A Retrospective on eIF2A—and Not the Alpha Subunit of eIF2
Abstract
1. Introduction
2. Eukaryotic Initiation Factor 2 (eIF2)
3. eIF2A—Discovery and Initial Characterization
4. Identification of the Mammalian eIF2A Gene and the Yeast Homologue
5. eIF2A Function in Yeast S. cerevisiae
6. eIF2A Function in Mammalian Cellular Systems
7. eIF2A Function in Mammalian Systems in Non-AUG Initiation Events
8. eIF2A Knockout Mouse
9. eIF2A Structure
10. eIF2A Interacting Partners
11. eIF2A Modifications
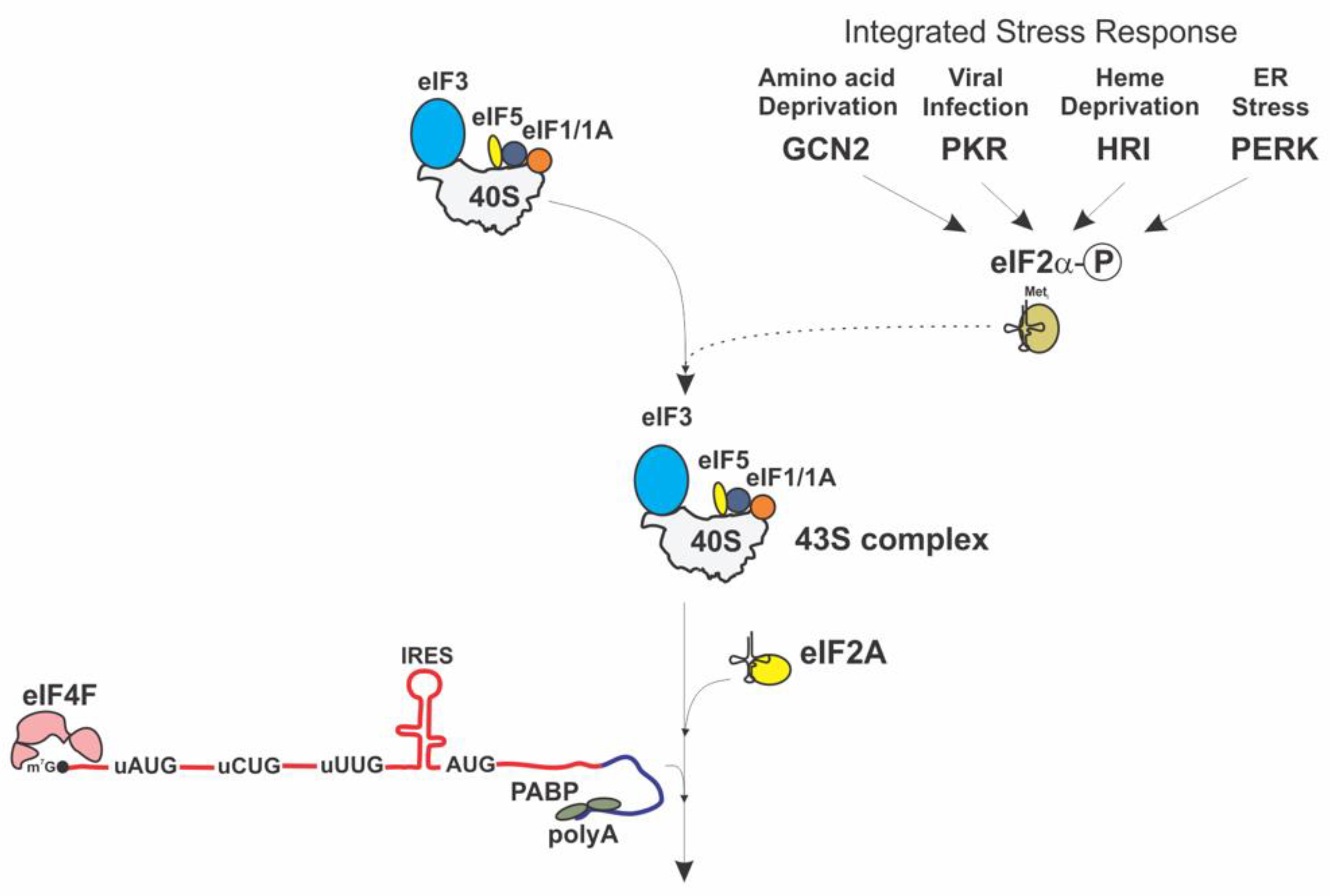
12. Alternate Initiation Pathways as a Response to Stress and Non-eIF2-Mediated Translation
13. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aa | amino acid |
| ABCE1 | ATP-Binding Cassette (ABC) protein E1 |
| ATF4 | Activating Transcription Factor 4 |
| Bcl-xL | B-cell lymphoma-extra Large protein |
| BiP | Binding immunoglobulin Protein |
| CTD | C-Terminal Domain |
| CM | CarboxyMethyl |
| CSFV | Classical Swine Fever Virus |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DDX3 | DEAD box RNA helicase 3 |
| DEAE | DiEthylAminoEthyl |
| DENR | DENsity-Regulated protein |
| eIF | Eukaryotic translation Initiation Factor |
| eEF | Eukaryotic translation Elongation Factor |
| ER | Endoplasmic Reticulum |
| FALS | Familial Amyotrophic Lateral Sclerosis |
| FTD | FrontoTemporal Dementia |
| fMet-tRNAi | Initiator N-Formylmethionine-tRNA |
| GCN4,GCN2 | General Control Protein 4 or 2 |
| GDP | Guanosine DiPhosphate |
| GIC1 | GTPase-Interacting Component 1 protein |
| GTP | Guanosine TriPhosphate |
| HCV | Hepatitis C Virus |
| HRE | Hexanucleotide Repeat |
| HRI | Heme Regulated Inhibitor protein |
| IC | Initiation Complex |
| IF | Initiation Factor (bacterial) |
| IRES | Internal Ribosomal Entry Site |
| ISR | Integrated Stress Response |
| KO | KnockOut |
| MCT-1 | Multiple Copies in T-cell Lymphoma-1 |
| MHC | Major Histocompatibility Complex |
| MW | Molecular Weight |
| nt | nucleotide |
| NTD | N-Terminal Domain |
| ORF | Open Reading Frame |
| PABP | Poly(A) Binding Protein |
| PERK | PKR-like Endoplasmic Reticulum (ER) Kinase |
| PIC | PreInitiation Complex |
| PKR | Protein Kinase R |
| PTEN | Phosphatase and TENsin homologue deleted on chromosome 10 protein |
| RACK1 | Receptor For Activated C Kinase 1 |
| SCC | Squamous Cell Carcinomas |
| SERP1 | Stress-associated Endoplasmic Reticulum Protein 1 |
| siRNA | Small Interfering RNA |
| SV | Sindbis Virus |
| SUMO | Small Ubiquitin Like Modifier |
| TC | Ternary Complex |
| TCGA | The Cancer Genome Atlas |
| tRNA | Transfer RiboNucleic Acid |
| uORF | Upstream Open Reading Frame |
| UTR | UnTranslated Region |
| XIAP | X-chromosome linked Inhibitor of Apoptosis |
References
- Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 2002, 108, 557–572. [Google Scholar] [CrossRef]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Voigts-Hoffmann, F.; Klinge, S.; Ban, N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr. Opin. Struct. Biol. 2012, 22, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Kong, J.; Lasko, P. Translational control in cellular and developmental processes. Nat. Rev. Genet. 2012, 13, 383–394. [Google Scholar] [CrossRef]
- Chu, J.; Cargnello, M.; Topisirovic, I.; Pelletier, J. Translation Initiation Factors: Reprogramming Protein Synthesis in Cancer. Trends Cell Biol. 2016, 26, 918–933. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006, 31, 553–562. [Google Scholar] [CrossRef]
- Cate, J.H. Human eIF3: From ‘blobology’ to biological insight. Philos. Trans. R. Soc. Lond. B BiolSci. 2017, 372, 1716. [Google Scholar] [CrossRef]
- Allen, E.H.; Schweet, R.S. Synthesis of hemoglobin in a cell free system: I. Properties of the complete system. J. Biol. Chem. 1962, 237, 760–767. [Google Scholar]
- Miller, R.L.; Schweet, R. Isolation of a protein fraction from reticulocyte ribosomes required for de novo synthesis of hemoglobin. Arch. Biochem. Biophys. 1968, 125, 632–646. [Google Scholar] [CrossRef]
