Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Cohort
2.2. Intergenerational Correlations of BMI, Insulin Sensitivity, and Lifestyle
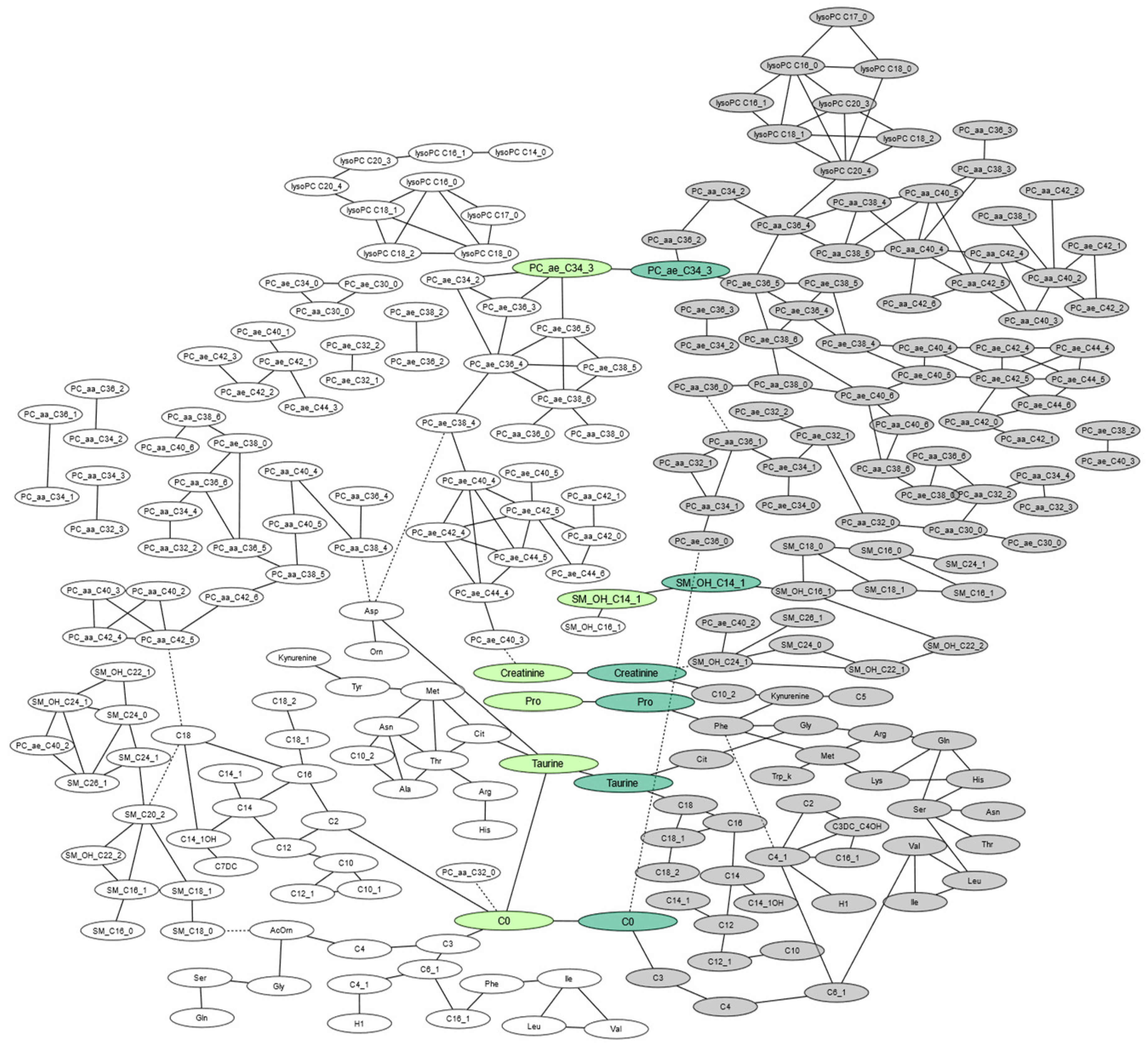
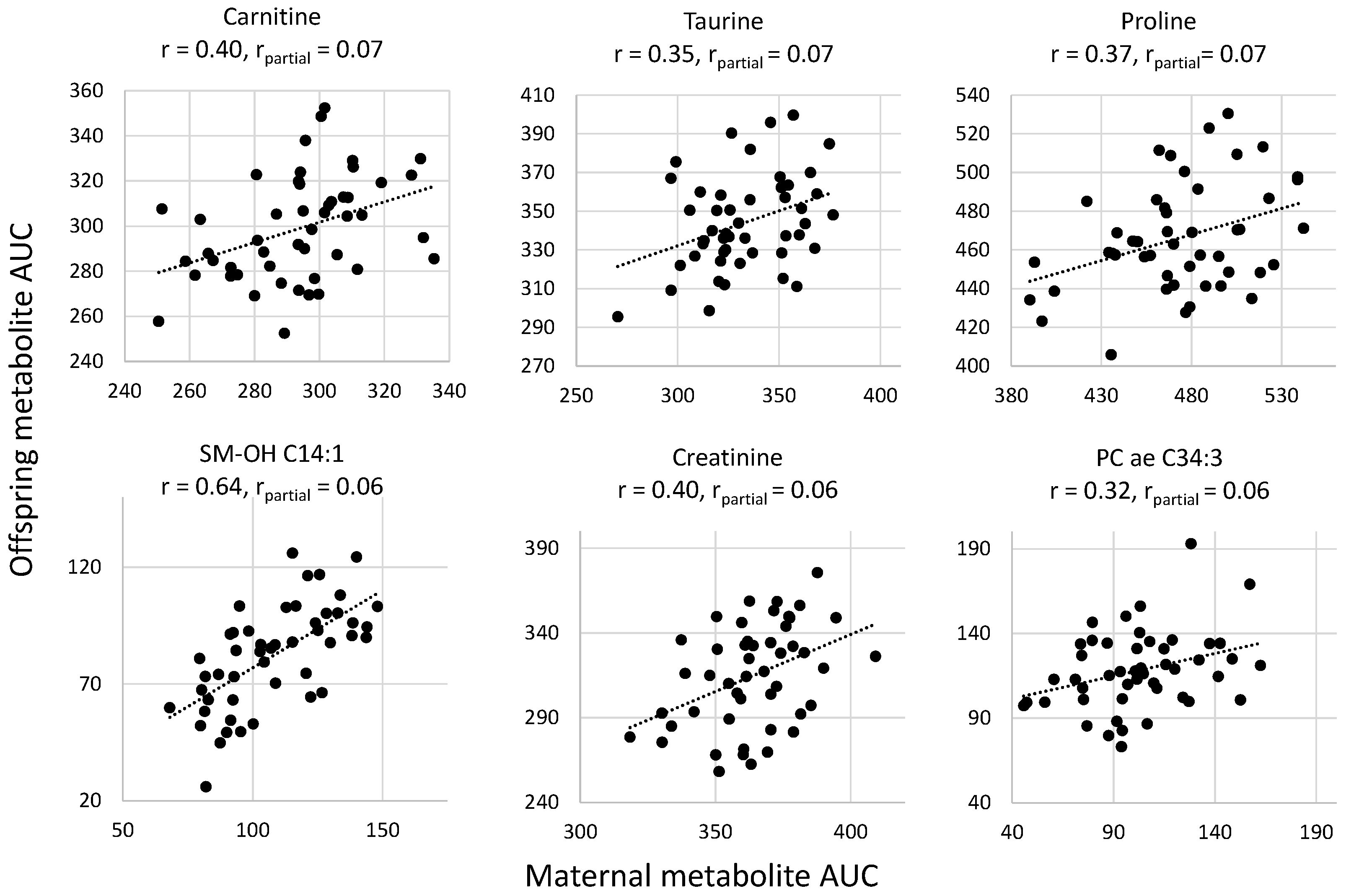
2.3. Intergenerational Correlations of AUC of Metabolites
2.4. Effect of Lifestyle on Intergenerationally Correlated Metabolites
3. Discussion
4. Materials and Methods
4.1. Postpartum Outcomes in Women with Gestational Diabetes and Their Offspring (POGO) Study
4.2. Measurement of Insulin and Plasma Glucose
4.3. Dietary Behavior
4.4. Physical Activity
4.5. Metabolomics Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GDM | Gestational diabetes mellitus |
| OGTT | Oral glucose tolerance test |
| SDS | Standard deviation score |
| ISI-M | Matsuda insulin sensitivity index |
| FFQ | Food frequency questionnaire |
| GI | Glycemic index |
| CHO | Carbohydrate |
| MET | Metabolic equivalent of task |
| GGM | Gaussian graphical model |
| FDR | False discovery rate |
| PC | Phosphatidylcholine |
| AC | Acylcarnitine |
| SM | Sphingomyelin |
| Lyso-PC | Acyl-lysophosphatidylcholine |
| PCaa | Diacyl-phosphatidylcholine |
| PCae | Acyl-alkyl-phosphatidylcholine |
| DASH | Dietary Approach to Stop Hypertension |
| AUC | Area under the curve |
Appendix A
| Included Mother–Offspring Pairs | Excluded Mother–Offspring Pairs | ||||
|---|---|---|---|---|---|
| N | N | p-Value | |||
| Country of origin Germany, n (%) | 48 | 40 (83.3%) | 70 | 57 (81.4%) | 0.40 |
| Offspring sex, female n (%) | 48 | 23 (48%) | 70 | 31 (44%) | 0.7 |
| Maternal age at follow-up (years), median (IQR) | 48 | 42.1 (38.0; 44.2) | 70 | 40.2 (35.2; 43.5) | 0.1 |
| Offspring age at follow-up (years), median (IQR) | 48 | 8.0 (5.4; 8.8) | 70 | 5.1 (4.1; 7.4) | <0.0001 |
| Maternal BMI at follow-up (kg/m2), median (IQR) | 48 | 26.4 (22.6; 32.0) | 70 | 25.3 (22.5; 30.4) | 0.5 |
| Offspring BMI-SDS, median (IQR) | 48 | 0.1 (−0.6; 0.8) | 47 | 0.04 (−0.8; 0.5) | 0.3 |
| Maternal fasting glucose (mg/dl), median (IQR) | 48 | 94 (87; 103) | 70 | 91 (87; 100) | 0.2 |
| Offspring fasting glucose (mg/dl), median (IQR) | 48 | 88 (84; 92) | 34 | 84 (80; 92) | 0.06 |
Appendix B
| Intergenerational Correlation | ||
|---|---|---|
| Correlation Estimate r | p-Value | |
| Energy and Nutrients | ||
| Energy (kcal/day) | −0.05 | 0.83 |
| Protein (% of total EI) | 0.17 | 0.47 |
| Carbohydrates (% of total EI) | 0.21 | 0.37 |
| Fiber (g/day) | −0.10 | 0.60 |
| Total Fat (% of total EI) | −0.06 | 0.78 |
| Polyunsaturated fatty acids (% of total EI) | 0.40 | 0.07 |
| Monounsaturated fatty acids (% of total EI) | 0.17 | 0.46 |
| Saturated fatty acids (% of total EI) | 0.08 | 0.74 |
| DASH score | 0.24 | 0.29 |
Appendix C
| Node1 | Node2 | Estimate r | P Raw | P FDR |
|---|---|---|---|---|
| Intergenerational correlations of same metabolites between mothers and offspring | ||||
| Carnitine | 0.070 | <0.00001 | <0.01 | |
| Taurine | 0.069 | <0.00001 | <0.01 | |
| Proline | 0.065 | <0.00001 | <0.01 | |
| SM –OH C14:1 | 0.061 | 0.0001 | 0.01 | |
| Creatinine | 0.061 | 0.0001 | 0.01 | |
| PC ae C34:3 | 0.060 | 0.0001 | <0.05 | |
| Offspring correlations | ||||
| Lyso PC a C16:0 | Lyso PC a C18:0 | 0.118 | <0.00001 | <0.0001 |
| Leucine | Valine | 0.115 | <0.00001 | <0.0001 |
| AC C5 | Kynurenine | 0.112 | <0.00001 | <0.0001 |
| PC aa C40:4 | PC aa C40:5 | 0.111 | <0.00001 | <0.0001 |
| Isoleucine | Leucine | 0.110 | <0.00001 | <0.0001 |
| AC C4:1 | AC C6:1 | 0.110 | <0.00001 | <0.0001 |
| SM C18:0 | SM C18:1 | 0.107 | <0.00001 | <0.0001 |
| PC ae C34:2 | PC ae C36:3 | 0.106 | <0.00001 | <0.0001 |
| PC aa C36:4 | PC aa C38:4 | 0.104 | <0.00001 | <0.0001 |
| Citrulline | Taurine | 0.103 | <0.00001 | <0.0001 |
| SM (-OH) C22:1 | SM (-OH) C24:1 | 0.103 | <0.00001 | <0.0001 |
| PC ae C42:4 | PC ae C44:4 | 0.101 | <0.00001 | <0.0001 |
| PC ae C32:1 | PC ae C32:2 | 0.101 | <0.00001 | <0.0001 |
| PC aa C34:2 | PC aa C36:2 | 0.101 | <0.00001 | <0.0001 |
| SM (-OH) C14:1c | SM (-OH) C16:1 | 0.100 | <0.00001 | <0.0001 |
| SM -C24:0 | SM (-OH) C22:1 | 0.094 | <0.00001 | <0.0001 |
| PC ae C44:5 | PC ae C44:6 | 0.093 | <0.00001 | <0.0001 |
| PC aa C38:6 | PC aa C40:6 | 0.092 | <0.00001 | <0.0001 |
| PC aa C34:1 | PC aa C36:1 | 0.091 | <0.00001 | <0.0001 |
| AC C6:1 | Valine | 0.090 | <0.00001 | <0.0001 |
