Electron Microscopic Recording of the Power and Recovery Strokes of Individual Myosin Heads Coupled with ATP Hydrolysis: Facts and Implications
Abstract
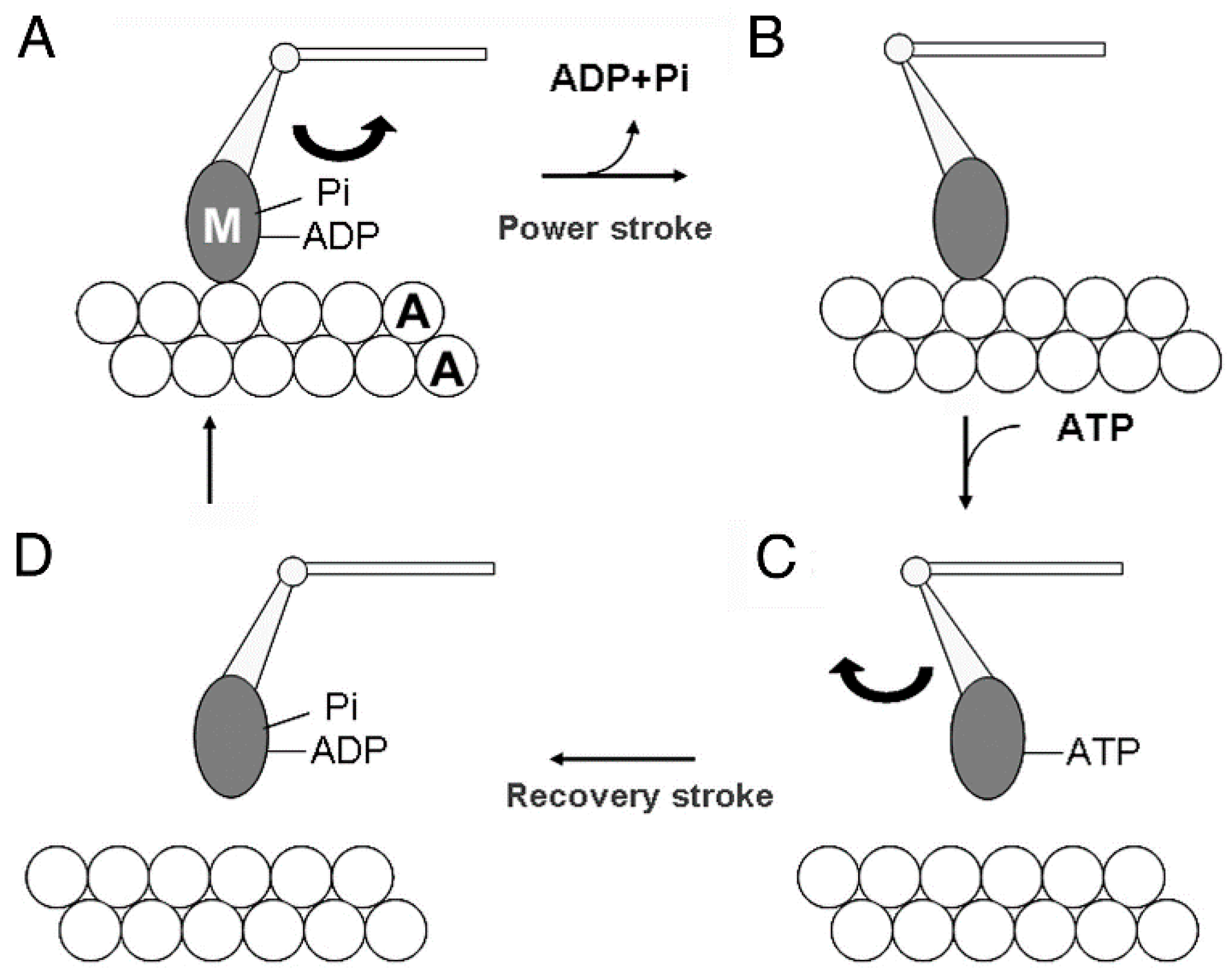
:1. Introduction
