The Osteoporosis/Microbiota Linkage: The Role of miRNA
Abstract
1. Introduction
1.1. General Consideration on Osteoporosis
1.2. The Microbiota
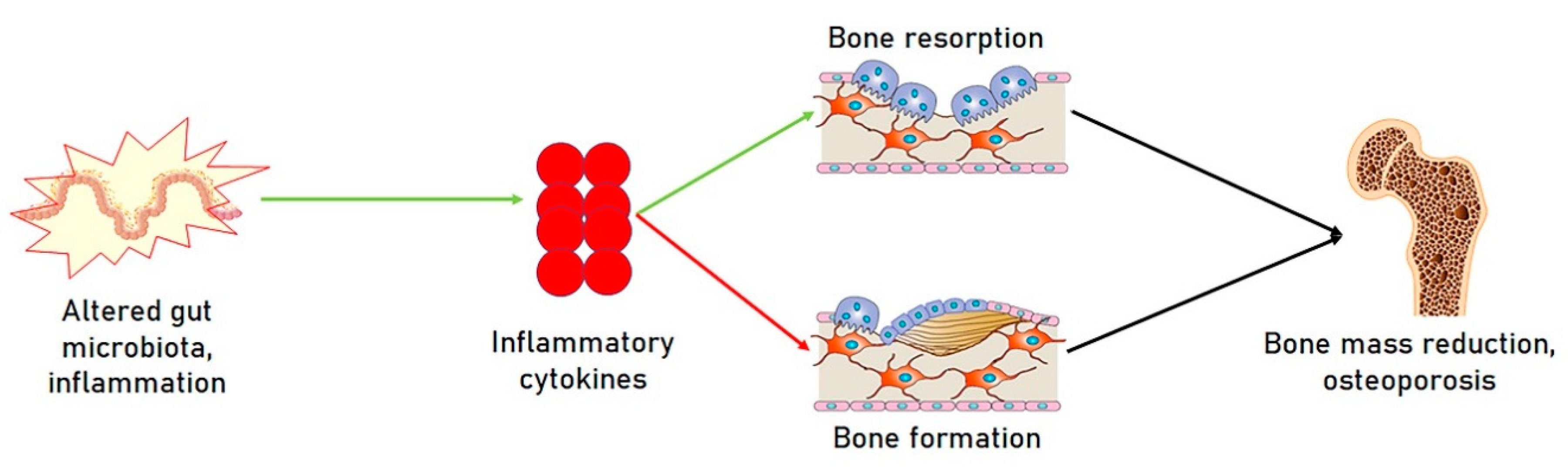
1.3. Microbiota and Osteoporosis
1.4. miRNAs and Osteoporosis
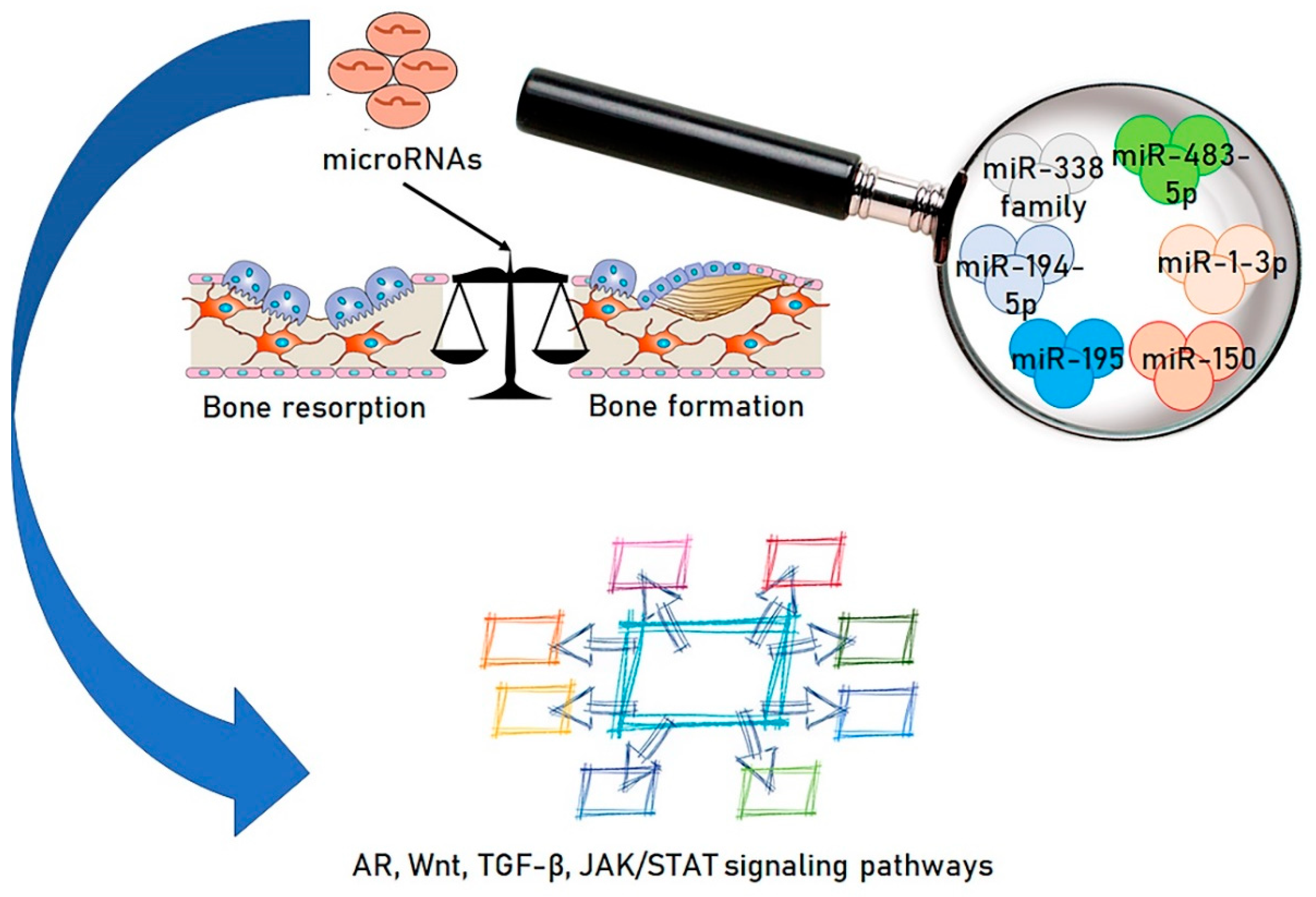
2. Microbiota and miRNAs; A Novel Functional Axis
3. Modifying the Microbiota/miRNAs Axis: A New Approach to Osteoporosis Therapy
4. Conclusions
Funding
Conflicts of Interest
References
- De Martinis, M.; Sirufo, M.M.; Ginaldi, L. Osteoporosis: Current and emerging therapies targeted to immunological checkpoints. Curr. Med. Chem. 2020, 27, 6356–6372. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Ginaldi, L.; Sirufo, M.M.; Bassino, E.M.; De Pietro, F.; Pioggia, G.; Gangemi, S. IL-33/Vitamin D Crosstalk in Psoriasis-associated Osteoporosis. Front. Immunol. 2020, in press. [Google Scholar]
- Ciccarelli, F.; De Martinis, M.; Ginaldi, L. Glucocorticoids in patients with rheumatic diseases: Friends or enemies of bone? Curr. Med. Chem. 2015, 22, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Sirufo, M.M.; Suppa, M.; Ginaldi, L.; De Martinis, M. Does Allergy Break Bones? Osteoporosis and Its Connection to Allergy. Int. J. Mol. Sci. 2020, 21, 712. [Google Scholar] [CrossRef]
- Ginaldi, L.; De Martinis, M.; Ciccarelli, F.; Saitta, S.; Imbesi, S.; Mannucci, C.; Gangemi, S. Increased levels of interleukin 31 (IL-31) in osteoporosis. BMC Immunol. 2015, 16, 60. [Google Scholar] [CrossRef]
- Irelli, A.; Sirufo, M.M.; D’Ugo, C.; Ginaldi, L.; De Martinis, M. Real-life use of denosumab 120 mg every 12 weeks in prolonged treatment over 2 years of patients with breast cancer bone metastases. J. Buon 2020, 25, 1799–1804. [Google Scholar]
- Irelli, A.; Sirufo, M.M.; Scipioni, T.; De Pietro, F.; Pancotti, A.; Ginaldi, L.; De Martinis, M. Breast cancer patients receiving denosumab during adjuvant aromatase inhibitors treatment: Who are the “inadequate responders” patients to denosumab? J. Buon 2020, 25, 648–654. [Google Scholar] [PubMed]
- Irelli, A.; Sirufo, M.M.; Scipioni, T.; De Pietro, F.; Pancotti, A.; Ginaldi, L.; De Martinis, M. Denosumab in breast cancer patients receiving aromatase inhibitors: A single-center observational study of effectiveness in adjuvant setting. Indian J. Cancer 2020. [Google Scholar] [CrossRef]
- Ginaldi, L.; De Martinis, M.; Saitta, S.; Sirufo, M.M.; Mannucci, C.; Casciaro, M.; Ciccarelli, F.; Gangemi, S. Interleukin-33 serum levels in postmenopausal women with osteoporosis. Sci. Rep. 2019, 9, 3786. [Google Scholar] [CrossRef]
- De Martinis, M.; Sirufo, M.M.; Suppa, M.; Ginaldi, L. IL-33/IL-31 Axis in Osteoporosis. Int. J. Mol. Sci. 2020, 21, 1239. [Google Scholar] [CrossRef]
- De Martinis, M.; Ginaldi, L.; Sirufo, M.M.; Pioggia, G.; Calapai, G.; Gangemi, S.; Mannucci, C. Alarmins in Osteoporosis, RAGE, IL-1, and IL-33 Pathways: A Literature Review. Medicina 2020, 56, 138. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Sirufo, M.M.; Polsinelli, M.; Placidi, G.; Di Silvestre, D.; Ginaldi, L. Gender differences in osteoporosis: A single-center observational study. WJMH 2020, in press. [Google Scholar]
- Hsu, E.; Pacifici, R. From osteoimmunology to osteomicrobiology: How the microbiota and the immune system regulate bone. Calcif. Tissue Int. 2018, 102, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Wang, G.; Zuo, X.; Qu, C.; Yao, B.; Wang, D. Gut microbiota: An overlooked factor that plays a significant role in osteoporosis. J. Int. Med. Res. 2019, 47, 4095–4103. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Innao, V.; Allegra, A.G.; Ettari, R.; Pugliese, M.; Pulvirenti, N.; Musolino, C. Role of the microbiota in hematologic malignancies. Neth. J. Med. 2019, 77, 67–80. [Google Scholar]
- Pacifici, R. Bone Remodeling and the Microbiome. Cold Spring Harb. Perspect. Med. 2018, 8, a031203. [Google Scholar] [CrossRef]