| Kynurenine | Phenylalanine | 0.089 | <0.00001 | <0.0001 |
| Histidine | Serine | 0.088 | <0.00001 | <0.0001 |
| PC ae C42:5 | PC ae C44:5 | 0.088 | <0.00001 | <0.0001 |
| PC ae C40:4 | PC ae C42:4 | 0.088 | <0.00001 | <0.0001 |
| Isoleucine | Valine | 0.087 | <0.00001 | <0.0001 |
| SM C24:0 | SM (-OH) C24:1 | 0.086 | <0.00001 | <0.0001 |
| Carnitine | AC C3 | 0.086 | <0.00001 | <0.0001 |
| SM (-OH) C22:1 | SM (-OH) C22:2 | 0.086 | <0.00001 | <0.0001 |
| LysoPC a C18:2 | LysoPC a C20:4 | 0.085 | <0.00001 | <0.0001 |
| SM C18:0 | SM (-OH) C16:1 | 0.085 | <0.00001 | <0.0001 |
| PC ae C38:4 | PC ae C40:4 | 0.084 | <0.00001 | <0.0001 |
| PC aa C42:5 | PC aa C42:6 | 0.083 | <0.00001 | <0.0001 |
| Serine | Threonine | 0.083 | <0.00001 | <0.0001 |
| AC C16 | AC C18 | 0.082 | <0.00001 | <0.0001 |
| PC ae C34:3 | PC ae C36:5 | 0.082 | <0.00001 | <0.0001 |
| PC aa C40:5 | PC aa C42:5 | 0.082 | <0.00001 | <0.0001 |
| PC aa C38:5 | PC aa C40:5 | 0.081 | <0.00001 | <0.0001 |
| PC ae C36:4 | PC ae C38:5 | 0.081 | <0.00001 | <0.0001 |
| PC ae C44:4 | PC ae C44:5 | 0.081 | <0.00001 | <0.0001 |
| AC C18:1 | AC C18:2 | 0.081 | <0.00001 | <0.0001 |
| LysoPC a C20:3 | LysoPC a C20:4 | 0.081 | <0.00001 | <0.001 |
| AC C4-OH | AC C4:1 | 0.080 | <0.00001 | <0.001 |
| AC C2 | AC C4-OH | 0.080 | <0.00001 | <0.001 |
| LysoPC a C18:1 | LysoPC a C18:2c | 0.079 | <0.00001 | <0.001 |
| PC aa C38:6 | PC ae C38:0 | 0.078 | <0.00001 | <0.001 |
| PC ae C42:5 | PC ae C44:6 | 0.078 | <0.00001 | <0.001 |
| LysoPC a C18:2 | LysoPC a C20:3 | 0.078 | <0.00001 | <0.001 |
| Glycine | Phenylalanine | 0.078 | <0.00001 | <0.001 |
| LysoPC a C18:1 | LysoPC a C20:3 | 0.078 | <0.00001 | <0.001 |
| PC ae C38:4 | PC ae C40:5 | 0.077 | <0.00001 | <0.001 |
| SM C26:1 | SM (-OH) C24:1 | 0.077 | <0.00001 | <0.001 |
| PC aa C38:0 | PC ae C38:6 | 0.077 | <0.00001 | <0.001 |
| PC aa C30:0 | PC ae C30:0 | 0.077 | <0.00001 | <0.001 |
| PC aa C30:0 | PC aa C32:0 | 0.077 | <0.00001 | <0.001 |
| PC ae C40:4 | PC ae C40:5 | 0.077 | <0.00001 | <0.001 |
| PC ae C42:4 | PC ae C44:5 | 0.077 | <0.00001 | <0.001 |
| AC C10:2 | Creatinine | 0.076 | <0.00001 | <0.001 |
| Glutamine | Lysine | 0.076 | <0.00001 | <0.001 |
| PC aaC40:4 | PC aaC42:5 | 0.076 | <0.00001 | <0.001 |
| Citrulline | Glycine | 0.076 | <0.00001 | <0.001 |
| AC C18 | Taurine | 0.076 | <0.00001 | <0.001 |
| PC ae C42:4 | PC ae C42:5 | 0.076 | <0.00001 | <0.001 |
| LysoPC a C16:0 | LysoPC a C17:0 | 0.075 | <0.00001 | <0.001 |
| PC aa C42:0 | PC aa C42:1 | 0.075 | <0.00001 | <0.001 |
| PC ae C40:4 | PC ae C42:5 | 0.075 | <0.00001 | <0.001 |
| Asparagine | Serine | 0.074 | <0.00001 | <0.001 |
| SM C16:1 | SM C18:1 | 0.074 | <0.00001 | <0.001 |
| PC aa C38:4 | PC aa C40:4 | 0.073 | <0.00001 | <0.001 |
| LysoPC a C18:1 | LysoPC a C20:4 | 0.073 | <0.00001 | 0.001 |
| PC ae C36:5 | PC ae C38:6 | 0.073 | <0.00001 | 0.001 |
| AC C16:1 | AC C4:1 | 0.073 | <0.00001 | 0.001 |
| PC aa C42:0 | PC ae C44:6 | 0.073 | <0.00001 | 0.001 |
| LysoPC a C18:0 | LysoPC a C20:4 | 0.073 | <0.00001 | 0.001 |
| PC aa C32:2 | PC aa C34:4 | 0.073 | <0.00001 | <0.01 |
| LysoPC a C17:0 | LysoPC a C18:0 | 0.072 | <0.00001 | <0.01 |
| PC aa C38:6 | PC ae C40:6 | 0.072 | <0.00001 | <0.01 |
| PC aa C40:4 | PC aa C42:6 | 0.072 | <0.00001 | <0.01 |
| AC C12 | AC C14 | 0.071 | <0.00001 | <0.01 |
| AC C16:1 | AC C4-OH (C3DC) | 0.071 | <0.00001 | <0.01 |
| PC aa C40:6 | PC ae C40:6 | 0.071 | <0.00001 | <0.01 |
| PC aa C38:0 | PC ae C40:6 | 0.071 | <0.00001 | <0.01 |
| SM C16:0 | SM C16:1 | 0.070 | <0.00001 | <0.01 |
| PC ae C40:5 | PC ae C40:6 | 0.070 | <0.00001 | <0.01 |
| PC aa C36:2 | PC ae C34:3 | 0.070 | <0.00001 | <0.01 |
| PC aa C40:3 | PC aa C42:5 | 0.070 | <0.00001 | <0.01 |
| PC aa C36:6 | PC ae C38:0 | 0.070 | <0.00001 | <0.01 |
| SM C16:0 | SM C18:0 | 0.070 | <0.00001 | <0.01 |
| PC aa C40:2 | PC aa C40:3 | 0.070 | <0.00001 | <0.01 |
| PC aa C38:5 | PC aa C40:4 | 0.069 | <0.00001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C16:1 | 0.069 | <0.00001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C20:4 | 0.069 | <0.00001 | <0.01 |
| AC C10:2 | Kynurenine | 0.068 | <0.00001 | <0.01 |
| PC ae C38:4 | PC ae C38:5 | 0.068 | <0.00001 | <0.01 |
| PC ae C42:5 | PC ae C44:4 | 0.067 | <0.00001 | <0.01 |
