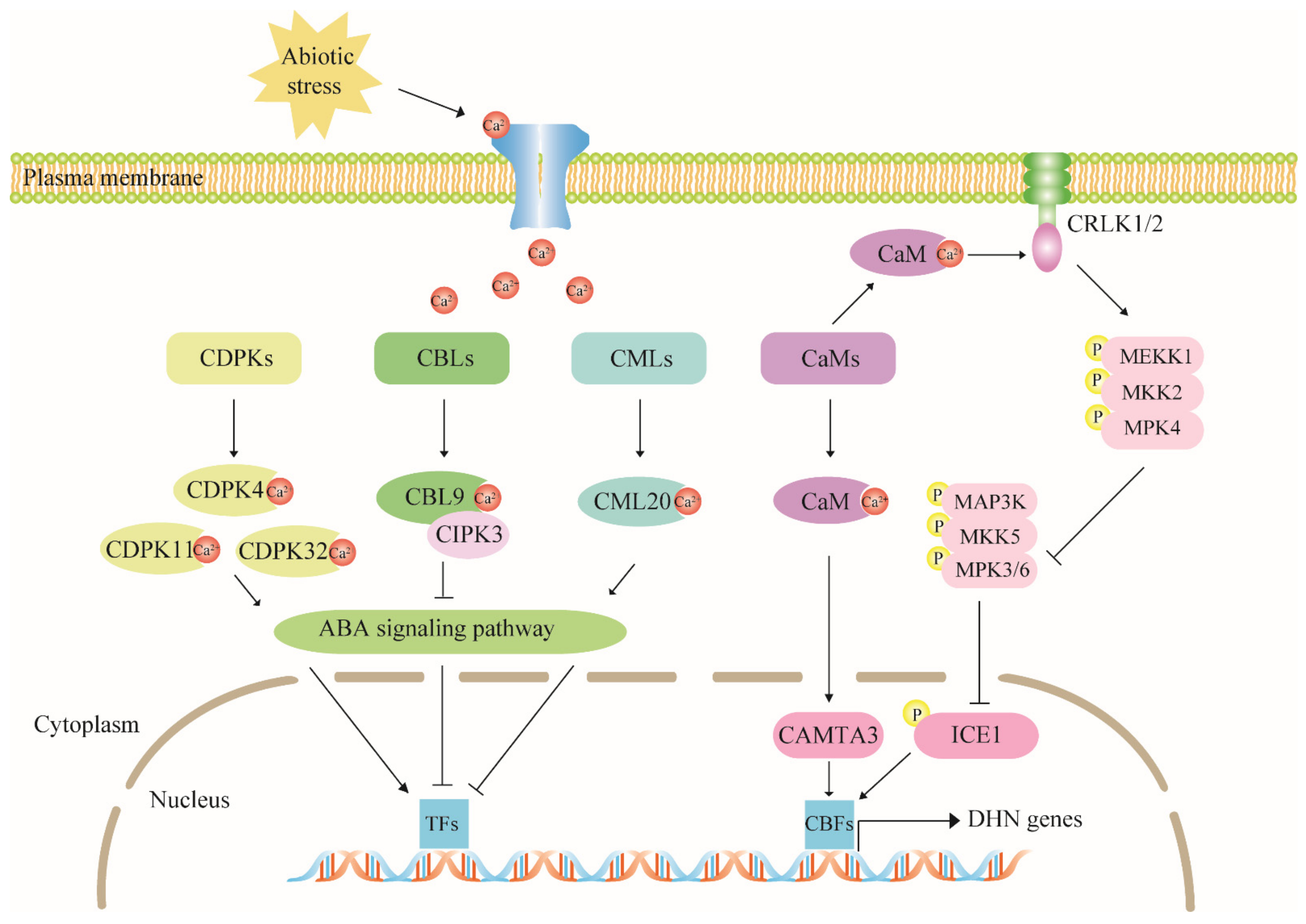
4.2. The Calcium Signaling Pathway Has Multiple Regulatory Effects on Dehydrin Expression
4.3. The MAPK Cascade Pathway Acts Upstream to Regulate Dehydrin Gene Expression
5. Functional Diversity of Dehydrins
5.1. Dehydrins Protect Seeds from Dehydration during Maturation
5.2. Dehydrins Stabilize Plasma Membranes
5.3. Dehydrins Protect Enzymatic Activity
5.4. Dehydrins Bind Metal Ions and DNA
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 21, 413. [Google Scholar] [CrossRef] [PubMed]
- Alam Syed, B.; Patel, M.; Patel, A.; Gami, B.; Patel, B. Regulation of antioxidant enzymes and osmo-protectant molecules by salt and drought responsive genes in Bambusa balcooa. J. Plant Res. 2021, 134, 165–175. [Google Scholar] [CrossRef]
- Kaneko, K.; Lazarus, M.; Miyamoto, C.; Oishi, Y.; Nagata, N.; Yang, S.; Yoshikawa, M.; Aritake, K.; Furuyashiki, T.; Narumiya, S.; et al. Orally administered rubiscolin-6, a delta opioid peptide derived from Rubisco, stimulates food intake via leptomeningeal lipocallin-type prostaglandin D synthase in mice. Mol. Nutr. Food Res. 2012, 56, 1315–1323. [Google Scholar] [CrossRef]
- Yoshikawa, M. Bioactive peptides derived from natural proteins with respect to diversity of their receptors and physiological effects. Peptides 2015, 72, 208–225. [Google Scholar] [CrossRef] [Green Version]
- Stefanucci, A.; Dimmito, M.P.; Tenore, G.; Pieretti, S.; Minosi, P.; Zengin, G.; Sturaro, C.; Calò, G.; Novellino, E.; Cichelli, A.; et al. Plant-derived peptides rubiscolin-6, soymorphin-6 and their c-terminal amide derivatives: Pharmacokinetic properties and biological activity. J. Funct. Foods 2020, 73, 104154. [Google Scholar] [CrossRef]
- Soba, D.; Müller, M.; Aranjuelo, I.; Munné-Bosch, S. Vitamin E in legume nodules: Occurrence and antioxidant function. Phytochemistry 2020, 172, 112261. [Google Scholar] [CrossRef]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses. Sci. Rep. 2019, 9, 14123. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Z.; Song, X.; Xu, J.; Jiang, C.; Zhao, Y.; Ma, C.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 2014, 86, 303–317. [Google Scholar] [CrossRef]
- Maryan, K.E.; Lahiji, H.S.; Farrokhi, N.; Komeleh, H.H. Analysis of Brassica napus dehydrins and their Co-Expression regulatory networks in relation to cold stress. Gene Expr. Patterns 2019, 31, 7–17. [Google Scholar] [CrossRef]
- Sánchez, I.E.H.; Maruri-López, I.; Graether, S.P.; Jiménez-Bremont, J.F. In vivo evidence for homo- and heterodimeric interactions of Arabidopsis thaliana dehydrins AtCOR47, AtERD10, and AtRAB18. Sci. Rep. 2017, 7, 17036. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Sánchez, I.E.; Maruri-López, I.; Molphe-Balch, E.P.; Becerra-Flora, A.; Jaimes-Miranda, F.; Jiménez-Bremont, J.F. Evidence for in vivo interactions between dehydrins and the aquaporin AtPIP2B. Biochem. Biophys. Res. Commun. 2019, 510, 545–550. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress response. Curr. Res. Biotechnol. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Eriksson, S.; Eremina, N.; Barth, A.; Danielsson, J.; Harryson, P. Membrane-Induced Folding of the Plant Stress Dehydrin Lti30. Plant Physiol. 2016, 171, 932–943. [Google Scholar]
- Hara, M.; Endo, T.; Kamiya, K.; Kameyama, A. The role of hydrophobic amino acids of K-segments in the cryoprotection of lactate dehydrogenase by dehydrins. J. Plant Physiol. 2017, 210, 18–23. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Zhang, T.; Yang, X.; Li, D. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 2017, 7, 7361. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Xu, S.; Jiang, L.; Liu, S. Identification and transcriptional analysis of dehydrin gene family in cucumber (Cucumis sativus). Acta Physiol. Plant. 2018, 40, 144. [Google Scholar] [CrossRef]