- Ginaldi, L.; De Martinis, M. Osteoimmunology and Beyond. Curr. Med. Chem. 2016, 23, 3754–3774. [Google Scholar] [CrossRef]
- Ohlsson, C.; Sjögren, K. Osteomicrobiology: A New Cross-Disciplinary Research Field. Calcif. Tissue Int. 2018, 102, 426–432. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 2017, 5, e1373208. [Google Scholar] [CrossRef]
- Yatsonsky, D.; Pan, K.; Shendge, V.B.; Jiayong, A.; Liu, J.; Ebraheim, N.A. Linkage of microbiota and osteoporosis: A mini literature review. World J. Orthop. 2019, 10, 123–127. [Google Scholar]
- Ding, K.; Hua, F.; Ding, W. Gut Microbiome and Osteoporosis. Aging Dis. 2020, 11, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Quach, D.; Britton, R.A. Gut Microbiota and Bone Health. Adv. Exp. Med. Biol. 2017, 1033, 47–58. [Google Scholar] [PubMed]
- Sjögren, K.; Engdahl, C.; Henning, P.; Lerner, U.H.; Tremaroli, V.; Lagerquist, M.K.; Bäckhed, F.; Ohlsson, C. The gut microbiota regulates bone mass in mice. J. Bone Miner. Res. 2012, 27, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jia, L.; Mo, L.; Zheng, L.; Yuan, Q.; Zhou, X. Intestinal microbiota: A potential target for the treatment of postmenopausal osteoporosis. Bone Res. 2017, 5, 17046. [Google Scholar] [CrossRef]
- Chen, Y.C.; Greenbaum, J.; Shen, H.; Deng, H.W. Association between Gut Microbiota and Bone Health: Potential Mechanisms and Prospective. J. Clin. Endocrinol. Metab. 2017, 102, 3635–3646. [Google Scholar] [CrossRef]
- Guss, J.D.; Taylor, E.; Rous, Z.; Roubert, S.; Higgins, C.H.; Thomas, C.J.; Baker, S.P.; Vashishth, D.; Donnelly, E.; Shea, M.K.; et al. The microbial metagenome and bone tissue composition in mice with microbiome-induced reductions in bone strength. Bone 2019, 127, 146–154. [Google Scholar] [CrossRef]
- D’Amelio, P.; Sassi, F. Gut Microbiota, Immune System, and Bone. Calcif. Tissue Int. 2018, 102, 415–442. [Google Scholar] [CrossRef]
- Li, L.; Rao, S.; Cheng, Y.; Zhuo, X.; Deng, C.; Xu, N.; Zhang, H.; Yang, L. Microbial osteoporosis: The interplay between the gut microbiota and bones via host metabolism and immunity. MicrobiologyOpen 2019, 8, e810. [Google Scholar] [CrossRef]
- Hernandez, C.J. The Microbiome and Bone and Joint Disease. Curr. Rheumatol. Rep. 2017, 19, 77. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. T reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.; Oshima, K.; Morita, H.; Fukuda, S.; Imaoka, A.; Kumar, N.; Sharma, V.K.; Kim, S.W.; Takahashi, M.; Saitou, N.; et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe 2011, 10, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Irelli, A.; Sirufo, M.M.; Scipioni, T.; De Pietro, F.; Pancotti, A.; Ginaldi, L.; De Martinis, M. mTOR Links Tumor Immunity and Bone Metabolism: What are the Clinical Implications? Int. J. Mol. Sci. 2019, 20, 5841. [Google Scholar] [CrossRef] [PubMed]
- Massimini, M.; Palmieri, C.; De Maria, R.; Romanucci, M.; Malatesta, D.; De Martinis, M.; Maniscalco, L.; Ciccarelli, A.; Ginaldi, L.; Buracco, P.; et al. 17-AAG and Apoptosis, Autophagy, and Mitophagy in Canine Osteosarcoma Cell Lines. Vet. Pathol. 2017, 54, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Herzog, J.W.; Tsang, K.; Brennan, C.A.; Bower, M.A.; Garrett, W.S.; Sartor, B.R.; Aliprantis, A.O.; Charles, J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. USA 2016, 113, 7554–7563. [Google Scholar] [CrossRef]
- Biver, E.; Berenbaum, F.; Valdes, A.M.; de Carvalho, I.A.; Bindels, L.B.; Brandi, M.L.; Calder, P.C.; Castronovo, V.; Cavalierj, E.; Cherubinik, A.; et al. Gut microbiota and osteoarthritis management: An expert consensus of the European society for clinical and economic aspects of osteoporosis, osteoarthritis and musculoskeletal diseases (ESCEO). Ageing Res. Rev. 2019, 55, 100946. [Google Scholar] [CrossRef]