| Glutamine | Serine | 0.067 | <0.00001 | <0.01 |
| PC aa C36:3 | PC aa C38:3 | 0.067 | <0.00001 | <0.01 |
| AC C3 | AC C4 | 0.067 | <0.00001 | <0.01 |
| PC aa C40:2 | PC aa C42:4 | 0.067 | <0.00001 | <0.01 |
| PC aa C40:2 | PC ae C42:1 | 0.067 | <0.00001 | <0.01 |
| AC C14 | AC C16 | 0.066 | <0.00001 | <0.01 |
| PC aa C38:3 | PC aa C40:4 | 0.066 | <0.00001 | <0.01 |
| PC aa C32:0 | PC ae C32:1 | 0.066 | <0.00001 | <0.01 |
| PC aa C38:4 | PC aa C40:5 | 0.066 | <0.00001 | <0.01 |
| Arginine | Methionine | 0.066 | <0.00001 | <0.01 |
| SM C16:0 | SM C24:1 | 0.065 | <0.00001 | <0.01 |
| AC C16 | AC C18:1 | 0.065 | <0.00001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C18:1 | 0.065 | <0.00001 | <0.01 |
| Leucine | Serine | 0.065 | <0.00001 | <0.01 |
| PC aa C38:4 | PC aa C38:5 | 0.065 | <0.00001 | <0.01 |
| PC ae C42:1 | PC ae C42:2 | 0.064 | <0.00001 | <0.01 |
| Glutamine | Histidine | 0.064 | <0.00001 | <0.01 |
| PC ae C36:4 | PC ae C36:5 | 0.064 | 0.0001 | <0.01 |
| Histidine | Lysine | 0.064 | 0.0001 | <0.01 |
| PC aa C42:4 | PC aa C42:5 | 0.064 | 0.0001 | <0.01 |
| PC aa C40:3 | PC aa C42:4 | 0.064 | 0.0001 | <0.01 |
| PC aa C34:2 | PC aa C36:4 | 0.064 | 0.0001 | <0.01 |
| PC ae C40:2 | SM (-OH) C24:1 | 0.064 | 0.0001 | <0.01 |
| Methionine | Tryptophan | 0.064 | 0.0001 | <0.01 |
| PC aa C38:3 | PC aa C40:5 | 0.064 | 0.0001 | <0.01 |
| PC aa C40:2 | PC ae C42:2 | 0.064 | 0.0001 | <0.01 |
| LysoPC a C20:4 | PC aa C36:4 | 0.063 | 0.0001 | <0.01 |
| PC aa C42:0 | PC ae C42:5 | 0.063 | 0.0001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C20:3 | 0.063 | 0.0001 | <0.01 |
| Methionine | Phenylalanine | 0.063 | 0.0001 | <0.01 |
| PC aa C32:1 | PC aa C36:1 | 0.063 | 0.0001 | <0.01 |
| Lysine | Methionine | 0.063 | 0.0001 | <0.01 |
| AC C14 | AC C14:1-OH | 0.063 | 0.0001 | 0.01 |
| PC ae C36:4 | PC ae C38:4 | 0.063 | 0.0001 | 0.01 |
| PC aa C36:1 | PC ae C34:1 | 0.062 | 0.0001 | 0.01 |
| PC aa C36:4 | PC aa C38:5 | 0.062 | 0.0001 | 0.01 |
| SM C18:1 | SM (OH) C16:1 | 0.062 | 0.0001 | <0.05 |
| PC ae C40:5 | PC ae C42:5 | 0.062 | 0.0001 | <0.05 |
| AC C2 | AC C4:1 | 0.062 | 0.0001 | <0.05 |
| LysoPC a C16:1 | LysoPC a C18:1 | 0.062 | 0.0001 | <0.05 |
| PC aa C32:2 | PC aa C36:6 | 0.062 | 0.0001 | <0.05 |
| PC aa C32:1 | PC aa C34:1 | 0.062 | 0.0001 | <0.05 |
| PC ae C36:5 | PC ae C38:5 | 0.062 | 0.0001 | <0.05 |
| PC ae C38:2 | PC ae C40:3 | 0.062 | 0.0001 | <0.05 |
| PC aa C36:0 | PC aa C38:0 | 0.061 | 0.0001 | <0.05 |
| AC C10 | AC C12:1 | 0.061 | 0.0001 | <0.05 |
| PC aa C34:1 | PC ae C36:0 | 0.061 | 0.0001 | <0.05 |
| Arginine | Glycine | 0.061 | 0.0001 | <0.05 |
| PC ae C34:0 | PC ae C34:1 | 0.061 | 0.0001 | <0.05 |
| Arginine | Glutamine | 0.061 | 0.0001 | <0.05 |
| AC C18 | AC C18:1 | 0.061 | 0.0001 | <0.05 |
| PC aa C38:1 | PC aa C40:2 | 0.061 | 0.0001 | <0.05 |
| AC C12 | AC C14:1 | 0.061 | 0.0001 | <0.05 |
| PC aa C36:4 | PC ae C36:5 | 0.061 | 0.0001 | <0.05 |
| PC ae C38:6 | PC ae C40:6 | 0.061 | 0.0001 | <0.05 |
| SM (OH) C16:1 | SM (OH) C22:2 | 0.061 | 0.0001 | <0.05 |
| Phenylalanine | Proline | 0.061 | 0.0001 | <0.05 |
| PC ae C36:4 | PC ae C38:6 | 0.061 | 0.0001 | <0.05 |
| PC aa C40:2 | PC aa C42:2 | 0.061 | 0.0001 | <0.05 |
| PC ae C32:1 | PC ae C34:1 | 0.060 | 0.0001 | <0.05 |
| PC aa C30:0 | PC aa C32:2 | 0.060 | 0.0001 | <0.05 |
| AC C4:1 | Hexoses | 0.060 | 0.0001 | <0.05 |
| PC aa C40:4 | PC aa C42:4 | 0.060 | 0.0001 | <0.05 |
| AC C4 | AC C6:1 | 0.060 | 0.0001 | <0.05 |
| PC aa C32:3 | PC aa C34:4 | 0.060 | 0.0001 | <0.05 |
| AC C12 | AC C12:1 | 0.060 | 0.0001 | <0.05 |
| PC aa C36:0 | PC aa C36:1 | −0.061 | 0.0001 | <0.05 |
| Carnitine | PC ae C36:0 | −0.062 | 0.0001 | <0.05 |
| AC C4:1 | Phenylalanine | −0.075 | <0.00001 | <0.001 |
| Creatinine | SM (OH) C24:1 | −0.078 | <0.00001 | <0.001 |
| Maternal correlations | ||||
| PC aa C40:2 | PC aa C40:3 | 0.112 | <0.00001 | <0.0001 |
| Methionine | Threonine | 0.111 | <0.00001 | <0.0001 |
| PC ae C34:2 | PC ae C36:3 | 0.107 | <0.00001 | <0.0001 |
| PC aa C40:3 | PC aa C42:5 | 0.099 | <0.00001 | <0.0001 |
| Aspartate | Taurine | 0.099 | <0.00001 | <0.0001 |
| PC ae C42:4 | PC ae C44:4 | 0.098 | <0.00001 | <0.0001 |
| PC ae C42:5 | PC ae C44:5 | 0.098 | <0.00001 | <0.0001 |
| SM C16:1 | SM C18:1 | 0.094 | <0.00001 | <0.0001 |
| SM C18:1 | SM C20:2 | 0.094 | <0.00001 | <0.0001 |