- Shafritz, D.A.; Pritchard, P.M.; Glibert, J.M.; Anderson, W.F. Separation of two factors, M1 and M2, required for poly(U)-dependent synthesis by rabbit reticulocyte ribosomes at low magnesium ion concentration. Biochem. Biophys. Res. Commun. 1970, 38, 721–727. [Google Scholar] [CrossRef]
- Shafritz, D.A.; Anderson, W.F. Isolation and partial characterization of reticulocyte factors M1 and M2. J. Biol. Chem. 1970, 245, 5553–5559. [Google Scholar] [PubMed]
- Culp, W.; Morrisey, J.; Hardesty, B. Initiator tRNA for the synthesis of globin peptides. Biochem. Biophys. Res. Commun. 1970, 40, 777–785. [Google Scholar] [CrossRef]
- Shafritz, D.A.; Laycock, D.G.; Anderson, W.F. Puromycin-peptide bond formation with reticulocyte initiation factors M1 and M2. Proc. Natl. Acad. Sci. USA 1971, 68, 496–499. [Google Scholar] [CrossRef]
- Anderson, W.F.; Shafritz, D.A. Methionine transfer RNAf: The initiator transfer RNA for hemoglobin biosynthesis. Cancer Res. 1971, 31, 701–703. [Google Scholar]
- Gasior, E.; Moldave, K. Evidence for a soluble protein factor specific for the interaction between aminoacylated transfer RNA’s and the 40S subunit of mammalian ribosomes. J. Mol. Biol. 1972, 66, 391–402. [Google Scholar] [CrossRef]
- Zasloff, M.; Ochoa, S. Polypeptide chain initiation in eukaryotes: Functional identity of supernatant factor from various sources. Proc. Natl. Acad. Sci. USA 1972, 69, 1796–1799. [Google Scholar] [CrossRef]
- Levin, D.H.; Kyner, D.; Acs, G. Protein initiation in eukaryotes: Formation and Function of a ternary complex composed of a partially purified ribosomal factor, Methionyl transfer RNA and Guanosine triphosphate. Proc. Natl. Acad. Sci. USA 1973, 70, 41–45. [Google Scholar] [CrossRef]
- Dettman, G.L.; Stanley, W.W., Jr. The ternary complex of initiation factor IF-1, Met-tRNAf and GTP: An aurintricarboxylate-sensitive intermediate in the initiation of eukaryotic protein synthesis. Biochem. Biophys. Acta 1973, 299, 142–147. [Google Scholar] [PubMed]
- Merrick, W.C.; Anderson, W.F. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J. Biol. Chem. 1975, 250, 1107–1206. [Google Scholar]
- Safer, B.; Adams, S.L.; Anderson, W.F.; Merrick, W.C. Binding of Met-tRNAf and GTP to homogenous initiation factor MP. J. Biol. Chem. 1975, 250, 9076–9082. [Google Scholar] [PubMed]
- Adams, S.L.; Safer, B.; Anderson, W.F.; Merrick, W.C. Eukaryotic initiation complex formation: Evidence for two distinct pathways. J. Biol. Chem. 1975, 250, 9083–9089. [Google Scholar]
- Cimadevilla, J.M.; Hardesty, B. Aminoacyl-tRNA specificity of a 40S ribosomal subunit binding factor from rabbit reticulocytes. Biochem. Biophys. Res. Commun. 1975, 63, 16–23. [Google Scholar] [CrossRef]
- Cimadevilla, J.M.; Hardesty, B. Isolation and partial characterization of a 40S ribosomal subunit-transfer Ribonucleic Acid binding factor from rabbit reticulocytes. J. Biol. Chem. 1975, 250, 4389–4397. [Google Scholar]
- Gupta, N.K.; Chatterjee, B.; Majumdar, A. Protein synthesis in rabbit reticulocytes XII: Requirement of mRNA (AUG codon) for Met-tRNAf binding to 40S ribosomes. Biochem. Biophys. Res. Commun. 1975, 65, 797–805. [Google Scholar] [CrossRef]
- Geisen, M.; Roman, R.; Seal, S.N.; Marcus, M. Formation of an 80S Methionyl-tRNA initiation complex with soluble factors from wheat germ. J. Biol. Chem. 1976, 251, 6075–6081. [Google Scholar]
- Benne, R.; Wong, C.; Leudi, M.; Hershey, J.W. Purification and characterization of initiation factor IF-E2 from rabbit reticulocytes. J. Biol. Chem. 1976, 215, 7675–7681. [Google Scholar]
- Trachsel, H.; Erni, B.; Schreier, M.H.; Staehelin, T. Initiation of mammalian protein synthesis: II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol. 1977, 116, 755–767. [Google Scholar] [CrossRef]