- Li, J.-Y.; Chassaing, B.; Tyagi, A.M.; Vaccaro, C.; Luo, T.; Adams, J.; Trevor, T.M.; Weitzmann, M.N.; Mulle, J.G.; Gewirtz, A.T.; et al. Sex steroid deficiency–associated bone loss is microbiota dependent and prevented by probiotics. J. Clin. Investig. 2016, 126, 2049–2063. [Google Scholar] [CrossRef]
- Sirufo, M.M.; De Pietro, F.; Bassino, E.M.; Ginaldi, L.; De Martinis, M. Osteoporosis in Skin Diseases. Int. J. Mol. Sci. 2020, 21, 4749. [Google Scholar] [CrossRef]
- Waterhouse, M.; Hope, B.; Krause, L.; Morrison, M.; Protani, M.M.; Zakrzewski, M.; Neale, R.E. Vitamin D and the gut microbiome: A systematic review of in vivo studies. Eur. J. Nutr. 2019, 58, 2895–2910. [Google Scholar] [CrossRef]
- De Martinis, M.; Sirufo, M.M.; Nocelli, C.; Fontanella, L.; Ginaldi, L. Hyperhomocysteinemia is Associated with Inflammation, Bone Resorption, Vitamin B12 and Folate Deficiency and MTHFR C677T Polymorphism in Postmenopausal Women with Decreased Bone Mineral Density. Int. J. Environ. Res. Public Health 2020, 17, 4260. [Google Scholar] [CrossRef] [PubMed]
- Ducy, P. 5-HT and bone biology. Curr. Opin. Pharmacol. 2011, 11, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Ranuh, R.; Athiyyah, A.F.; Darma, A.; Risky, V.P.; Riawan, W.; Surono, I.S.; Sudarmo, S.M. Effect of the probiotic Lactobacillus plantarum IS-10506 on BDNF and 5HT stimulation: Role of intestinal microbiota on the gut-brain axis. Iran. J. Microbiol. 2019, 11, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Hata, T.; Asano, Y.; Yoshihara, K.; Kimura-Todani, T.; Miyata, N.; Zhang, X.T.; Takakura, S.; Aiba, Y.; Koga, Y.; Sudo, N. Regulation of gut luminal serotonin by commensal microbiota in mice. PLoS ONE 2017, 12, e0180745. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef]
- Jin, D.; Wu, S.; Zhang, Y.G.; Lu, R.; Xia, Y.; Dong, H.; Sun, J. Lack of vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin. Ther. 2015, 37, 996–1009. [Google Scholar] [CrossRef]
- Wang, J.; Xu, J.; Wang, B.; Shu, F.R.; Chen, K.; Mi, M.T. Equol promotes rat osteoblast proliferation and differentiation through activating estrogen receptor. Genet. Mol. Res. 2014, 13, 5055–5063. [Google Scholar] [CrossRef]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; Di Padova, F.; Lin, S.C.; Gram, H.; Han, J. Involvement of microRNA in AU-rich element-mediated mRNA stability. Cell 2005, 120, 623–634. [Google Scholar] [CrossRef]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer. Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar] [CrossRef]
- Batra, S.K.; Heier, C.R.; Diaz-Calderon, L.; Tully, C.B.; Fiorillo, A.A.; van den Anker, J.; Conklin, L.S. Serum miRNAs are Pharmacodynamic Biomarkers Associated With Therapeutic Response in Pediatric Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2020, 26, 1597–1606. [Google Scholar] [CrossRef]
- Musolino, C.; Oteri, G.; Allegra, A.; Mania, M.; D’Ascola, A.; Avenoso, A.; Innao, V.; Allegra, A.G.; Campo, S. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann. Hematol. 2018, 97, 1259–1269. [Google Scholar] [CrossRef]
- Cui, Q.; Xing, J.; Yu, M.; Wang, Y.; Xu, J.; Gu, Y.; Nan, X.; Ma, W.; Liu, H.; Zhao, H. Mmu-miR-185 depletion promotes osteogenic differentiation and suppresses bone loss in osteoporosis through the Bgn-mediated BMP/Smad pathway. Cell Death Dis. 2019, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-D.; Han, W.-X.; Liu, Y.-X. Suppression of miR-451a accelerates osteogenic differentiation and inhibits bone loss via Bmp6 signaling during osteoporosis. Biomed. Pharmacother. 2019, 120, 109378. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Wu, L.; Chen, H.; Huang, Z.; Xu, J.; Zhou, K.; Zhang, Y.; Chen, J.; Xia, J.; Yin, X. Identification of differentially expressed microRNAs in the bone marrow of osteoporosis patients. Am. J. Transl. Res. 2019, 11, 2940. [Google Scholar] [PubMed]