- Da Falavigna, V.S.; Malabarba, J.; Silveira, C.P.; Buffon, V.; de Mariath, J.E.A.; Pasquali, G.; Margis-Pinheiro, M.; Revers, L.F. Characterization of the nucellus-specific dehydrin MdoDHN11 demonstrates its involvement in the tolerance to water deficit. Plant Cell Rep. 2019, 38, 1099–1107. [Google Scholar] [CrossRef]
- Goday, A.; Jensen, A.B.; Culiáñez-Macià, F.A.; Mar Albà, M.; Figueras, M.; Serratosa, J.; Torrent, M.; Pagès, M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 1994, 6, 351–360. [Google Scholar]
- Kalemba, E.M.; Bagniewska-Zadworna, A.; Ratajczak, E. Multiple subcellular localizations of dehydrin-like proteins in the embryonic axes of common beech (Fagus sylvatica L.) seeds during maturation and dry storage. J. Plant Growth Regul. 2015, 34, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Wang, Y.; Yu, T.; Chen, S.; Chen, Y.; Lu, C. Heterologous expression of three ammopiptanthus mongolicus dehydrin genes confers abiotic stress tolerance in Arabidopsis thaliana. Plants 2020, 9, 193. [Google Scholar] [CrossRef] [Green Version]
- Sena, J.S.; Giguère, I.; Rigault, P.; Bousquet, J.; Mackay, J. Expansion of the dehydrin gene family in the Pinaceae is associated with considerable structural diversity and drought-responsive expression. Tree Physiol. 2018, 38, 442–456. [Google Scholar] [CrossRef]
- Lv, A.; Fan, N.; Xie, J.; Yuan, S.; An, Y.; Zhou, P. Expression of CdDHN4, a novel YSK2-type dehydrin gene from bermudagrass, responses to drought stress through the ABA-dependent signal pathway. Front. Plant Sci. 2017, 8, 748. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhang, L.S.; Liu, L.; Zhu, W.N.; Yang, W.B. Changes of dehydrin profiles induced by drought in winter wheat at different developmental stages. Biol. Plant. 2013, 57, 797–800. [Google Scholar] [CrossRef]
- Vítámvás, P.; Kosová, K.; Musilová, J.; Holková, L.; Mařík, P.; Smutná, P.; Klíma, M.; Prášil, I.T. Relationship between dehydrin accumulation and winter survival in winter wheat and barley grown in the field. Front. Plant Sci. 2019, 10, 7. [Google Scholar] [CrossRef] [Green Version]
- Kiyosue, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 1994, 35, 225–231. [Google Scholar] [CrossRef]
- Welin, B.V.; Olson, A.; Palva, E.T. Structure and organization of two closely related low-temperature-induced dhn/lea/rab-like genes in Arabidopsis thaliana L. Heynh. Plant Mol. Biol. 1995, 29, 391–395. [Google Scholar] [CrossRef]
- Gupta, A.; Marzinek, J.K.; Jefferies, D.; Bond, P.J.; Harryson, P.; Wohland, T. The disordered plant dehydrin Lti30 protects the membrane during water-related stress by cross-linking lipids. J. Biol. Chem. 2019, 294, 6468–6482. [Google Scholar] [CrossRef] [Green Version]
- Burrieza, H.P.; Koyro, H.-W.; Tosar, L.M.; Kobayashi, K.; Maldonado, S. High salinity induces dehydrin accumulation in Chenopodium quinoa Willd. cv. Hualhuas embryos. Plant Soil 2011, 354, 69–79. [Google Scholar] [CrossRef]
- Jing, H.; Li, C.; Ma, F.; Ma, J.-H.; Khan, A.; Wang, X.; Zhao, L.-Y.; Gong, Z.-H.; Chen, R.-G. Genome-wide identification, expression diversication of dehydrin gene family and characterization of CaDHN3 in pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef]