- Anderson, W.F.; Bosch, L.; Cohn, W.E.; Lodish, H.; Merrick, W.C.; Weissbach, H.; Wittmann, H.G.; Wool, I.G. International symposium on protein synthesis. FEBS Lett. 1977, 76, 1–10. [Google Scholar] [CrossRef]
- Merrick, W.C. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979, 60, 108–123. [Google Scholar] [PubMed]
- Benne, R.; Amesz, H.; Hershey, J.W.; Voorma, H.O. The activity of eukaryotic initiation factor eIF-2 in ternary complex formation with GTP and Met-tRNA. J. Biol. Chem. 1979, 254, 3201–3205. [Google Scholar] [PubMed]
- Seal, S.N.; Schmidt, A.; Marcus, A. Fractionation and partial characterization of the protein synthesis system of wheat germ. I. Resolution of two elongation factors and five initiation factors. J. Biol. Chem. 1983, 258, 859–865. [Google Scholar] [PubMed]
- Lax, R.R.; Lauer, S.J.; Browning, K.S.; Ravel, J.M. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol. 1986, 118, 109–128. [Google Scholar] [PubMed]
- Lengyel, P.; Speyer, J.F.; Ochoa, S. Synthetic polynucleotides and the amino acid code. Proc. Natl. Acad. Sci. USA 1961, 47, 1936–1942. [Google Scholar] [CrossRef] [PubMed]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef]
- Mukundan, M.A.; Hershey, J.W.; Dewey, K.F.; Thach, R.E. Binding of formylmethioniyl-tRNA to 30S ribosomal subunits. Nature 1968, 217, 1013–1016. [Google Scholar] [CrossRef]
- Iwasaki, K.; Sabol, S.; Wahba, A.J.; Ochoa, S. Translation of the genetic message: VII. Role of initiation factors in formation of the chain initiation complex with Escherichia coli ribosomes. Arch. Biochem. Biophys. 1968, 125, 542–547. [Google Scholar] [CrossRef]
- Thach, R.E.; Hershey, J.W.; Kolakofsky, D.; Dewey, K.F.; Remold-O’Donnell, E. Purification and properties of initiation factors F1 and F2. Cold Spring Harb. Symp. Quant. Biol. 1969, 34, 277–284. [Google Scholar] [CrossRef]
- Wahba, A.J.; Miller, M.J. Chain initiation. Factors from E. coli. Methods Enzymol. 1974, 30, 3–18. [Google Scholar]
- Clark, B.F.C.; Grunberg-Manago, M.; Gupta, N.K.; Hershey, J.W.B.; Hinnebusch, A.G.; Jackson, R.J.; Maitra, U.; Mathews, M.B.; Merrick, W.C.; Rhoads, R.E.; et al. Prokaryotic and eukaryotic translation factors. Ad Hoc Nomenclature Subcommittee Report. Biochimie 1996, 78, 1119–1122. [Google Scholar] [CrossRef]
- Browning, K.S.; Gallie, D.R.; Hershey, J.W.B.; Hinnebusch, A.G.; Maitra, U.; Merrick, W.C.; Norbury, C. Unified nomenclature of the subunits of eukaryotic initiation factor 3. Trends Biochem. Sci. 2001, 26, 284. [Google Scholar] [CrossRef]
- Benne, R.; Brown-Luedi, M.L.; Hershey, J.W. Protein synthesis initiation factors from rabbit reticulocytes: Purification, characterization, and radiochemical labeling. Methods Enzymol. 1979, 60, 15–35. [Google Scholar]
- Majumdar, A.; Dasgupta, A.; Chatterjee, B.; Das, H.K.; Gupta, N.K. Purification and properties of rabbit reticulocyte protein synthesis initiation factors EIF-1, EIF-2, and EIF-3. Methods Enzymol. 1979, 60, 35–52. [Google Scholar]
- Staehelin, T.; Erni, B.; Schreier, M.H. Purification and characterization of seven initiation factors for mammalian protein synthesis. Methods Enzymol. 1979, 60, 136–165. [Google Scholar]
- Merrick, W.C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 1992, 56, 291–315. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J. Biol. Chem. 2005, 280, 23425–23428. [Google Scholar] [CrossRef]
- Pestova, T.V.; de Breyne, S.; Pisarev, A.V.; Abaeva, I.S.; Hellen, C.U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J. 2008, 27, 1060–1072. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar] [CrossRef]
- Komar, A.A.; Mazumder, B.; Merrick, W.C. A new framework for understanding IRES-mediated translation. Gene 2012, 502, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.; Pestova, T.V. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 2013, 51, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Akulich, K.A.; Andreev, D.E.; Terenin, I.M.; Smirnova, V.V.; Anisimova, A.S.; Makeeva, D.S.; Arkhipova, V.I.; Stolboushkina, E.A.; Garber, M.B.; Prokofjeva, M.M.; et al. Four translation initiation pathways employed by the leaderless mRNA in eukaryotes. Sci. Rep. 2016, 6, 37905. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.G.; Wilusz, J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017, 31, 1717–1731. [Google Scholar] [CrossRef] [PubMed]