- Shao, M. Construction of a miRNA-regulated pathway network reveals candidate biomarkers for postmenopausal osteoporosis. Comput. Math. Methods Med. 2017, 2017, 9426280. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Karpinski, K.; Haug, A.T.; Vester, H.; Schmitt, A.; Bauer, J.S.; van Griensven, M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014, 29, 1718–1728. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, P.; Li, X.; Liang, Y.; Ouyang, K.; Xiong, J.; Wang, D.; Duan, L. MicroRNA expression profiling in an ovariectomized rat model of postmenopausal osteoporosis before and after estrogen treatment. Am. J. Transl. Res. 2020, 12, 4251–4263. [Google Scholar]
- Weilner, S.; Skalicky, S.; Salzer, B.; Keider, V.; Wagner, M.; Hildner, F.; Gabriel, C.; Dovjak, P.; Pietschmann, P.; Grillari-Voglauer, R.; et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone 2015, 79, 43–51. [Google Scholar] [CrossRef]
- Hensley, A.P.; McAlinden, A. The role of microRNAs in bone development. Bone 2020, 115760. [Google Scholar] [CrossRef]
- Ramírez-Salazar, E.G.; Carrillo-Patiño, S.; Hidalgo-Bravo, A.; Rivera-Paredez, B.; Quiterio, M.; Ramírez-Palacios, P.; Patiño, N.; Valdés-Flores, M.; Salmerón, J.; Velázquez-Cruz, R. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene 2018, 679, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kocijan, R.; Muschitz, C.; Geiger, E.; Skalicky, S.; Baierl, A.; Dormann, R.; Plachel, F.; Feichtinger, X.; Heimel, P.; Fahrleitner-Pammer, A.; et al. Circulating microRNA Signatures in Patients With Idiopathic and Postmenopausal Osteoporosis and Fragility Fractures. J. Clin. Endocrinol. Metab. 2016, 101, 4125–4134. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, X.; Muschitz, C.; Heimel, P.; Baierl, A.; Fahrleitner-Pammer, A.; Redl, H.; Resch, H.; Geiger, E.; Skalicky, S.; Dormann, R.; et al. Bone-related Circulating MicroRNAs miR-29b-3p, miR-550a-3p, and miR-324-3p and their Association to Bone Microstructure and Histomorphometry. Sci. Rep. 2018, 8, 4867. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; De Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019, 122, 52–75. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, Z.; Feng, S. Systematic analysis of miRNAs in patients with postmenopausal osteoporosis. Gynecol. Endocrinol. 2020, 28, 1–5. [Google Scholar] [CrossRef]
- Nakashima, H.; Ando, K.; Kobayashi, K.; Seki, T.; Ishizuka, S.; Fujii, R.; Takegami, Y.; Yamada, H.; Ando, Y.; Suzuki, K.; et al. Associations of Serum MicroRNA with Bone Mineral Density in Community-Dwelling Subjects: The Yakumo Study. BioMed Res. Int. 2020, 2020, 5047243. [Google Scholar] [CrossRef]
- Long, G.; Wang, F.; Duan, Q.; Yang, S.; Chen, F.; Gong, W.; Yang, X.; Wang, Y.; Chen, C.; Wang, D.W. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS ONE 2012, 7, e50926. [Google Scholar] [CrossRef]
- Ren, Y.; Li, H.; Xie, W.; Wei, N.; Liu, M. MicroRNA-195 triggers neuroinflammation in Parkinson’s disease in a Rho-associated kinase 1-dependent manner. Mol. Med. Rep. 2019, 19, 5153–5161. [Google Scholar] [CrossRef]
- Yu, W.; Liang, X.; Li, X.; Zhang, Y.; Sun, Z.; Liu, Y.; Wang, J. MicroRNA-195: A review of its role in cancers. OncoTargets Ther. 2018, 11, 7109–7123. [Google Scholar] [CrossRef]
- Ma, L. MicroRNA and Metastasis. Adv. Cancer Res. 2016, 132, 165–207. [Google Scholar] [CrossRef]
- Gu, Y.L.; Rong, X.X.; Wen, L.T.; Zhu, G.X.; Qian, M.Q. miR-195 inhibits the proliferation and migration of chondrocytes by targeting GIT1. Mol. Med. Rep. 2017, 15, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Grünhagen, J.; Bhushan, R.; Degenkolbe, E.; Jäger, M.; Knaus, P.; Mundlos, S.; Robinson, P.N.; Ott, C.E. MiR-497~195 cluster microRNAs regulate osteoblast differentiation by targeting BMP signaling. J. Bone Miner. Res. 2015, 30, 796–808. [Google Scholar] [CrossRef]