- Luo, D.; Hou, X.; Zhang, Y.; Meng, Y.; Zhang, H.; Liu, S.; Wang, X.; Chen, R. CaDHN5, a dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int. J. Mol. Sci. 2019, 20, 1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-F.; Liu, S.-Y.; Ma, J.-H.; Wang, X.-K.; Haq, S.U.; Meng, Y.-C.; Zhang, Y.-M.; Chen, R.-G. CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper decreases abscisic acid sensitivity in Arabidopsis. Int. J. Mol. Sci. 2019, 21, 26. [Google Scholar] [CrossRef] [Green Version]
- Kirungu, J.N.; Magwanga, R.O.; Pu, L.; Cai, X.; Xu, Y.; Hou, Y.; Zhou, Y.; Cai, Y.; Hao, F.; Zhou, Z.; et al. Knockdown of Gh_A05G1554 (GhDHN_03) and Gh_D05G1729 (GhDHN_04) dehydrin genes, reveals their potential role in enhancing osmotic and salt tolerance in cotton. Genomics 2020, 112, 1902–1915. [Google Scholar] [CrossRef]
- Zolotarov, Y.; Strömvik, M. De novo regulatory motif discovery identifies significant motifs in promoters of five classes of plant dehydrin genes. PLoS ONE 2015, 10, e0129016. [Google Scholar] [CrossRef]
- Mundy, J.; Chua, N.H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef]
- Lång, V.; Palva, E.T. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1992, 20, 951–962. [Google Scholar] [CrossRef]
- Mantyla, E.; Lang, V.; Palva, E.T. Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 1995, 107, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Rouse, D.T.; Marotta, R.; Parish, R.W. Promoter and expression studies on an Arabidopsis thaliana dehydrin gene. FEBS Lett. 1996, 381, 252–256. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Ward, R.W.; Thomashow, M.F. Characterization of a cold-regulated wheat gene related to Arabidopsis cor47. Plant Physiol. 1992, 100, 915–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xu, H.; Zhu, H.; Tao, Y.; Zhang, G.; Zhang, L.; Zhang, C.; Zhang, Z.; Ma, Z. Classification and expression diversification of wheat dehydrin genes. Plant Sci. 2014, 214, 113–120. [Google Scholar] [CrossRef]
- Miyazono, K.-I.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.-J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Soon, F.-F.; Ng, L.-M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Li, Q.; Wu, Q.; Wang, A.; Lv, B.; Dong, Q.; Yao, Y.; Wu, Q.; Zhao, H.; Li, C.; Chen, H.; et al. Tartary buckwheat transcription factor FtbZIP83 improves the drought/salt tolerance of Arabidopsis via an ABA-mediated pathway. Plant Physiol. Biochem. 2019, 144, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Indoliya, Y.; Singh, P.K.; Singh, P.C.; Chauhan, P.S.; Pande, V.; Chakrabarty, D. Role of dehydrin-FK506-binding protein complex in enhancing drought tolerance through the ABA-mediated signaling pathway. Environ. Exp. Bot. 2019, 158, 136–149. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Zhang, L. Identification of upstream transcription factors and an interacting PP2C protein of dehydrin WZY2 gene in wheat. Plant Signal. Behav. 2019, 14, 1678370. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhang, L.; Lv, H.; Zhang, H.; Zhang, D.; Wang, X.; Chen, J. The dehydrin wzy2 promoter from wheat defines its contribution to stress tolerance. Funct. Integr. Genom. 2014, 14, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wan, S.; Liu, X.; He, J.; Cheng, L.; Duan, M.; Liu, H.; Wang, W.; Yu, Y. Overexpression of CsSnRK2.5 increases tolerance to drought stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 150, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chu, J.; Ma, C.; Jiang, Y.; Ma, Y.; Xiong, J.; Cheng, Z.-M. Overexpression of an ABA-dependent grapevine bZIP transcription factor, VvABF2, enhances osmotic stress in Arabidopsis. Plant Cell Rep. 2019, 38, 587–596. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, X.; Yang, Y.; Kim, H.S.; Jia, X.; Yu, H.; Kwak, S.