- Shatsky, I.N.; Terenin, I.M.; Smirnova, V.V.; Andreev, D.E. Cap-independent translation: What’s in a name? Trends Biochem. Sci. 2018, 43, 882–895. [Google Scholar] [CrossRef]
- James, C.C.; Smyth, J.W. Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease. Life Sci. 2018, 212, 138–144. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. In Translational Control in Biology and Medicine; Sonenberg, N., Hershey, J.W.H., Matthews, M., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2018; pp. 23–44. [Google Scholar]
- Pelletier, J.; Sonenberg, N. The organizing principles of eukaryotic ribosome recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef]
- Skabkin, M.A.; Skabkina, O.V.; Dhote, V.; Komar, A.A.; Hellen, C.U.; Pestova, T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010, 24, 1787–1801. [Google Scholar] [CrossRef]
- Dmitriev, S.E.; Terenin, I.M.; Andreev, D.E.; Ivanov, P.A.; Dunaevsky, J.E.; Merrick, W.C.; Shatsky, I.N. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 2010, 285, 26779–26787. [Google Scholar] [CrossRef]
- Kimball, S.R. Eukaryotic initiation factor eIF2. Int. J. Biochem. Cell. Biol. 1999, 31, 25–29. [Google Scholar] [CrossRef]
- Schmitt, E.; Naveau, M.; Mechulam, Y. Eukaryotic and archaeal translation initiation factor 2: A heterotrimeric tRNA carrier. FEBS Lett. 2010, 584, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, N.; Gorman, A.M.; Gupta, S.; Samali, A. The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 2013, 70, 3493–3511. [Google Scholar] [CrossRef] [PubMed]
- Wek, R.C. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol. 2018, 10, a032870. [Google Scholar] [CrossRef] [PubMed]
- Llácer, J.L.; Hussain, T.; Marler, L.; Aitken, C.E.; Thakur, A.; Lorsch, J.R.; Hinnebusch, A.G.; Ramakrishnan, V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol. Cell 2015, 59, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Adomavicius, T.; Guaita, M.; Zhou, Y.; Jennings, M.D.; Latif, Z.; Roseman, A.M.; Pavitt, G.D. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 2019, 10, 2136. [Google Scholar] [CrossRef]
- Kapp, L.D.; Lorsch, J.R. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 2004, 335, 923–936. [Google Scholar] [CrossRef]
- Jennings, M.D.; Kershaw, C.J.; Adomavicius, T.; Pavitt, G.D. Fail-safe control of translation initiation by dissociation of eIF2alpha phosphorylated ternary complexes. Elife 2017, 6, e24542. [Google Scholar] [CrossRef]
- Unbehaun, A.; Borukhov, S.I.; Hellen, C.U.; Pestova, T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004, 18, 3078–3093. [Google Scholar] [CrossRef]
- Singh, C.R.; Lee, B.; Udagawa, T.; Mohammad-Qureshi, S.S.; Yamamoto, Y.; Pavitt, G.D.; Asano, K. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006, 25, 4537–4546. [Google Scholar] [CrossRef]
- Jennings, M.D.; Pavitt, G.D. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 2010, 465, 378–381. [Google Scholar] [CrossRef]
- Jennings, M.D.; Zhou, Y.; Mohammad-Qureshi, S.S.; Bennett, D.; Pavitt, G.D. eIF2B promotes eIF5 dissociation from eIF2•GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 2013, 27, 2696–2707. [Google Scholar] [CrossRef] [PubMed]
- Wortham, N.C.; Proud, C.G. eIF2B: Recent structural and functional insights into a key regulator of translation. Biochem. Soc. Trans. 2015, 43, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Lee, J.H.; Zoll, W.L.; Merrick, W.C.; Dever, T.E. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 1998, 280, 1757–1760. [Google Scholar] [CrossRef] [PubMed]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Shatsky, I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008, 15, 836–841. [Google Scholar] [CrossRef]