- Gu, H.; Shi, S.; Xiao, F.; Huang, Z.; Xu, J.; Chen, G.; Zhou, K.; Lu, L.; Yin, X. MiR-1-3p regulates the differentiation of mesenchymal stem cells to prevent osteoporosis by targeting secreted frizzled-related protein 1. Bone 2020, 137, 115444. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Lian, J.X.; Meng, S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell Mol. Biol. Lett. 2019, 24, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jia, L.; Zheng, Y.; Li, W. Involvement of Noncoding RNAs in the Differentiation of Osteoclasts. Stem Cells Int. 2020, 2020, 4813140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Song, H.Y.; Gao, L.L.; Yang, L.Y.; Mu, S.; Fu, Q. MicroRNA1005p inhibits osteoclastogenesis and bone resorption by regulating fibroblast growth factor 21. Int. J. Mol. Med. 2019, 43, 727–738. [Google Scholar] [PubMed]
- Lin, C.; Yu, S.; Jin, R.; Xiao, Y.; Pan, M.; Pei, F.; Zhu, X.; Huang, H.; Zhang, Z.; Chen, S.; et al. Circulating miR-338 cluster activities on osteoblast differentiation: Potential diagnostic and therapeutic targets for postmenopausal osteoporosis. Theranostics 2019, 9, 3780–3797. [Google Scholar] [CrossRef]
- Li, K.; Chen, S.; Cai, P.; Chen, K.; Li, L.; Yang, X.; Yi, J.; Luo, X.; Du, Y.; Zheng, H. MiRNA-483-5p is involved in the pathogenesis of osteoporosis by promoting osteoclast differentiation. Mol. Cell. Probes 2020, 49, 101479. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, D.; Pan, N.; Sun, N.; Wang, Q.; Fan, J.; Zhou, P.; Zhu, W.; Jiang, L. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. PeerJ 2015, 3, e971. [Google Scholar] [CrossRef]
- Anastasilakis, A.D.; Makras, P.; Pikilidou, M.; Tournis, S.; Makris, K.; Bisbinas, I.; Tsave, O.; Yovos, J.G.; Yavropoulou, M.P. Changes of Circulating MicroRNAs in Response to Treatment With Teriparatide or Denosumab in Postmenopausal Osteoporosis. J. Clin. Endocrinol. Metab. 2018, 103, 1206–1213. [Google Scholar] [CrossRef]
- Ma, J.; Lin, X.; Chen, C.; Li, S.; Zhang, S.; Chen, Z.; Li, D.; Zhao, F.; Yang, C.; Qiu, W.; et al. Circulating miR-181c-5p and miR-497-5p are potential biomarkers for prognosis and diagnosis of osteoporosis. J. Clin. Endocrinol. Metab. 2019, 105, dgz300. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Huang, G.; Na, N.; Chen, L. MicroRNA-214-5p/TGF-b/Smad2 signaling alters adipogenic differentiation of bone marrow stem cells in postmenopausal osteoporosis. Mol. Med. Rep. 2018, 17, 6301–6310. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Chen, W.-D.; Wang, Y.-D. Gut Microbiota: An Integral Moderator in Health and Disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Blanton, L.V.; Cao, S.; Zhao, G.; Manary, M.; Trehan, I.; Smith, M.I.; Wang, D.; Virgin, H.W.; Rohwer, F.; et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. USA 2015, 112, 11941–11946. [Google Scholar] [CrossRef]
- Blasco-Baque, V.; Coupé, B.; Fabre, A.; Handgraaf, S.; Gourdy, P.; Arnal, J.F.; Courtney, M.; Schuster-Klein, C.; Guardiola, B.; Tercé, F.; et al. Associations between hepatic miRNA expression, liver triacylglycerols and gut microbiota during metabolic adaptation to high-fat diet in mice. Diabetologia 2017, 60, 690–700. [Google Scholar] [CrossRef]
- Kelch, S.; Balmayor, E.R.; Seeliger, C.; Vester, H.; Kirschke, J.S.; van Griensven, M. miRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci. Rep. 2017, 7, 15861. [Google Scholar] [CrossRef]
- Panach, L.; Mifsut, D.; Tarin, J.J.; Cano, A.; Garcia-Perez, M.A. Serum Circulating MicroRNAs as Biomarkers of Osteoporotic Fracture. Calcif. Tissue Int. 2015, 97, 495–505. [Google Scholar] [CrossRef]
- Chen, R.; Liao, X.; Chen, F.; Wang, B.; Huang, J.; Jian, G.; Huang, Z.; Yin, G.; Liu, H.; Jin, D. Circulating microRNAs, miR-10b-5p, miR-328-3p, miR-100 and let-7, are associated with osteoblast differentiation in osteoporosis. Int. J. Clin. Exp. Pathol. 2018, 11, 1383–1390. [Google Scholar]
- Suarjana, I.N.; Isbagio, H.; Soewondo, P.; Rachman, I.A.; Sadikin, M.; Prihartono, J.; Malik, S.G.; Soeroso, J. The Role of Serum Expression Levels of Microrna-21 on Bone Mineral Density in Hypostrogenic Postmenopausal Women with Osteoporosis: Study on Level of RANKL, OPG, TGFβ-1, Sclerostin, RANKL/OPG Ratio, and Physical Activity. Acta Med. Indones. 2019, 51, 245–252. [Google Scholar]