| PC ae C36:4 | PC ae C36:5 | 0.094 | <0.00001 | <0.0001 |
| SM C24:0 | SM (OH) C22:1 | 0.093 | <0.00001 | <0.0001 |
| PC ae C36:4 | PC ae C38:5 | 0.093 | <0.00001 | <0.0001 |
| Alanine | Threonine | 0.093 | <0.00001 | <0.0001 |
| PC aa C40:4 | PC aa C40:5 | 0.092 | <0.00001 | <0.0001 |
| Carnitine | AC C2 | 0.092 | <0.00001 | <0.0001 |
| SM C18:0 | SM C18:1 | 0.091 | <0.00001 | <0.0001 |
| Asparagine | Threonine | 0.091 | <0.00001 | <0.0001 |
| Isoleucine | Leucine | 0.090 | <0.00001 | <0.0001 |
| Leucine | Valine | 0.089 | <0.00001 | <0.0001 |
| PC ae C34:3 | PC ae C36:5 | 0.089 | <0.00001 | <0.0001 |
| PC aa C36:4 | PC aa C38:4 | 0.089 | <0.00001 | <0.0001 |
| SM (OH) C22:1 | SM (OH) C24:1 | 0.088 | <0.00001 | <0.0001 |
| PC ae C40:4 | PC ae C42:4 | 0.088 | <0.00001 | <0.0001 |
| SM C26:1 | SM (OH) C24:1 | 0.088 | <0.00001 | <0.0001 |
| Methionine | Tyrosine | 0.088 | <0.00001 | <0.0001 |
| PC ae C40:2 | SM C26:1 | 0.087 | <0.00001 | <0.0001 |
| AC C16 | AC C18 | 0.086 | <0.00001 | <0.0001 |
| SM (OH) C14:1 | SM (OH) C16:1 | 0.086 | <0.00001 | <0.0001 |
| PC aa C36:6 | PC ae C38:0 | 0.086 | <0.00001 | <0.0001 |
| PC ae C40:2 | SM (OH) C24:1 | 0.085 | <0.00001 | <0.0001 |
| Glycine | Serine | 0.085 | <0.00001 | <0.0001 |
| PC aa C38:6 | PC aa C40:6 | 0.085 | <0.00001 | <0.0001 |
| PC ae C44:4 | PC ae C44:5 | 0.084 | <0.00001 | <0.0001 |
| PC aa C36:5 | PC aa C36:6 | 0.084 | <0.00001 | <0.0001 |
| PC ae C36:4 | PC ae C38:4 | 0.084 | <0.00001 | <0.0001 |
| Citrulline | Threonine | 0.083 | <0.00001 | <0.0001 |
| Asparagine | Methionine | 0.083 | <0.00001 | <0.0001 |
| PC ae C36:2 | PC ae C38:2 | 0.082 | <0.00001 | <0.0001 |
| Isoleucine | Valine | 0.082 | <0.00001 | 0.0001 |
| SM C16:1 | SM C20:2 | 0.081 | <0.00001 | 0.0001 |
| AC C3 | AC C4 | 0.081 | <0.00001 | 0.0001 |
| SM C24:0 | SM C24:1 | 0.081 | <0.00001 | 0.0001 |
| PC ae C32:1 | PC ae C32:2 | 0.080 | <0.00001 | <0.001 |
| PC ae C38:4 | PC ae C40:4 | 0.079 | <0.00001 | <0.001 |
| AC C12 | AC C2 | 0.079 | <0.00001 | <0.001 |
| AC C4:1 | AC C6:1 | 0.078 | <0.00001 | <0.001 |
| LysoPC a C16:0 | LysoPC a C18:0 | 0.077 | <0.00001 | <0.001 |
| Citrulline | Methionine | 0.076 | <0.00001 | <0.001 |
| PC aa C32:2 | PC aa C34:4 | 0.076 | <0.00001 | <0.001 |
| SM C24:0 | SM (OH) C24:1 | 0.075 | <0.00001 | <0.001 |
| PC ae C34:2 | PC ae C34:3 | 0.075 | <0.00001 | <0.001 |
| Alanine | AC C10:2 | 0.075 | <0.00001 | <0.001 |
| AC C16 | AC C18:1 | 0.075 | <0.00001 | <0.001 |
| PC ae C42:2 | PC ae C42:3 | 0.075 | <0.00001 | <0.001 |
| PC ae C34:3 | PC ae C36:3 | 0.075 | <0.00001 | <0.001 |
| PC aa C36:5 | PC ae C38:0 | 0.075 | <0.00001 | <0.001 |
| PC aa C34:4 | PC aa C36:6c | 0.074 | <0.00001 | <0.001 |
| PC aa C38:5 | PC aa C40:5 | 0.074 | <0.00001 | <0.001 |
| PC ae C40:1 | PC ae C42:1 | 0.074 | <0.00001 | <0.001 |
| LysoPC a C18:1 | LysoPC a C18:2 | 0.074 | <0.00001 | <0.001 |
| AC C14 | AC C16 | 0.073 | <0.00001 | <0.001 |
| PC ae C42:4 | PC ae C44:5 | 0.073 | <0.00001 | <0.001 |
| LysoPC a C17:0 | LysoPC a C18:0 | 0.073 | <0.00001 | <0.001 |
| SM C20:2 | SM (OH) C22:2 | 0.072 | <0.00001 | 0.001 |
| SM C24:1 | SM C26:1 | 0.071 | <0.00001 | 0.001 |
| AC C10 | AC C12:1 | 0.071 | <0.00001 | <0.01 |
| PC aa C40:2 | PC aa C42:5 | 0.071 | <0.00001 | <0.01 |
| AC C10 | AC C10:1 | 0.071 | <0.00001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C17:0 | 0.071 | <0.00001 | <0.01 |
| PC ae C36:3 | PC ae C36:4 | 0.070 | <0.00001 | <0.01 |
| PC ae C36:5 | PC ae C38:6 | 0.070 | <0.00001 | <0.01 |
| AC C18:1 | AC C18:2 | 0.070 | <0.00001 | <0.01 |
| PC aa C38:0 | PC ae C38:6 | 0.070 | <0.00001 | <0.01 |
| AC C14 | AC C14:1-OH | 0.069 | <0.00001 | <0.01 |
| AC C10:1 | AC C12:1 | 0.069 | <0.00001 | <0.01 |
| PC aa C34:1 | PC aa C36:1 | 0.069 | <0.00001 | <0.01 |
| PC aa C36:0 | PC ae C38:6 | 0.069 | <0.00001 | <0.01 |
| PC ae C36:5 | PC ae C38:5 | 0.069 | <0.00001 | <0.01 |
| PC aa C40:2 | PC aa C42:4 | 0.068 | <0.00001 | <0.01 |
| PC aa C34:2 | PC aa C36:2 | 0.068 | <0.00001 | <0.01 |
| PC ae C40:5 | PC ae C42:5 | 0.068 | <0.00001 | <0.01 |
| LysoPC a C16:1 | LysoPC a C20:3 | 0.068 | <0.00001 | <0.01 |
| PC ae C34:2 | PC ae C36:4 | 0.068 | <0.00001 | <0.01 |
| PC ae C40:4 | PC ae C44:5 | 0.068 | <0.00001 | <0.01 |
| LysoPC a C16:0c | LysoPC a C18:1 | 0.068 | <0.00001 | <0.01 |