- Zoll, W.L.; Horton, L.E.; Komar, A.A.; Hensold, J.O.; Merrick, W.C. Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 2002, 277, 37079–37087. [Google Scholar] [CrossRef]
- Komar, A.A.; Gross, S.R.; Barth-Baus, D.; Strachan, R.; Hensold, J.O.; Kinzy, T.G.; Merrick, W.C. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J. Biol. Chem. 2005, 280, 15601–15611. [Google Scholar] [CrossRef]
- Davey, N.E.; Cowan, J.L.; Shields, D.C.; Gibson, T.J.; Coldwell, M.J.; Edwards, R.J. SLiMPrints: Conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res. 2012, 40, 10628–10641. [Google Scholar] [CrossRef]
- Kim, E.; Kim, J.H.; Seo, K.; Hong, K.Y.; An, S.W.A.; Kwon, J.; Lee, S.V.; Jang, S.K. eIF2A, an initiator tRNA carrier refractory to eIF2α kinases, functions synergistically with eIF5B. Cell. Mol. Life Sci. 2018, 75, 4287–4300. [Google Scholar] [CrossRef]
- Komar, A.A.; Lesnik, T.; Cullin, C.; Merrick, W.C.; Trachsel, H.; Altmann, M. Internal initiaition drives the synthesis of Ure2 protein lacking the prion domain and affects [URE3] propagation in yeast cells. EMBO J. 2003, 22, 1199–1209. [Google Scholar] [CrossRef]
- Reineke, L.C.; Komar, A.A.; Caprara, M.G.; Merrick, W.C. A small stem loop element directs internal initiation of the Ure2 internal ribosome entry site in Saccharomyces cerevisiae. J. Biol. Chem. 2008, 283, 19011–19025. [Google Scholar] [CrossRef]
- Reineke, L.C.; Merrick, W.C. Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA 2009, 15, 2264–2277. [Google Scholar] [CrossRef] [PubMed]
- Reineke, L.C.; Cao, Y.; Baus, D.; Hossain, N.M.; Merrick, W.C. Insights into the role of yeast eIF2A in IRES-mediated translation. PLoS ONE 2011, 6, e24492. [Google Scholar] [CrossRef] [PubMed]
- Ventoso, I.; Sanz, M.A.; Molina, S.; Berlanga, J.J.; Carrasco, L.; Esteban, M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006, 20, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Barth-Baus, D.; Bhasker, C.R.; Zoll, W.L.; Merrick, W.C. Influence of translation factor activities on start site selection in six different mRNAs. Translation 2013, 1, e24419. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, L.; Sanz, M.A.; González-Almela, E. The Regulation of Translation in Alphavirus-Infected Cells. Viruses 2018, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; González Almela, E.; Carrasco, L. Translation of Sindbis Subgenomic mRNA is Independent of eIF2, eIF2A and eIF2D. Sci. Rep. 2017, 7, 43876. [Google Scholar] [CrossRef]
- Sanz, M.A.; Almela, E.G.; García-Moreno, M.; Marina, A.I.; Carrasco, L. A viral RNA motif involved in signaling the initiation of translation on non-AUG codons. RNA 2019, 25, 431–452. [Google Scholar] [CrossRef]
- Niepmann, M. Hepatitis C virus RNA translation. Curr. Top. Microbiol. Immunol. 2013, 369, 143–166. [Google Scholar]
- Kim, J.H.; Park, S.M.; Park, J.H.; Keum, S.J.; Jang, S.K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011, 30, 2454–2464. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Oguro, A.; Nakamura, Y.; Kieft, J.S. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife 2016, 5, e21198. [Google Scholar] [CrossRef]
- González-Almela, E.; Williams, H.; Sanz, M.A.; Carrasco, L. The Initiation Factors eIF2, eIF2A, eIF2D, eIF4A, and eIF4G are Not Involved in Translation Driven by Hepatitis C Virus IRES in Human Cells. Front. Microbiol. 2018, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.S.; An, S.; Kim, E.; Yu, J.; Hong, K.Y.; Lee, J.S.; Jang, S.K. An mRNA-specific tRNAi carrier eIF2A plays a pivotal role in cell proliferation under stress conditions: Stress-resistant translation of c-Src mRNA is mediated by eIF2A. Nucleic Acids Res. 2017, 45, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Starck, S.R.; Jiang, V.; Pavon-Eternod, M.; Prasad, S.; McCarthy, B.; Pan, T.; Shastri, N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 2012, 336, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; He, S.; Yang, J.; Jia, X.; Wang, P.; Chen, X.; Zhang, Z.; Zou, X.; McNutt, M.A.; Shen, W.H.; et al. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab. 2014, 19, 836–848. [Google Scholar] [CrossRef]