- Zhang, D.; Lee, H.; Wang, X.; Groot, M.; Sharma, L.; Cruz, C.S.D.; Jin, Y. A potential role of microvesicle-containing miR-223/142 in lung inflammation. Thorax 2019, 74, 865–874. [Google Scholar] [CrossRef]
- Yavropoulou, M.P.; Anastasilakis, A.D.; Makras, P.; Tsalikakis, D.G.; Grammatiki, M.; Yovos, J.G. Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur. J. Endocrinol. 2017, 176, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Breunig, I.R.; Ohse, M.C.; Roubrocks, S.; Kleinfeld, S.; Roy, S.; Streetz, K.; Trautwein, C.; Roderburg, C.; Sellge, G. Faecal Micro-RNAs in Inflammatory Bowel Diseases. J. Crohns Colitis 2020, 14, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, H.; Cichon, C.; Ölschläger, T.A.; Sonnenborn, U.; Schmidt, M.A. Interleukin-8, CXCL1, and MicroRNA miR-146a Responses to Probiotic Escherichia coli Nissle 1917 and Enteropathogenic E. coli in Human Intestinal Epithelial T84 and Monocytic THP-1 Cells after Apical or Basolateral Infection. Infect. Immun. 2016, 84, 2482–2492. [Google Scholar] [CrossRef]
- Kuang, W.; Zheng, L.; Xu, X.; Lin, Y.; Lin, J.; Wu, J.; Tan, J. Dysregulation of the miR-146a-Smad4 axis impairs osteogenesis of bone mesenchymal stem cells under inflammation. Bone Res. 2017, 5, 17037. [Google Scholar] [CrossRef]
- Ahn, T.K.; Kim, J.O.; Kumar, H.; Choi, H.; Jo, M.J.; Sohn, S.; Ropper, A.E.; Kim, N.K.; Han, I.B. Polymorphisms of miR-146a, miR-149, miR-196a2, and miR-499 are associated with osteoporotic vertebral compression fractures in Korean postmenopausal women. J. Orthop. Res. 2018, 36, 244–253. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, M.; Zhang, X.; Xu, J.; Hu, G.; Zhao, X.; Cui, P.; Zhang, X. MiR-146a Deletion Protects From Bone Loss in OVX Mice by Suppressing RANKL/OPG and M-CSF in Bone Microenvironment. J. Bone Miner. Res. 2019, 34, 2149–2161. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Fu, Q.; Zhang, J. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers 2014, 19, 553–556. [Google Scholar] [CrossRef]
- Aguilar, C.; Cruz, A.R.; Rodrigues Lopes, I.; Maudet, C.; Sunkavalli, U.; Silva, R.J.; Sharan, M.; Lisowski, C.; Zaldívar-López, S.; Garrido, J.J.; et al. Functional screenings reveal different requirements for host microRNAs in Salmonella and Shigella infection. Nat. Microbiol. 2020, 5, 192–205. [Google Scholar] [CrossRef]
- Wang, Y.W.; Zhao, S.; Yuan, X.Y.; Liu, Y.; Zhang, K.; Wang, J.; Zhu, J.; Ma, R. miR-4732-5p promotes breast cancer progression by targeting TSPAN13. J. Cell Mol. Med. 2019, 23, 2549–2557. [Google Scholar] [CrossRef]
- Das, M.; Cronin, O.; Keohane, D.M.; Cormac, E.M.; Nugent, H.; Nugent, M.; Molloy, C.; Paul, W.; O’Toole, P.W.; Shanahan, F.; et al. Gut microbiota alterations associated with reduced bone mineral density in older adults. Rheumatology 2019, 58, 2295–2304. [Google Scholar] [CrossRef] [PubMed]
- De Ceulaer, J.; Tacconelli, E.; Vandecasteele, S.J. Actinomyces osteomyelitis in bisphosphonate-related osteonecrosis of the jaw (BRONJ): The missing link? Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1873–1880. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Fordham, J.B.; Khan, A.; Nares, S. MicroRNAs responsive to Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis LPS modulate expression of genes regulating innate immunity in human macrophages. Innate Immun. 2014, 20, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Heydari, Z.; Rahaie, M.; Alizadeh, A.M.; Agah, S.; Khalighfard, S.; Bahmani, S. Effects of Lactobacillus acidophilus and Bifidobacterium bifidum Probiotics on the Expression of MicroRNAs 135b, 26b, 18a and 155, and Their Involving Genes in Mice Colon Cancer. Probiotics Antimicrob. Proteins 2019, 11, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bai, Y.; Zhang, Z.; Lu, J. A validated miRNA signature for the diagnosis of osteoporosis related fractures using SVM algorithm classification. Exp. Ther. Med. 2020, 20, 2209–2217. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Zhu, Y.; Hao, W.; Chu, C.; Su, H. MicroRNA-155 inhibition up-regulates LEPR to inhibit osteoclast activation and bone resorption via activation of AMPK in alendronate-treated osteoporotic mice. IUBMB Life 2019, 71, 1916–1928. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; He, J.; Zeng, Z.; Yang, H.; Liu, Z.; Cao, Z.; Yu, H.; Zhao, W.; Pan, X. MiR-155 inhibition alleviates suppression of osteoblastic differentiation by high glucose and free fatty acids in human bone marrow stromal cells by upregulating SIRT1. Pflügers Archiv Euro. J. Physiol. 2020, 472, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhao, C.; Zhang, P.; Liu, Y.; Jiang, Y.; Wu, E.; Xue, H.; Liu, C.; Li, Z. miR-26b modulates OA induced BMSC osteogenesis through regulating GSK3β/β-catenin pathway. Exp. Mol. Pathol. 2019, 107, 158–164. [Google Scholar] [CrossRef]
- Campisi, G.; Chiappelli, M.; De Martinis, M.; Franco, V.; Ginaldi, L.; Guiglia, R.; Licastro, F.; Lio, D. Pathophysiology of age-related diseases. Immun. Ageing 2009, 6, 2. [Google Scholar] [CrossRef]
- Collins, F.L.; Rios-Arce, N.D.; Schepper, J.D.; Parameswaran, N.; McCabe, L.R. The potential of probiotics as a therapy for osteoporosis. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Weaver, C.M. Diet, Gut Microbiome, and Bone Health. Curr. Osteoporos. Rep. 2015, 13, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.L.; Irwin, R.; Bierhalter, H.; Schepper, J.; Britton, R.A.; Parameswaran, N.; McCabe, L.R. Lactobacillus reuteri 6475 Increases Bone Density in Intact Females Only under an Inflammatory Setting. PLoS ONE 2016, 11, e0153180. [Google Scholar] [CrossRef] [PubMed]
- Karius, T.; Schnekenburger, M.; Dicato, M.; Dieterich, M. MicroRNAs in cancer management and their modulation by dietary agents. Biochem. Pharmacol. 2012, 83, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.T.; Ye, X.L.; Li, R.R.; Chen, H.; Wang, Y.Y.; Yong, H.J.; Pan, M.L.; Lu, W.; Tang, Y.; Miao, H.; et al. Resveratrol Modulates the Gut Microbiota and Inflammation to Protect Against Diabetic Nephropathy in Mice. Front. Pharmacol. 2020, 11, 1249. [Google Scholar] [CrossRef]
- Latruffe, N.; Lançon, A.; Frazzi, R.; Aires, V.; Delmas, D.; Michaille, J.J.; Djouadi, F.; Bastin, J.; Cherkaoui-Malki, M. Exploring new ways of regulation by resveratrol involving miRNAs, with emphasis on inflammation. Ann. N. Y. Acad. Sci. 2015, 1348, 97–106. [Google Scholar] [CrossRef]
- Allegra, A.; Innao, V.; Russo, S.; Gerace, D.; Alonci, A.; Musolino, C. Anticancer Activity of Curcumin and Its Analogues: Preclinical and Clinical Studies. Cancer Investig. 2017, 35, 1–22. [Google Scholar] [CrossRef]
- Allegra, A.; Speciale, A.; Molonia, M.S.; Guglielmo, L.; Musolino, C.; Ferlazzo, G.; Costa, G.; Saija, A.; Cimino, F. Curcumin ameliorates the in vitro efficacy of carfilzomib in human multiple myeloma U266 cells targeting p53 and NF-κB pathways. Toxicol. In Vitro 2018, 47, 186–194. [Google Scholar] [CrossRef]
- Scazzocchio, B.; Minghetti, L.; D’Archivio, M. Interaction between Gut Microbiota and Curcumin: A New Key of Understanding for the Health Effects of Curcumin. Nutrients 2020, 12, 2499. [Google Scholar] [CrossRef]
- Li, J.; Wei, H.; Liu, Y.; Li, Q.; Guo, H.; Guo, Y.; Chang, Z. Curcumin Inhibits Hepatocellular Carcinoma via Regulating miR-21/TIMP3 Axis. Evid. Based Complement. Altern. Med. 2020, 17, 2892917. [Google Scholar] [CrossRef]