| PC ae C40:4 | PC ae C44:4 | 0.067 | <0.00001 | <0.01 |
| Kynurenine | Tyrosine | 0.067 | <0.00001 | <0.01 |
| PC aa C42:0 | PC aa C42:1 | 0.067 | <0.00001 | <0.01 |
| PC ae C40:4 | PC ae C42:5 | 0.066 | <0.00001 | <0.01 |
| PC ae C42:1 | PC ae C44:3 | 0.066 | <0.00001 | <0.01 |
| AC C16 | AC C2 | 0.066 | <0.00001 | <0.01 |
| Asparagine | AC C10:2 | 0.066 | <0.00001 | <0.01 |
| PC aa C42:0 | PC ae C44:6 | 0.066 | <0.00001 | <0.01 |
| Carnitine | Taurine | 0.066 | <0.00001 | <0.01 |
| Carnitine | AC C3 | 0.066 | <0.00001 | <0.01 |
| PC aa C42:5 | PC aa C42:6 | 0.066 | <0.00001 | <0.01 |
| Aspartate | Ornithine | 0.066 | <0.00001 | <0.01 |
| Acetylornithine | Glycine | 0.065 | <0.00001 | <0.01 |
| PC aa C30:0 | PC ae C30:0 | 0.065 | <0.00001 | <0.01 |
| Acetylornithine | AC C4 | 0.065 | <0.00001 | <0.01 |
| PC aa C32:3 | PC aa C34:3 | 0.065 | <0.00001 | <0.01 |
| Glutamine | Serine | 0.065 | <0.00001 | <0.01 |
| LysoPC a C18:1 | LysoPC a C20:4 | 0.065 | <0.00001 | <0.01 |
| PC aa C42:4 | PC aa C42:5 | 0.065 | <0.00001 | <0.01 |
| PC ae C42:4 | PC ae C42:5 | 0.065 | <0.00001 | <0.01 |
| AC C4:1 | Hexoses | 0.065 | <0.00001 | <0.01 |
| Arginine | Histidine | 0.064 | <0.00001 | <0.01 |
| LysoPC a C16:0 | LysoPC a C18:2 | 0.064 | <0.00001 | <0.01 |
| PC aa C38:5 | PC aa C42:6 | 0.064 | 0.0001 | <0.01 |
| LysoPC a C14:0 | LysoPC a C16:1 | 0.064 | 0.0001 | <0.01 |
| PC aa C38:6 | PC ae C38:0 | 0.064 | 0.0001 | <0.01 |
| Arginine | Threonine | 0.064 | 0.0001 | <0.01 |
| AC C3 | AC C6:1 | 0.064 | 0.0001 | <0.01 |
| PC ae C42:1 | PC ae C42:2 | 0.064 | 0.0001 | <0.01 |
| AC C10 | AC C12 | 0.064 | 0.0001 | <0.01 |
| Alanine | Asparagine | 0.063 | 0.0001 | <0.01 |
| PC ae C40:3 | PC ae C44:4 | 0.063 | 0.0001 | <0.01 |
| SM C16:1 | SM (OH) C22:2 | 0.063 | 0.0001 | <0.01 |
| PC ae C36:4 | PC ae C38:6 | 0.063 | 0.0001 | <0.01 |
| PC aa C42:0 | PC ae C42:5 | 0.063 | 0.0001 | 0.01 |
| LysoPC a C20:3 | LysoPC a C20:4 | 0.062 | 0.0001 | 0.01 |
| PC aa C30:0 | PC ae C34:0 | 0.062 | 0.0001 | 0.01 |
| AC C16:1 | AC C6:1 | 0.062 | 0.0001 | 0.01 |
| PC aa C38:4 | PC aa C40:4 | 0.062 | 0.0001 | 0.01 |
| SM C24:0 | SM C26:1 | 0.062 | 0.0001 | 0.01 |
| SM C20:2 | SM C24:1 | 0.062 | 0.0001 | 0.01 |
| AC C12 | AC C14 | 0.061 | 0.0001 | 0.01 |
| PC ae C42:5 | PC ae C44:6 | 0.061 | 0.0001 | 0.01 |
| LysoPC a C18:0 | LysoPC a C18:2 | 0.061 | 0.0001 | 0.01 |
| Isoleucine | Phenylalanine | 0.061 | 0.0001 | 0.01 |
| AC C14:1-OH | AC C7-DC | 0.061 | 0.0001 | 0.01 |
| AC C14:1-OH | AC C18 | 0.061 | 0.0001 | 0.01 |
| AC C16:1 | Phenylalanine | 0.061 | 0.0001 | 0.01 |
| PC aa C40:3 | PC aa C42:4 | 0.061 | 0.0001 | 0.01 |
| PC ae C30:0 | PC ae C34:0 | 0.061 | 0.0001 | 0.01 |
| AC C14 | AC C14:1 | 0.061 | 0.0001 | 0.01 |
| SM C16:0 | SM C16:1 | 0.061 | 0.0001 | 0.01 |
| Citrulline | Taurine | 0.060 | 0.0001 | <0.05 |
| PC ae C38:5 | PC ae C38:6 | 0.060 | 0.0001 | <0.05 |
| PC aa C36:5 | PC aa C38:5 | 0.060 | 0.0001 | <0.05 |
| LysoPC a C18:0 | LysoPC a C18:1 | 0.060 | 0.0001 | <0.05 |
| AC C18 | PC aa C42:5 | −0.060 | 0.0001 | <0.05 |
| Creatinine | PC ae C40:3 | −0.062 | 0.0001 | 0.01 |
| Carnitine | PC aa C32:0 | −0.062 | 0.0001 | 0.01 |
| Aspartate | PC aa C38:4 | −0.062 | 0.0001 | 0.01 |
| AC C18 | SM C20:2 | −0.067 | 0.0000 | <0.01 |
| Acetylornitine | SM C18:0 | −0.068 | 0.0000 | <0.01 |
| Aspartate | PC ae C38:4 | −0.070 | 0.0000 | <0.01 |
Appendix D
| Metabolites (AUC) | Mothers with Impaired Fasting Glucose or Impaired Glucose Tolerance | Mothers with Normal Glucose Tolerance | p-Value * | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | (SD) | ||
| Carnitine (C0) | 294.1 | 20.6 | 293.3 | 20.4 | 0.7 |
| Creatinine | 368.1 | 19.2 | 361.5 | 17.3 | 0.2 |
| PC ae C34:3 | 95.9 | 28.1 | 105.9 | 27.5 | 0.2 |
| 476.1 | 35.9 | 472.0 | 39.1 | 0.6 | |
| 116.3 | 22.3 | 103.4 | 19.4 | 0.05 | |
| 330.1 | 27.4 | 334.9 | 22.0 | 0.6 | |
Appendix E

References
- Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; Naghavi, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar]
- Melchior, H.; Kurch-Bek, D.; Mund, M. The Prevalence of Gestational Diabetes. Dtsch. Arztebl. Int. 2017, 114, 412–418. [Google Scholar] [PubMed]