- Masson, G.R.; Williams, R.L. Structural Mechanisms of PTEN Regulation. Cold Spring Harb. Perspect. Med. 2019, 10, a036152. [Google Scholar] [CrossRef]
- Starck, S.R.; Tsai, J.C.; Chen, K.; Shodiya, M.; Wang, L.; Yahiro, K.; Martins-Green, M.; Shastri, N.; Walter, P. Translation from the 5′ untranslated region shapes the integrated stress response. Science 2016, 351, aad3867. [Google Scholar] [CrossRef]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef]
- Guan, B.J.; van Hoef, V.; Jobava, R.; Elroy-Stein, O.; Valasek, L.S.; Cargnello, M.; Gao, X.H.; Krokowski, D.; Merrick, W.C.; Kimball, S.R.; et al. A Unique ISR Program Determines Cellular Responses to Chronic Stress. Mol. Cell 2017, 68, 885–900. [Google Scholar] [CrossRef]
- Sendoel, A.; Dunn, J.G.; Rodriguez, E.H.; Naik, S.; Gomez, N.C.; Hurwitz, B.; Levorse, J.; Dill, B.D.; Schramek, D.; Molina, H.; et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature 2017, 541, 494–499. [Google Scholar] [CrossRef]
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11, a032896. [Google Scholar] [CrossRef]
- Chen, L.; He, J.; Zhou, J.; Xiao, Z.; Ding, N.; Duan, Y.; Li, W.; Sun, L.Q. EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J. Cell. Mol. Med. 2019, 23, 6060–6071. [Google Scholar] [CrossRef] [PubMed]
- Sonobe, Y.; Ghadge, G.; Masaki, K.; Sendoel, A.; Fuchs, E.; Roos, R.P. Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol. Dis. 2018, 116, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Golovko, A.; Kojukhov, A.; Guan, B.J.; Morpurgo, B.; Merrick, W.C.; Mazumder, B.; Hatzoglou, M.; Komar, A.A. The eIF2A knockout mouse. Cell Cycle 2016, 15, 3115–3120. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, K.; Ito, T.; Yokoyama, S. Crystal structure of the eukaryotic translation initiation factor 2A from Schizosaccharomyces pombe. J. Struct. Funct. Genom. 2014, 15, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Chan, N.L.; Wang, A.H. The many blades of the β-propeller proteins: Conserved but versatile. Trends Biochem. Sci. 2011, 36, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Jain, B.P.; Pandey, S. WD40 Repeat Proteins: Signalling Scaffold with Diverse Functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef]
- Nielsen, M.H.; Flygaard, R.K.; Jenner, L.B. Structural analysis of ribosomal RACK1 and its role in translational control. Cell. Signal. 2017, 35, 272–281. [Google Scholar] [CrossRef]
- Liu, Y.; Neumann, P.; Kuhle, B.; Monecke, T.; Schell, S.; Chari, A.; Ficner, R. Translation initiation factor eIF3b contains a nine-bladed β-propeller and interacts with the 40S ribosomal subunit. Structure 2014, 22, 923–930. [Google Scholar] [CrossRef]
- Ross, J.A.; Bressler, K.R.; Thakor, N. Eukaryotic Initiation Factor 5B (eIF5B) Cooperates with eIF1A and eIF5 to Facilitate uORF2-Mediated Repression of ATF4 Translation. Int. J. Mol. Sci. 2018, 19, 4032. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, A.R.; Gsponer, J.; Foster, L.J. A high-throughput approach for measuring temporal changes in the interactome. Nat. Methods 2012, 9, 907–909. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Borgeson, B.; Phanse, S.; Tu, F.; Drew, K.; Clark, G.; Xiong, X.; Kagan, O.; Kwan, J.; Bezginov, A.; et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015, 525, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef]
- Gonen, H.; Smith, C.E.; Siegel, N.R.; Kahana, C.; Merrick, W.C.; Chakraburtty, K.; Schwartz, A.L.; Ciechanover, A. Protein synthesis elongation factor EF-1alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc. Natl. Acad. Sci. USA 1994, 91, 7648–7652. [Google Scholar] [CrossRef]