-S. Sweetpotato bZIP transcription factor IbABF4 confers tolerance to multiple abiotic stresses. Front. Plant Sci. 2019, 10, 630. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Zhang, L.; Wang, X.; Zhao, Z.; Tao, Z.; Wang, J.; Wang, J.; Lin, M.; Li, X.; et al. Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. Int. J. Mol. Sci. 2014, 15, 8473–8490. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yang, Z.; Ding, Y.; Liu, L.; Han, X.; Zhan, J.; Wei, X.; Diao, Y.; Qin, W.; Wang, P.; et al. Over-expression of an R2R3 MYB gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019, 286, 28–36. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Zhao, L.; Ren, Y.; Yang, S.; Zhu, J.-K.; Zhao, C. The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 2020, 62, 258–263. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Lee, C.-M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef] [Green Version]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef]
- Yang, X.; Wang, R.; Hu, Q.; Li, S.; Mao, X.; Jing, H.; Zhao, J.; Hu, G.; Fu, J.; Liu, C. DlICE1, a stress-responsive gene from Dimocarpus longan, enhances cold tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2019, 142, 490–499. [Google Scholar] [CrossRef]
- Zhou, L.; He, Y.J.; Li, J.; Li, L.; Liu, Y.; Chen, H.Y. An eggplant SmICE1a gene encoding MYC-type ICE1-like transcription factor enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Biol. 2020, 22, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.-H.; Lin, C.-H.; Tsai, H.-W.; Lin, T.-Y. Cold response in Phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1. Plant Cell Physiol. 2014, 55, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-induced CBF–PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. 2006, 27, 141–177. [Google Scholar]
- Rubio, S.; Noriega, X.; Pérez, F.J. Abscisic acid (ABA) and low temperatures synergistically increase the expression of CBF/DREB1 transcription factors and cold-hardiness in grapevine dormant buds. Ann. Bot. 2019, 123, 681–689. [Google Scholar] [CrossRef]
- Li, J.; Wang, N.; Xin, H.; Li, S. Overexpression of VaCBF4, a transcription factor from Vitis amurensis, improves cold tolerance accompanying increased resistance to drought and salinity in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 1518–1528. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ormancey, M.; Thuleau, P.; Mazars, C.; Cotelle, V. CDPKs and 14-3-3 proteins: Emerging duo in signaling. Trends Plant Sci. 2017, 22, 263–272. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Lei, L.; Yao, L.; Wang, L.; Hao, X.; Li, N.; Wang, Y.; Yin, P.; Guo, G.; Yang, Y.; et al. The involvements of calcium-dependent protein kinases and catechins in tea plant [Camellia sinensis (L.) O. Kuntze] cold responses. Plant Physiol. Biochem. 2019, 143, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Hou, X.; Xia, Z.; Yan, Y.; Wei, Y.; Wang, L.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava. Mol. Genet. Genom. 2016, 291, 241–253. [Google Scholar] [CrossRef]
- Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on calcium-dependent protein kinases (CPKs) signaling for abiotic stress tolerance in plants. Int. J. Mol. Sci. 2019, 20, 5298. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xie, J.; Wang, L.; Si, L.; Zheng, S.; Yang, Y.; Yang, H.; Tian, S. Wheat TabZIP8, 9, 13 participate in ABA biosynthesis in NaCl-stressed roots regulated by TaCDPK9-1. Plant Physiol. Biochem. 2020, 151, 650–658. [Google Scholar] [CrossRef]