- Hughes, A.E.; Nodzenski, M.; Beaumont, R.N.; Talbot, O.; Shields, B.M.; Scholtens, D.M.; Knight, B.A.; Lowe, W.L.; Hattersley, A.T.; Freathy, R.M. Fetal Genotype and Maternal Glucose Have Independent and Additive Effects on Birth Weight. Diabetes 2018, 67, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Ruchat, S.-M.; Hivert, M.-F.; Bouchard, L. Epigenetic programming of obesity and diabetes by in utero exposure to gestational diabetes mellitus. Nutr. Rev. 2013, 71, S88–S94. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Howard, A.G.; Herring, A.H.; Thompson, A.L.; Adair, L.S.; Popkin, B.M.; Aiello, A.E.; Zhang, B.; Gordon-Larsen, P. Longitudinal associations of away-from-home eating, snacking, screen time, and physical activity behaviors with cardiometabolic risk factors among Chinese children and their parents. Am. J. Clin. Nutr. 2017, 106, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Mei, H.; Xiu, L.; Svensson, V.; Xiong, Y.; Marcus, C.; Zhang, J.; Hagstromer, M. Physical activity in young children and their parents–An Early STOPP Sweden–China comparison study. Sci. Rep. 2016, 6, 29595. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Perng, W.; Ringham, B.M.; Smith, H.A.; Michelotti, G.; Kechris, K.M.; Dabelea, D. A prospective study of associations between in utero exposure to gestational diabetes mellitus and metabolomic profiles during late childhood and adolescence. Diabetol. 2019, 63, 296–312. [Google Scholar] [CrossRef]
- Marchlewicz, E.H.; Dolinoy, D.C.; Tang, L.; Milewski, S.; Jones, T.R.; Goodrich, J.M.; Soni, T.; Domino, S.E.; Song, P.X.K.; Burant, C.F.; et al. Lipid metabolism is associated with developmental epigenetic programming. Sci. Rep. 2016, 6, 34857. [Google Scholar] [CrossRef]
- Lowe, W.L., Jr.; Bain, J.R.; Nodzenski, M.; Reisetter, A.C.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Maternal BMI and Glycemia Impact the Fetal Metabolome. Diabetes Care 2017, 40, 902–910. [Google Scholar] [CrossRef]
- Voerman, E.; Jaddoe, V.W.V.; Uhl, O.; Shokry, E.; Horak, J.; Felix, J.F.; Koletzko, B.; Gaillard, R. A population-based resource for intergenerational metabolomics analyses in pregnant women and their children: The Generation R Study. Metabolomics 2020, 16, 1–14. [Google Scholar] [CrossRef]
- Nowak, C.; Hetty, S.; Salihovic, S.; Castillejo-López, C.; Ganna, A.; Cook, N.; Broeckling, C.D.; Prenni, J.E.; Shen, X.; Giedraitis, V.; et al. Glucose challenge metabolomics implicates medium-chain acylcarnitines in insulin resistance. Sci. Rep. 2018, 8, 8691. [Google Scholar] [CrossRef]
- Krug, S.; Kastenmüller, G.; Stückler, F.; Rist, M.J.; Skurk, T.; Sailer, M.; Raffler, J.; Römisch-Margl, W.; Adamski, J.; Prehn, C.; et al. The dynamic range of the human metabolome revealed by challenges. FASEB J. 2012, 26, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Ejigu, B.A.; Valkenborg, D.; Baggerman, G.; Vanaerschot, M.; Witters, E.; Dujardin, J.-C.; Burzykowski, T.; Berg, M. Evaluation of Normalization Methods to Pave the Way Towards Large-Scale LC-MS-Based Metabolomics Profiling Experiments. OMICS A J. Integr. Biol. 2013, 17, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, Á. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics 2019, 15, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Merino, J.; Leong, A.; Liu, C.-T.; Porneala, B.; Walford, G.A.; Von Grotthuss, M.; Wang, T.J.; Flannick, J.; Dupuis, J.; Levy, D.; et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetol. 2018, 61, 1315–1324. [Google Scholar] [CrossRef]
- Cho, K.; Moon, J.S.; Kang, J.-H.; Jang, H.B.; Lee, H.-J.; Park, S.I.; Yu, K.-S.; Cho, J.-Y. Combined untargeted and targeted metabolomic profiling reveals urinary biomarkers for discriminating obese from normal-weight adolescents. Pediatr. Obes. 2016, 12, 93–101. [Google Scholar] [CrossRef]
- Mihalik, S.J.; Goodpaster, B.H.; Kelley, D.E.; Chace, D.H.; Vockley, J.; Toledo, F.G.; Delany, J.P. Increased Levels of Plasma Acylcarnitines in Obesity and Type 2 Diabetes and Identification of a Marker of Glucolipotoxicity. Obesity 2010, 18, 1695–1700. [Google Scholar] [CrossRef]
- Murakami, S. The physiological and pathophysiological roles of taurine in adipose tissue in relation to obesity. Life Sci. 2017, 186, 80–86. [Google Scholar] [CrossRef]