- Polevoda, B.; Sherman, F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 2003, 325, 595–622. [Google Scholar] [CrossRef]
- Soppa, J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010, 2010, 820681. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Dephoure, N.; Zhou, C.; Villén, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.; Mohammed, S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef]
- Rechsteiner, M. PEST sequences are signals for rapid intracellular proteolysis. Semin. Cell Biol. 1990, 1, 433–440. [Google Scholar] [PubMed]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Xing, H.; Hong, Y.; Sarge, K.D. PEST sequences mediate heat shock factor 2 turnover by interacting with the Cul3 subunit of the Cul3-RING ubiquitin ligase. Cell Stress Chaperones 2010, 15, 301–308. [Google Scholar] [CrossRef]
- Li, Y.; Jin, K.; Bunker, E.; Zhang, X.; Luo, X.; Liu, X.; Hao, B. Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2018, 115, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Nixon, P.J.; Komenda, J.; Barber, J.; Deak, Z.; Vass, I.; Diner, B.A. Deletion of the PEST-like region of photosystem two modifies the QB-binding pocket but does not prevent rapid turnover of D1. J. Biol. Chem. 1995, 270, 14919–14927. [Google Scholar] [CrossRef] [PubMed]
- Bies, J.; Markus, J.; Wolff, L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 2002, 277, 8999–9009. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Northup, J.K.; Ray, K. Large putative PEST-like sequence motif at the carboxyl tail of human calcium receptor directs lysosomal degradation and regulates cell surface receptor level. J. Biol. Chem. 2012, 287, 4165–4176. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Shirokikh, N.E.; Hellen, C.U. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. Comptes Rendus Biol. 2005, 328, 589–605. [Google Scholar] [CrossRef]
- Hassanzadeh, G.; Naing, T.; Graber, T.; Jafarnejad, S.M.; Stojdl, D.F.; Alain, T.; Holcik, M. Characterizing Cellular Responses during Oncolytic Maraba Virus Infection. Int. J. Mol. Sci. 2019, 20, 580. [Google Scholar] [CrossRef] [PubMed]
- Thakor, N.; Holcik, M. IRES-mediated translation of cellular messenger RNA operates in eIF2α- independent manner during stress. Nucleic Acids Res. 2012, 40, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Schleich, S.; Strassburger, K.; Janiesch, P.C.; Koledachkina, T.; Miller, K.K.; Haneke, K.; Cheng, Y.S.; Kuechler, K.; Stoecklin, G.; Duncan, K.E.; et al. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 2014, 512, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Castelo-Szekely, V.; De Matos, M.; Tusup, M.; Pascolo, S.; Ule, J.; Gatfield, D. Charting DENR-dependent translation reinitiation uncovers predictive uORF features and links to circadian timekeeping via Clock. Nucleic Acids Res. 2019, 47, 5193–5209. [Google Scholar] [CrossRef]
- Young, D.J.; Makeeva, D.S.; Zhang, F.; Anisimova, A.S.; Stolboushkina, E.A.; Ghobakhlou, F.; Shatsky, I.N.; Dmitriev, S.E.; Hinnebusch, A.G.; Guydosh, N.R. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR Recycle Post-termination 40S Subunits In Vivo. Mol. Cell 2019, 71, 761–774. [Google Scholar] [CrossRef]
- Makeeva, D.S.; Lando, A.S.; Anisimova, A.; Egorov, A.A.; Logacheva, M.D.; Penin, A.A.; Andreev, D.E.; Sinitcyn, P.G.; Terenin, I.M.; Shatsky, I.N.; et al. Translatome and transcriptome analysis of TMA20 (MCT-1) and TMA64 (eIF2D) knockout yeast strains. Data Brief 2019, 23, 103701. [Google Scholar] [CrossRef]
- Weisser, M.; Schäfer, T.; Leibundgut, M.; Böhringer, D.; Aylett, C.H.S.; Ban, N. Structural and Functional Insights into Human Re-initiation Complexes. Mol. Cell 2017, 67, 447–456. [Google Scholar] [CrossRef]
- Lomakin, I.B.; Dmitriev, S.E.; Steitz, T.A. Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation. Proc. Natl. Acad. Sci. USA 2019, 116, 528–533. [Google Scholar] [CrossRef]