- Ma, F.; Liu, F.; Ding, L.; You, M.; Yue, H.; Zhou, Y.; Hou, Y. Anti-inflammatory effects of curcumin are associated with down regulating microRNA-155 in LPS-treated macrophages and mice. Pharm. Biol. 2017, 55, 1263–1273. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 2018, 24, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Teodori, L.; Petrignani, I.; Giuliani, A.; Prattichizzo, F.; Gurău, F.; Matacchione, G.; Olivieri, F.; Coppari, S.; Albertini, M.C. Inflamm-aging microRNAs may integrate signals from food and gut microbiota by modulating common signalling pathways. Mech. Ageing Dev. 2019, 182, 111127. [Google Scholar] [CrossRef] [PubMed]
- Ellur, G.; Sukhdeo, S.V.; Khan, M.T.; Sharan, K. Maternal high protein-diet programs impairment of offspring’s bone mass through miR-24-1-5p mediated targeting of SMAD5 in osteoblasts. Cell Mol. Life Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, M.; Sirufo, M.M.; Viscido, A.; Ginaldi, L. Food Allergies and Ageing. Int. J. Mol. Sci. 2019, 20, 5580. [Google Scholar] [CrossRef]
- De Martinis, M.; Sirufo, M.M.; Suppa, M.; Ginaldi, L. New Perspectives in Food Allergy. Int. J. Mol. Sci. 2020, 21, 1474. [Google Scholar] [CrossRef]
- De Martinis, M.; Sirufo, M.M.; Viscido, A.; Ginaldi, L. Food Allergy Insights: A Changing Landscape. Arch. Immunol. Ther. Exp. 2020, 68, 8–15. [Google Scholar] [CrossRef]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Z.; Wang, Y.; Hu, Z.; Zhou, H.; Zhang, L.; Hong, B.; Zhang, S.; Cao, X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci. Rep. 2016, 6, 23170–23178. [Google Scholar] [CrossRef]
- Xue, N.; Qi, L.; Zhang, G.; Zhang, Y. miRNA-125b regulates osteogenic differentiation of periodontal ligament cells through NKIRAS2/NF-κB pathway. Cell Physiol. Biochem. 2018, 48, 1771–1781. [Google Scholar] [CrossRef]
- Yang, Y.; Yujiao, W.; Fang, W.; Linhui, Y.; Ziqi, G.; Zhichen, W.; Zirui, W.; Shengwang, W. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol. Res. 2020, 53, 40. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Chen, L.; Xu, L. The roles of long non-coding RNA in osteoporosis. Curr. Stem Cell Res. Ther. 2020, 15, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Ai, L.; Qian, J.; Fang, J.Y.; Xu, J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 2015, 5, 11763. [Google Scholar] [CrossRef] [PubMed]
- De Nigris, F.; Ruosi, C.; Colella, G.; Napoli, C. Epigenetic therapies of osteoporosis. Bone 2020, 5, 115680. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Target(s) | Function(s) | Reference(s) |
|---|---|---|---|
| miR-1-3p | SFRP1 | Osteogenesis, adipogenesis, bone formation regulation | [73] |
| miR-21 | RANKL, TGF-Beta 1, OPG | Bone reabsorption | [88] |
| miR-26b | Wnt pathway | Osteogenesis | [107] |
| miR-100-5p | FGF-21 | Avoids bone loss | [75] |
| miR-125a-5p | TNFRSF1B | Increased osteoclast differentiation | [74] |
| miR-146a | Smad4 | Osteogenesis blockade | [94,95] |
| miR-155 | KRAS, TNF-α, RANK, IL-1beta, M-CSF, TRAP, and Bcl-2 | Stimulation of cell proliferation | [105,106] |
| miR-195 | GIT1 | Blocks the growth of chondrocytes | [71,72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Martinis, M.; Ginaldi, L.; Allegra, A.; Sirufo, M.M.; Pioggia, G.; Tonacci, A.; Gangemi, S. The Osteoporosis/Microbiota Linkage: The Role of miRNA. Int. J. Mol. Sci. 2020, 21, 8887. https://doi.org/10.3390/ijms21238887
De Martinis M, Ginaldi L, Allegra A, Sirufo MM, Pioggia G, Tonacci A, Gangemi S. The Osteoporosis/Microbiota Linkage: The Role of miRNA. International Journal of Molecular Sciences. 2020; 21(23):8887. https://doi.org/10.3390/ijms21238887
Chicago/Turabian StyleDe Martinis, Massimo, Lia Ginaldi, Alessandro Allegra, Maria Maddalena Sirufo, Giovanni Pioggia, Alessandro Tonacci, and Sebastiano Gangemi. 2020. "The Osteoporosis/Microbiota Linkage: The Role of miRNA" International Journal of Molecular Sciences 21, no. 23: 8887. https://doi.org/10.3390/ijms21238887
APA StyleDe Martinis, M., Ginaldi, L., Allegra, A., Sirufo, M. M., Pioggia, G., Tonacci, A., & Gangemi, S. (2020). The Osteoporosis/Microbiota Linkage: The Role of miRNA. International Journal of Molecular Sciences, 21(23), 8887. https://doi.org/10.3390/ijms21238887