- Wahl, S.; Yu, Z.; Kleber, M.; Singmann, P.; Holzapfel, C.; He, Y.; Mittelstrass, K.; Polonikov, A.; Prehn, C.; Römisch-Margl, W.; et al. Childhood Obesity Is Associated with Changes in the Serum Metabolite Profile. Obes. Facts 2012, 5, 660–670. [Google Scholar] [CrossRef]
- Bentley-Lewis, R.; Huynh, J.; Xiong, G.; Lee, H.; Wenger, J.; Clish, C.; Nathan, D.; Thadhani, R.; Gerszten, R. Metabolomic profiling in the prediction of gestational diabetes mellitus. Diabetologia 2015, 58, 1329–1332. [Google Scholar] [CrossRef] [PubMed]
- Reinehr, T.; Wolters, B.; Knop, C.; Lass, N.; Hellmuth, C.; Harder, U.; Peissner, W.; Wahl, S.; Grallert, H.; Adamski, J.; et al. Changes in the serum metabolite profile in obese children with weight loss. Eur. J. Nutr. 2015, 54, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and Creatinine Metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Peter, A.; Fritsche, J.; Elcnerova, M.; Fritsche, A.; Häring, H.-U.; Schleicher, E.D.; Xu, G.; Lehmann, R. Changes of the plasma metabolome during an oral glucose tolerance test: Is there more than glucose to look at? Am. J. Physiol. Metab. 2009, 296, E384–E393. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Dietrich, S.; Wittenbecher, C.; Krumsiek, J.; Kühn, T.; Lacruz, M.E.; Kluttig, A.; Prehn, C.; Adamski, J.; Von Bergen, M.; et al. Comparison of metabolite networks from four German population-based studies. Int. J. Epidemiol. 2018, 47, 2070–2081. [Google Scholar] [CrossRef]
- Hummel, S.; Much, D.; Rossbauer, M.; Ziegler, A.-G.; Beyerlein, A. Postpartum Outcomes in Women with Gestational Diabetes and their Offspring: POGO Study Design and First-Year Results. Rev. Diabet. Stud. 2013, 10, 49–57. [Google Scholar] [CrossRef]
- Kromeyer-Hauschild, K.; Wabitsch, M.; Kunze, D.; Geller, F.; Geiß, H.C.; Hesse, V.; Von Hippel, A.; Jaeger, U.; Johnsen, D.; Korte, W.; et al. Perzentile für den Body-mass-Index für das Kindes- und Jugendalter unter Heranziehung verschiedener deutscher Stichproben. Mon. Kinderheilkd. 2001, 149, 807–818. [Google Scholar] [CrossRef]
- Agueusop, I.; Musholt, P.B.; Klaus, B.; Hightower, K.; Kannt, A. Short-term variability of the human serum metabolome depending on nutritional and metabolic health status. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Henderson, M.; Rabasa-Lhoret, R.; Bastard, J.-P.; Chiasson, J.-L.; Baillargeon, J.-P.; Hanley, J.; Lambert, M. Measuring insulin sensitivity in youth: How do the different indices compare with the gold-standard method? Diabetes Metab. 2011, 37, 72–78. [Google Scholar] [CrossRef]
- Yeckel, C.W.; Weiss, R.; Dziura, J.; Taksali, S.E.; Dufour, S.; Burgert, T.S.; Tamborlane, W.V.; Caprio, S. Validation of Insulin Sensitivity Indices from Oral Glucose Tolerance Test Parameters in Obese Children and Adolescents. J. Clin. Endocrinol. Metab. 2004, 89, 1096–1101. [Google Scholar] [CrossRef]
- Matsuda, M.; DeFronzo, R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: Comparison with the euglycemic insulin clamp. Diabetes Care 1999, 22, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Toeller, M.; Frisch, A.; Müller-Wieland, D. Fragebogen zur Erfassung der Nahrungsaufnahme in Risikogruppen (NARI). Diabetol. Stoffwechs. 2010, 5, 309–314. [Google Scholar] [CrossRef]
- Aston, L.M.; Jackson, D.; Monsheimer, S.; Whybrow, S.; Handjieva-Darlenska, T.; Kreutzer, M.; Kohl, A.; Papadaki, A.; Martinez, J.A.; Kunova, V.; et al. Developing a methodology for assigning glycaemic index values to foods consumed across Europe. Obes. Rev. 2010, 11, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.T.; Chiuve, S.E.; McCullough, M.L.; Rexrode, K.M.; Logroscino, G.; Hu, F.B. Adherence to a DASH-Style Diet and Risk of Coronary Heart Disease and Stroke in Women. Arch. Intern. Med. 2008, 168, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Frey, I.; Berg, A.; Grathwohl, D.; Keul, J. Freiburger Fragebogen zur körperlichen Aktivität—Entwicklung, Prüfung und Anwendung (Freiburg Questionnaire of physical activity—Development, evaluation and application). Soz. Prav. 1999, 44, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Opper, E.; Worth, A.; Wagner, M.; Bös, K. Motorik-Modul (MoMo) im Rahmen des Kinder- und Jugendgesundheitssurveys (KiGGS). Motorische Leistungsfähigkeit und körperlich-sportliche Aktivität von Kindern und Jugendlichen in Deutschland (The module “Motorik” in the German Health Interview and Examination Survey for Children and Adolescents (KiGGS). Motor fitness and physical activity of children and young people). Bundesgesundheitsblatt Gesundh. Gesundh. 2007, 50, 879–888. [Google Scholar]