| Merrick/Anderson Rabbit Reticulocytes Ref. [22] | Ochoa A. salina Ref. [19] | Hardesty Rabbit Reticulocytes Ref. [25] | Moldave Rat Liver Ref. [18] |
|---|---|---|---|
| IF-M1 MW ~65,000 Da | aIF-2 MW ~(74,000)2 Da | Binding factor MW ~50,000 Da (~30,000+20,000 Da) | RL IF-1 MW ~70,000 Da |
| Factor | eIF2A | eIF2D | MCT-1/DENR | eIF5B | |
|---|---|---|---|---|---|
| Assay | Ref. [22,24] | Ref. [59,60] | Ref. [59,60] | Ref. [49,74,75] | |
| codon-directed binding to 40S | Met-tRNAi Phe-tRNAE.coli | Met-tRNAi Phe-tRNA tRNAMeti (non-acylated) | Met-tRNAi Phe-tRNA | Met-tRNAi | |
| aa-puromycin | Met-tRNAi | ||||
| poly(U)-directed synthesis of polyphenylalanine | Phe-tRNAE.coli | Phe-tRNA | Phe-tRNAE.coli | ||
| toe-printing | Met-tRNAi | Met-tRNAi | Met-tRNAi | ||
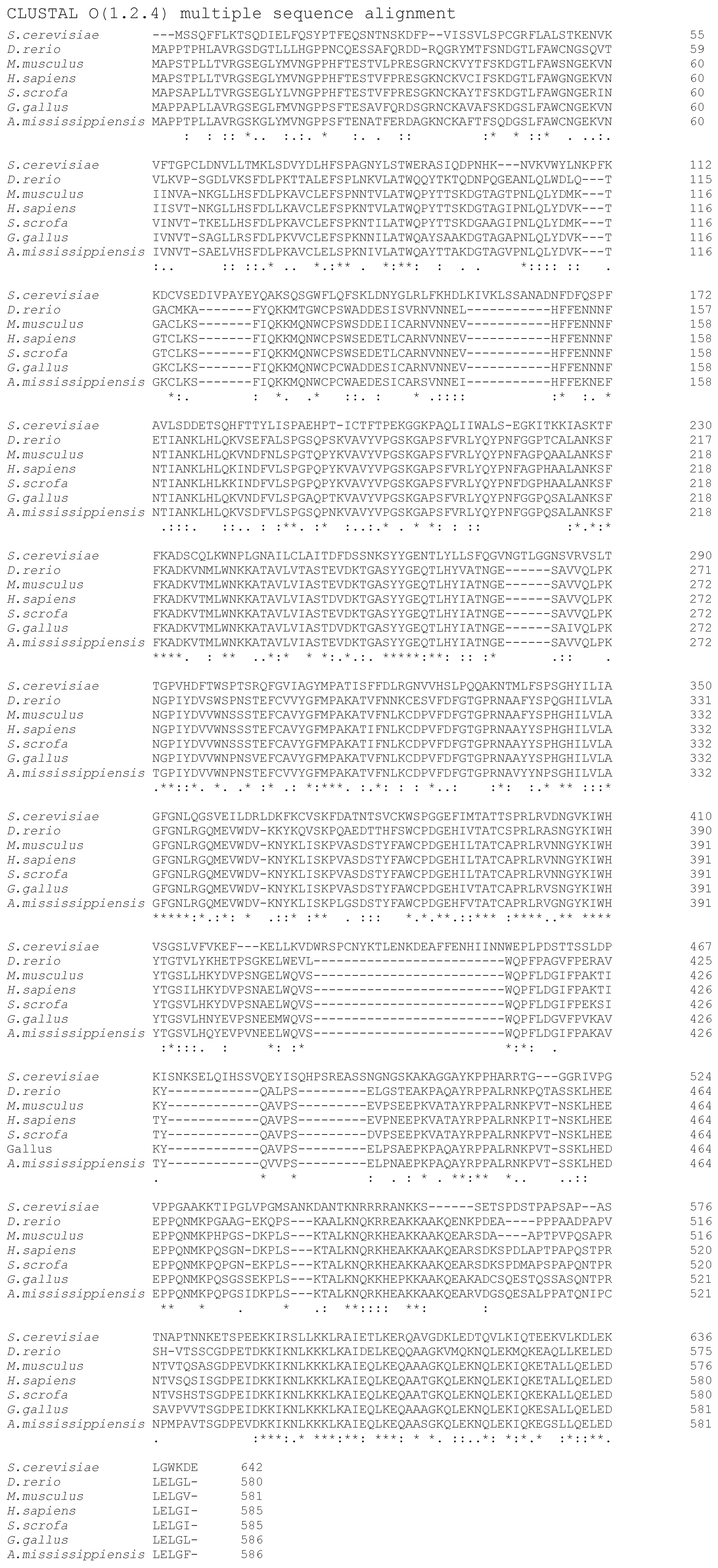
| IF-M1 Rabbit Ref. [22] Number of aa: 585 | eIF2A Rabbit (UNIPROT: G1TAW7) Number of aa: 585 | eIF2D Rabbit (UNIPROT: P0CL18) Number of aa: 566 | ||||||
|---|---|---|---|---|---|---|---|---|
| MW: ~65000.00 pI: ND in Ref. [22] | MW (predicted): 64839.70 Theoretical pI: 9.03 | MW (predicted): 62208.50 Theoretical pI: 6.97 | ||||||
| Ala (A) | 42.6 | 7.3% | Ala (A) | 45 | 7.7% | Ala (A) | 40 | 7.1% |
| Arg (R) | 20.3 | 3.5% | Arg (R) | 13 | 2.2% | Arg (R) | 22 | 3.9% |
| Asn (N) | Asn (N) | 35 | 6.0% | Asn (N) | 15 | 2.7% | ||
| Asp (D) | 54.3 | 9.3% | Asp (D) | 23 | 3.9% | Asp (D) | 30 | 5.3% |
| Cys (C) | 9 | 1.5% | Cys (C) | 9 | 1.5% | Cys (C) | 11 | 1.9% |
| Gln (Q) | Gln (Q) | 27 | 4.6% | Gln (Q) | 36 | 6.4% | ||
| Glu (E) | 61.4 | 10.5% | Glu (E) | 32 | 5.5% | Glu (E) | 34 | 6.0% |
| Gly (G) | 38.9 | 6.6% | Gly (G) | 36 | 6.2% | Gly (G) | 37 | 6.5% |
| His (H) | 12.2 | 2.1% | His (H) | 12 | 2.1% | His (H) | 16 | 2.8% |
| Ile (I) | 22.4 | 4 3.8% | Ile (I) | 22 | 3.8% | Ile (I) | 20 | 3.5% |
| Leu (L) | 46 | 7.9%1 | Leu (L) | 46 | 7.9% | Leu (L) | 68 | 12.0% |
| Lys (K) | 61.4 | 10.5% | Lys (K) | 55 | 9.4% | Lys (K) | 41 | 7.2% |
| Met (M) | 7.9 | 1.3% | Met (M) | 7 | 1.2% | Met (M) | 10 | 1.8% |
| Phe (F) | 19.4 | 3.3% | Phe (F) | 24 | 4.1% | Phe (F) | 12 | 2.1% |
| Pro (P) | 40.4 | 6.9% | Pro (P) | 45 | 7.7% | Pro (P) | 39 | 6.9% |
| Ser (S) | 42.7 | 7.3% | Ser (S) | 45 | 7.7% | Ser (S) | 40 | 7.1% |
| Thr (T) | 36.9 | 6.3% | Thr (T) | 39 | 6.7% | Thr (T) | 29 | 5.1% |
| Trp (W) | 9.3 | 1.6% | Trp (W) | 11 | 1.9% | Trp (W) | 5 | 0.9% |
| Tyr (Y) | 16.2 | 2.7% | Tyr (Y) | 22 | 3.8% | Tyr (Y) | 13 | 2.3% |
| Val (V) | 44.4 | 7.6% | Val (V) | 37 | 6.3% | Val (V) | 48 | 8.5% |
| Process Effect | IRES-Mediated Translation | Re-Initiation | Standard AUG and uAUG Translation | uCUG, uUUG Translation |
|---|---|---|---|---|
| Positive— (eIF2A facilitates) | HCV: Ref. [90] s-Src kinase: Ref. [93] | BiP (potentially): Ref. [97] | Sindbis virus (SV): Ref. [84]. Oncogenes: Ref. [100] | MHC I-peptides: Ref. [94] PTENα: Ref. [95] BiP: Ref. [97] Oncogenes: Ref. [100,102] C9ORF72: Ref. [103] |
| Negative— (eIF2A suppresses) | URE2, GIC1, PAB1: Ref. [77,81,82,83] | |||
| Does not affect | HCV: Ref. [91,92] | GCN4: Ref. [76] | SV: Ref. [87,88] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komar, A.A.; Merrick, W.C. A Retrospective on eIF2A—and Not the Alpha Subunit of eIF2. Int. J. Mol. Sci. 2020, 21, 2054. https://doi.org/10.3390/ijms21062054
Komar AA, Merrick WC. A Retrospective on eIF2A—and Not the Alpha Subunit of eIF2. International Journal of Molecular Sciences. 2020; 21(6):2054. https://doi.org/10.3390/ijms21062054
Chicago/Turabian StyleKomar, Anton A., and William C. Merrick. 2020. "A Retrospective on eIF2A—and Not the Alpha Subunit of eIF2" International Journal of Molecular Sciences 21, no. 6: 2054. https://doi.org/10.3390/ijms21062054
APA StyleKomar, A. A., & Merrick, W. C. (2020). A Retrospective on eIF2A—and Not the Alpha Subunit of eIF2. International Journal of Molecular Sciences, 21(6), 2054. https://doi.org/10.3390/ijms21062054