- Choi, H.-I.; Park, H.-J.; Park, J.H.; Kim, S.; Im, M.-Y.; Seo, H.-H.; Kim, Y.-W.; Hwang, I.; Kim, S.Y. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005, 139, 1750–1761. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.-Y.; Yu, X.-C.; Wang, X.-J.; Zhao, R.; Li, Y.; Fan, R.-C.; Shang, Y.; Du, S.-Y.; Wang, X.-F.; Wu, F.-Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Tang, R.-J.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, M.-M.; Ren, L.; Liu, Y.; Chen, H.-Y. Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genom. 2016, 291, 1769–1781. [Google Scholar] [CrossRef]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.; Zhao, C.; Zhao, M.; Li, Y.; Wen, G. Phylogeny and evolution of Calcineurin B-Like (CBL) gene family in grass and functional analyses of rice CBLs. J. Plant Biol. 2020, 63, 117–130. [Google Scholar] [CrossRef]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, J.-K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, T.; John, S.J.; Chen, M.; Chang, J.; Yang, G.; He, G. A CBL-interacting protein kinase TaCIPK27 confers drought tolerance and exogenous ABA sensitivity in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 123, 103–113. [Google Scholar] [CrossRef]
- Cui, X.-Y.; Du, Y.-T.; Fu, J.-D.; Yu, T.-F.; Wang, C.-T.; Chen, M.; Chen, J.; Ma, Y.-Z.; Xu, Z.-S. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018, 18, 93. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-N.; Cheong, Y.H.; Grant, J.J.; Pandey, G.K.; Luan, S. CIPK3, a calcium sensor–associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 2003, 15, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Wu, X.-Y.; Li, H.; Song, J.-H.; Liu, J.-Y. A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis aba response network. Mol. Plant 2016, 9, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, S.K.; Kanwar, P.; Yadav, A.K.; Sharma, C.; Kumar, A.; Pandey, G.K. Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 2017, 254, 48–59. [Google Scholar] [CrossRef]
- Sun, Z.; Qi, X.; Li, P.; Wu, C.; Zhao, Y.; Zhang, H.; Wang, Z. Overexpression of aThellungiella halophila CBl9 homolog, ThCBL9, confers salt and osmotic tolerances in transgenicarabidopsis thaliana. J. Plant Biol. 2008, 51, 25–34. [Google Scholar] [CrossRef]
- Ogunrinde, A.; Munro, K.; Davidson, A.; Ubaid, M.; Snedden, W.A. Arabidopsis calmodulin-like proteins, CML15 and CML16 possess biochemical properties distinct from calmodulin and show non-overlapping tissue expression patterns. Front. Plant Sci. 2017, 8, 2175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yue, D.; Wei, W.; Hu, Y.; Feng, J.; Zou, Z. Characterization and functional analysis of calmodulin and calmodulin-like genes in fragaria vesca. Front. Plant Sci. 2016, 7, 1820. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, X.; Liu, X.; Wang, J.; Yu, J.; Hu, D.; Hao, Y. Genome-wide identification, characterization, and expression analysis of calmodulin-like proteins (CMLs) in apple. Hortic. Plant J. 2017, 3, 219–231. [Google Scholar] [CrossRef]
- Wu, X.; Qiao, Z.; Liu, H.; Acharya, B.R.; Li, C.; Zhang, W. CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 2017, 8, 824. [Google Scholar] [CrossRef] [Green Version]