- Shephard, R. 2011 Compendium of Physical Activities: A Second Update of Codes and MET Values. Yearb. Sports Med. 2012, 2012, 126–127. [Google Scholar] [CrossRef]
- Zukunft, S.; Prehn, C.; Röhring, C.; Möller, G.; De Angelis, M.H.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 2018, 14, 1–12. [Google Scholar] [CrossRef]
- Do, K.T.; Wahl, S.; Raffler, J.; Molnos, S.; Laimighofer, M.; Adamski, J.; Suhre, K.; Strauch, K.; Peters, A.; Gieger, C.; et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics 2018, 14, 1–18. [Google Scholar] [CrossRef]
- Benedetti, E.; Gerstner, N.; Pučić-Baković, M.; Keser, T.; Reiding, K.R.; Ruhaak, L.R.; Štambuk, T.; Selman, M.H.J.; Rudan, I.; Polašek, O.; et al. Systematic Evaluation of Normalization Methods for Glycomics Data Based on Performance of Network Inference. Metabolites 2020, 10, 271. [Google Scholar] [CrossRef]
- Krumsiek, J.; Suhre, K.; Illig, T.; Adamski, J.; Theis, F.J. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol. 2011, 5, 21. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| Mothers | Offspring | |||
|---|---|---|---|---|
| N | N | |||
| Female n (%) | 48 | 48 (100) | 48 | 23 (48) |
| Age at postpartum follow-up (years), median (IQR) | 48 | 42.1 (38.0; 44.2) | 48 | 8.0 (5.4; 8.8) |
| BMI (kg/m2, mothers), BMI-SDS (offspring), median (IQR) | 48 | 26.4 (22.6; 32.0) | 48 | 0.1 (−0.6; 0.8) |
| Overweight * n (%) | 48 | 28 (58) | 48 | 7 (15) |
| Glycemic load, median (IQR) | 47 | 145.1 (114.8; 179.3) | 31 | 100.3 (84.4; 131.1) |
| Physical activity (MET-h/week), median (IQR) | 37 | 29.0 (18.0; 43.4) | 20 | 71.0 (45.1; 103.6) |
| ISI-Matsuda median (IQR) | 48 | 5.9 (3.6; 9.0) | 43 | 10.3 (6.5; 13.8) |
| Mothers | Offspring | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | BMI | ISI-M | Glycemic Load | MET-h/Week | BMI-SDS | ISI-M | Glycemic Load | MET-h/Week |
| BMI/BMI-SDS | 1 | −0.69 * | 0.16 | −0.10 | 1 | −0.32* | 0.21 | −0.05 |
| ISI-M | −0.69 * | 1 | −0.22 | 0.05 | −0.32 * | 1 | −0.03 | −0.07 |
| Glycemic load | 0.16 | −0.22 | 1 | 0.2 | 0.21 | −0.03 | 1 | 0.51 * |
| MET-h/week | −0.10 | 0.05 | 0.2 | 1 | −0.05 | −0.07 | 0.51 * | 1 |
| Metabolite (AUC) | Dietary Glycemic Load | Physical Activity (MET-h/Week) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mothers | Offspring | Mothers | Offspring | |||||
| r * | p-Value | r * | p-Value | r * | p-Value | r * | p-Value | |
| Carnitine (C0) | −0.02 | 0.9 | 0.28 | 0.12 | 0.06 | 0.72 | 0.20 | 0.40 |
| Taurine | 0.01 | 0.93 | −0.21 | 0.25 | 0.07 | 0.81 | 0.30 | 0.20 |
| Proline | −0.06 | 0.69 | 0.07 | 0.72 | 0.04 | 0.81 | 0.30 | 0.20 |
| SM –(OH) C14:1 | 0.26 | 0.08 | −0.15 | 0.42 | 0.32 | 0.06 | −0.27 | 0.26 |
| Creatinine | 0.19 | 0.20 | 0.05 | 0.77 | 0.50 | <0.01 | 0.10 | 0.84 |
| PC ae C34:3 | −0.01 | 0.51 | 0.001 | 1.0 | 0.16 | 0.35 | 0.15 | 0.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ott, R.; Pawlow, X.; Weiß, A.; Hofelich, A.; Herbst, M.; Hummel, N.; Prehn, C.; Adamski, J.; Römisch-Margl, W.; Kastenmüller, G.; et al. Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring. Int. J. Mol. Sci. 2020, 21, 9647. https://doi.org/10.3390/ijms21249647
Ott R, Pawlow X, Weiß A, Hofelich A, Herbst M, Hummel N, Prehn C, Adamski J, Römisch-Margl W, Kastenmüller G, et al. Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring. International Journal of Molecular Sciences. 2020; 21(24):9647. https://doi.org/10.3390/ijms21249647
Chicago/Turabian StyleOtt, Raffael, Xenia Pawlow, Andreas Weiß, Anna Hofelich, Melanie Herbst, Nadine Hummel, Cornelia Prehn, Jerzy Adamski, Werner Römisch-Margl, Gabi Kastenmüller, and et al. 2020. "Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring" International Journal of Molecular Sciences 21, no. 24: 9647. https://doi.org/10.3390/ijms21249647
APA StyleOtt, R., Pawlow, X., Weiß, A., Hofelich, A., Herbst, M., Hummel, N., Prehn, C., Adamski, J., Römisch-Margl, W., Kastenmüller, G., Ziegler, A.-G., & Hummel, S. (2020). Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring. International Journal of Molecular Sciences, 21(24), 9647. https://doi.org/10.3390/ijms21249647