- McCormack, E.; Tsai, Y.-C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Li, C.; Meng, D.; Zhang, J.; Cheng, L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malusxdomestica). Plant Physiol. Biochem. 2019, 139, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nagar, P.; Goel, P.; Singh, A.K.; Kumari, S.; Mustafiz, A. Second messengers: Central regulators in plant abiotic stress response. In Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective; Springer: Singapore, 2018; pp. 47–94. [Google Scholar]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Heo, W.D.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef] [Green Version]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, V.E.; Poppenberger, B. MAP kinase signaling turns to ICE. Dev. Cell 2017, 43, 545–546. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Ali, G.S.; Yang, L.; Du, L.; Reddy, A.S.; Poovaiah, B.W. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef]
- Townley, H.E.; Knight, M.R. Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 2002, 128, 1169–1172. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-H.; Sharrocks, A.D.; Whitmarsh, A. MAP kinase signalling cascades and transcriptional regulation. Gene 2013, 513, 1–13. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642.e4. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.-C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danquah, A.; de Zelicourt, A.; Boudsocq, M.; Neubauer, J.; Frey, N.F.D.; Leonhardt, N.; Pateyron, S.; Gwinner, F.; Tamby, J.-P.; Ortiz-Masia, M.D.; et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015, 82, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Zhang, J.; Hsu, P.-K.; Ceciliato, P.H.O.; Zhang, L.; Dubeaux, G.; Munemasa, S.; Ge, C.; Zhao, Y.; Hauser, F.; et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jin, Y.; Huang, W.; Sun, Q.; Liu, F.; Huang, X. Full-length transcriptome sequences of ephemeral plant Arabidopsis pumila provides insight into gene expression dynamics during continuous salt stress. BMC Genom. 2018, 19, 717. [Google Scholar] [CrossRef]
- Mitula, F.; Tajdel, M.; Cieśla, A.; Kasprowicz-Maluśki, A.; Kulik, A.; Babula-Skowronska, D.; Michalak, M.; Dobrowolska, G.; Sadowski, J.; Ludwików, A. Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin–proteasome pathway. Plant Cell Physiol. 2015, 56, 2351–2367. [Google Scholar] [CrossRef] [PubMed]
- Azarkovich, M.I. Dehydrins in orthodox and recalcitrant seeds. Russ. J. Plant Physiol. 2020, 67, 221–230. [Google Scholar] [CrossRef]
- Bojórquez-Velázquez, E.; Barrera-Pacheco, A.; Espitia-Rangel, E.; Herrera-Estrella, A.; De La Rosa, A.P.B. Protein analysis reveals differential accumulation of late embryogenesis abundant and storage proteins in seeds of wild and cultivated amaranth species. BMC Plant Biol. 2019, 19, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalemba, E.M.; Litkowiec, M. Functional characterization of a dehydrin protein from Fagus sylvatica seeds using experimental and in silico approaches. Plant Physiol. Biochem. 2015, 97, 246–254. [Google Scholar] [CrossRef]
- Eriksson, S.K.; Kutzer, M.; Procek, J.; Gröbner, G.; Harryson, P. Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 2011, 23, 2391–2404. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.-J.; Xu, S.-C.; Liu, N.; Zhang, G.-W.; Hu, Q.-Z.; Xu, Z.-S.; Gong, Y.-M. Identification of the AQP members involved in abiotic stress responses from Arabidopsis. Gene 2018, 646, 64–73. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Song, S.; Hou, X.; Jia, C.; Li, J.; Miao, H.; Wang, Z.; Tie, W.; et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Tossounian, M.-A.; Kovacs, D.S.; Thu, T.T.; Stijlemans, B.; Vertommen, D.; Pauwels, J.; Gevaert, K.; Angenon, G.; Messens, J.; et al. Dehydrin ERD14 activates glutathione transferase Phi9 in Arabidopsis thaliana under osmotic stress. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129506. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Monna, S.; Murata, T.; Nakano, T.; Amano, S.; Nachbar, M.; Wätzig, H. The Arabidopsis KS-type dehydrin recovers lactate dehydrogenase activity inhibited by copper with the contribution of His residues. Plant Sci. 2016, 245, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, T.; Kameyama, A.; Kamiya, K.; Kondo, M.; Hara, M. F-segments of Arabidopsis dehydrins show cryoprotective activities for lactate dehydrogenase depending on the hydrophobic residues. Phytochemistry 2020, 173, 112300. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sheng, J.; Lv, K.; Ren, L.; Zhang, D. Y2SK2 and SK3 type dehydrins from Agapanthus praecox can improve plant stress tolerance and act as multifunctional protectants. Plant Sci. 2019, 284, 143–160. [Google Scholar] [CrossRef]
- Kumar, M.; Lee, S.-C.; Kim, J.-Y.; Kim, S.-J.; Aye, S.S.; Kim, S.-R. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Saibi, W.; Feki, K.; Ben Mahmoud, R.; Brini, F. Durum wheat dehydrin (DHN-5) confers salinity tolerance to transgenic Arabidopsis plants through the regulation of proline metabolism and ROS scavenging system. Planta 2015, 242, 1187–1194. [Google Scholar] [CrossRef]
- Boddington, K.F.; Graether, S.P. Binding of a Vitis riparia dehydrin to DNA. Plant Sci. 2019, 287, 110172. [Google Scholar] [CrossRef]
- Kim, S.Y.; Nam, K.H. Physiological roles of ERD10 in abiotic stresses and seed germination of Arabidopsis. Plant Cell Rep. 2010, 29, 203–209. [Google Scholar] [CrossRef]
- Kovacs, D.; Kalmár, É.; Torok, Z.; Tompa, P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, T.; Ren, L. Y2SK2- and SK3-type dehydrins from Agapanthus praecox act as protectants to improve plant cell viability during cryopreservation. Plant Cell Tissue Organ Cult. 2021, 144, 271–279. [Google Scholar] [CrossRef]
- Meng, Y.-C.; Zhang, H.-F.; Pan, X.-X.; Chen, N.; Hu, H.-F.; Haq, S.; Khan, A.; Chen, R.-G. CaDHN3, a pepper (Capsicum annuum L.) dehydrin gene enhances the tolerance against salt and drought stresses by reducing ROS accumulation. Int. J. Mol. Sci. 2021, 22, 3205. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Kim, H.-Y.; Kim, I.-S.; Kim, J.-J.; Kim, Y.-S.; Yoon, H.-S. The dehydrin gene of the Arctic plant Cerastium arcticum, CaDHN, increases tolerance to multiple stresses in Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 387–395. [Google Scholar] [CrossRef]
- Poku, S.A.; Chukwurah, P.N.; Aung, H.H.; Nakamura, I. Over-expression of a melon Y3SK2-Type LEA gene confers drought and salt tolerance in transgenic tobacco plants. Plants 2020, 9, 1749. [Google Scholar] [CrossRef]
- Zhou, Y.; He, P.; Xu, Y.; Liu, Q.; Yang, Y.; Liu, S. Overexpression of CsLEA11, a Y3SK2-type dehydrin gene from cucumber (Cucumis sativus), enhances tolerance to heat and cold in Escherichia coli. AMB Express 2017, 7, 182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, J.; Su, H.; Xia, K.; Jian, S.; Zhang, M. Molecular cloning and functional characterization of the dehydrin (IpDHN) gene from ipomoea pes-caprae. Front. Plant Sci. 2018, 9, 1454. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.; Li, D.; Li, D.; Yang, X.; Liu, Y. Overexpression of ZmDHN11 could enhance transgenic yeast and tobacco tolerance to osmotic stress. Plant Cell Rep. 2021, 40, 1723–1733. [Google Scholar] [CrossRef]
- Koag, M.-C.; Wilkens, S.; Fenton, R.D.; Resnik, J.; Vo, E.; Close, T.J. The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 2009, 150, 1503–1514. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Zhang, R.; Qu, Y.; Miao, Z.; Zhang, Y.; Shen, X.; Wang, T.; Dong, J. Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 2012, 195, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, L.; Wang, X.; Zhang, M.; Xi, Y.; Wang, A.; Zhu, J. Overexpression of Saussurea involucrata dehydrin gene SiDHN promotes cold and drought tolerance in transgenic tomato plants. PLoS ONE 2019, 14, e0225090. [Google Scholar] [CrossRef]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and beta-glucosidase activities in vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef]
| Species | Dehydrin | Abiotic Stress Responses | Functions | References |
|---|---|---|---|---|
| Arabidopsis thaliana | RAB18 | Cold/drought/salinity | Freezing tolerance | [37] |
| COR47 | Cold | LDH cryoprotection | [123] | |
| Lti30 | Cold | Membrane stabilization | [14,28,117] | |
| ERD10 | Cold/drought/salinity | Oxidoreductase protection, membrane stabilization, chaperone activity | [121,128,129] | |
| ERD14 | Cold/drought/salinity | Same as above | [121,128,129] | |
| Agapanthus praecox | DHN (Y2SK2) | Cold/drought/salinity | Metal ion binding, enzyme activity protection, membrane cryoprotection | [124,130] |
| DHN (SK3) | Cold/drought/salinity | Same as above | [124,130] | |
| Ammopiptanthus mongolicus | DHN132 | Cold/salinity | Membrane-protection | [21] |
| DHN154 | Cold/salinity | Same as above | [21] | |
| DHN200 | Cold/salinity | Same as above | [21] | |
| Capsicum annuum | DHN3 | Drought/salinity | Upregulation of antioxidant enzyme system | [131] |
| DHN4 | Cold/salinity | Cell membrane stabilization, prevention of lipid peroxidation, inhibition of ROS accumulation | [32] | |
| DHN5 | Salinity | Antioxidant capacity improvement | [31] | |
| Cerastium arcticum | DHN | Cold/drought/salinity | Oxidative stress tolerance | [132] |
| Cucumis melo | LEA-S | drought/salinity | APX and CAT activity enhancement | [133] |
| Cucumis sativus | DHN4 | Cold/ salinity/ heat | LDH protection | [17,134] |
| Ipomoea pescaprae | DHN | Cold/drought/salinity | Antioxidant enzyme system upregulation | [135] |
| Maize | DHN11 | Cold/drought/salinity | antioxidant enzymes activity protection | [136] |
| DHN13 | Cold | LDH protection | [16] | |
| Medicago truncatula | DHN1 | Cold/salinity | Membrane stabilization | [137] |
| CAS31 | Drought | Stomatal density and root water loss reduction | [120,138] | |
| Rice | DHN1 | drought/salinity | Enhancement of ROS scavenging capacity | [125] |
| Saussurea involucrata | DHN | Cold/drought | Inhibition of cell membrane damage, chloroplast protection, enhancement of ROS scavenging capacity | [139] |
| Wheat | COR410 | Cold/drought | Plasma membrane protection against freezing and dehydration stress | [140] |
| DHN5 | Cold/salinity | LDH and bglG protection, and regulation of proline metabolism and ROS scavenging system | [126,141] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Li, S.; Chen, W.; Zhang, J.; Zhang, L.; Sun, W.; Wang, Z. Plant Dehydrins: Expression, Regulatory Networks, and Protective Roles in Plants Challenged by Abiotic Stress. Int. J. Mol. Sci. 2021, 22, 12619. https://doi.org/10.3390/ijms222312619
Sun Z, Li S, Chen W, Zhang J, Zhang L, Sun W, Wang Z. Plant Dehydrins: Expression, Regulatory Networks, and Protective Roles in Plants Challenged by Abiotic Stress. International Journal of Molecular Sciences. 2021; 22(23):12619. https://doi.org/10.3390/ijms222312619
Chicago/Turabian StyleSun, Zhenping, Shiyuan Li, Wenyu Chen, Jieqiong Zhang, Lixiao Zhang, Wei Sun, and Zenglan Wang. 2021. "Plant Dehydrins: Expression, Regulatory Networks, and Protective Roles in Plants Challenged by Abiotic Stress" International Journal of Molecular Sciences 22, no. 23: 12619. https://doi.org/10.3390/ijms222312619