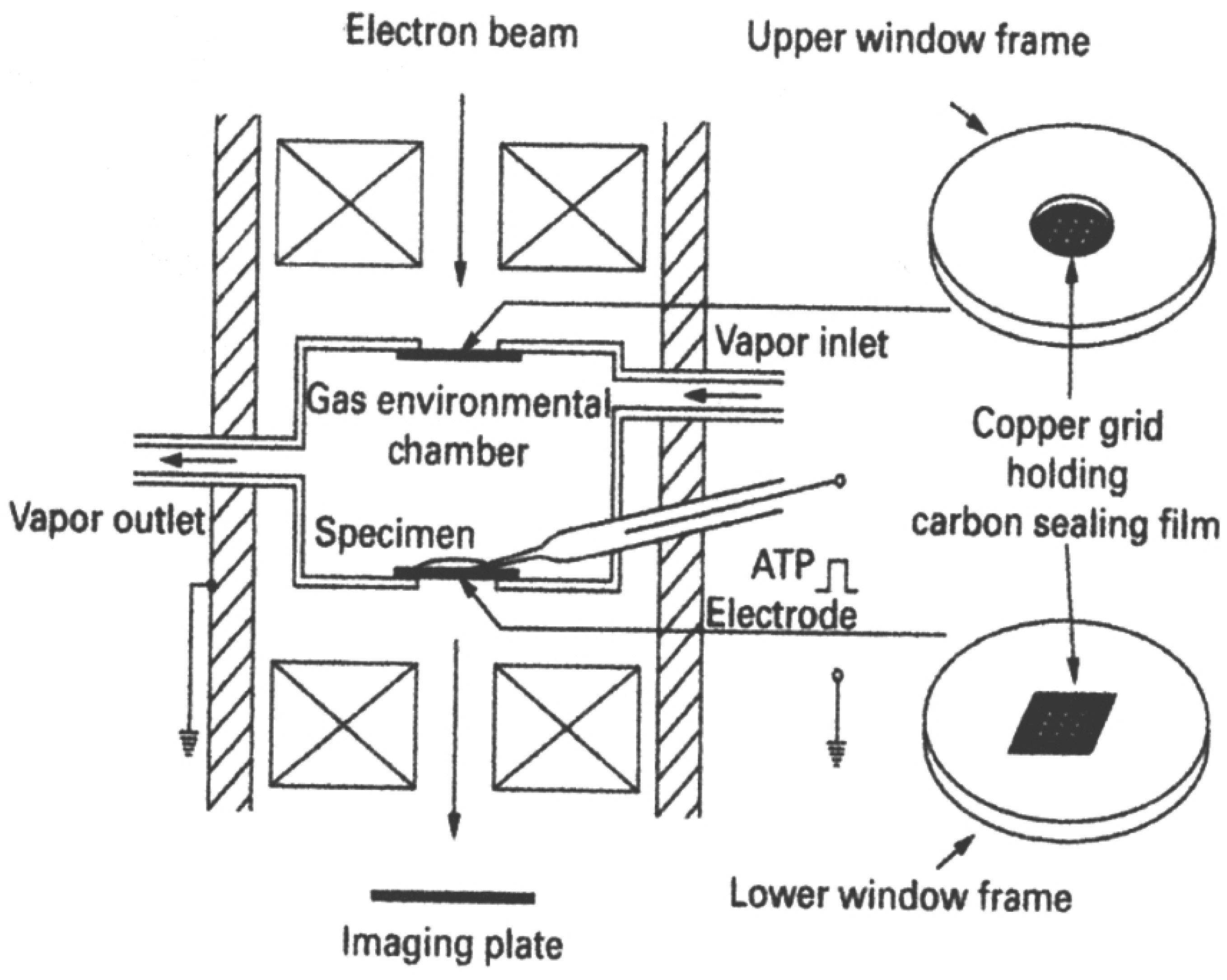
2. Experiments with the EC System
2.1. The EC System
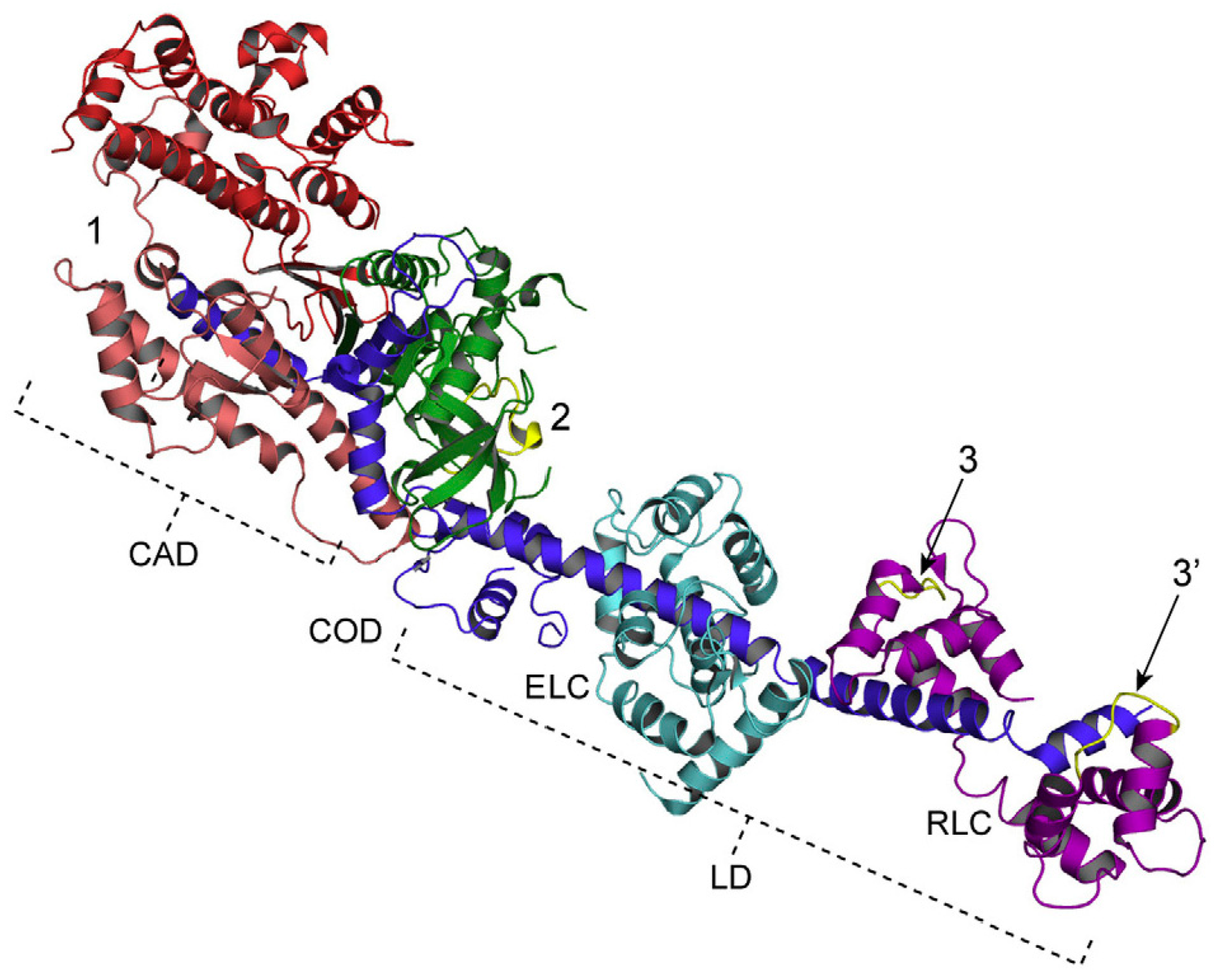
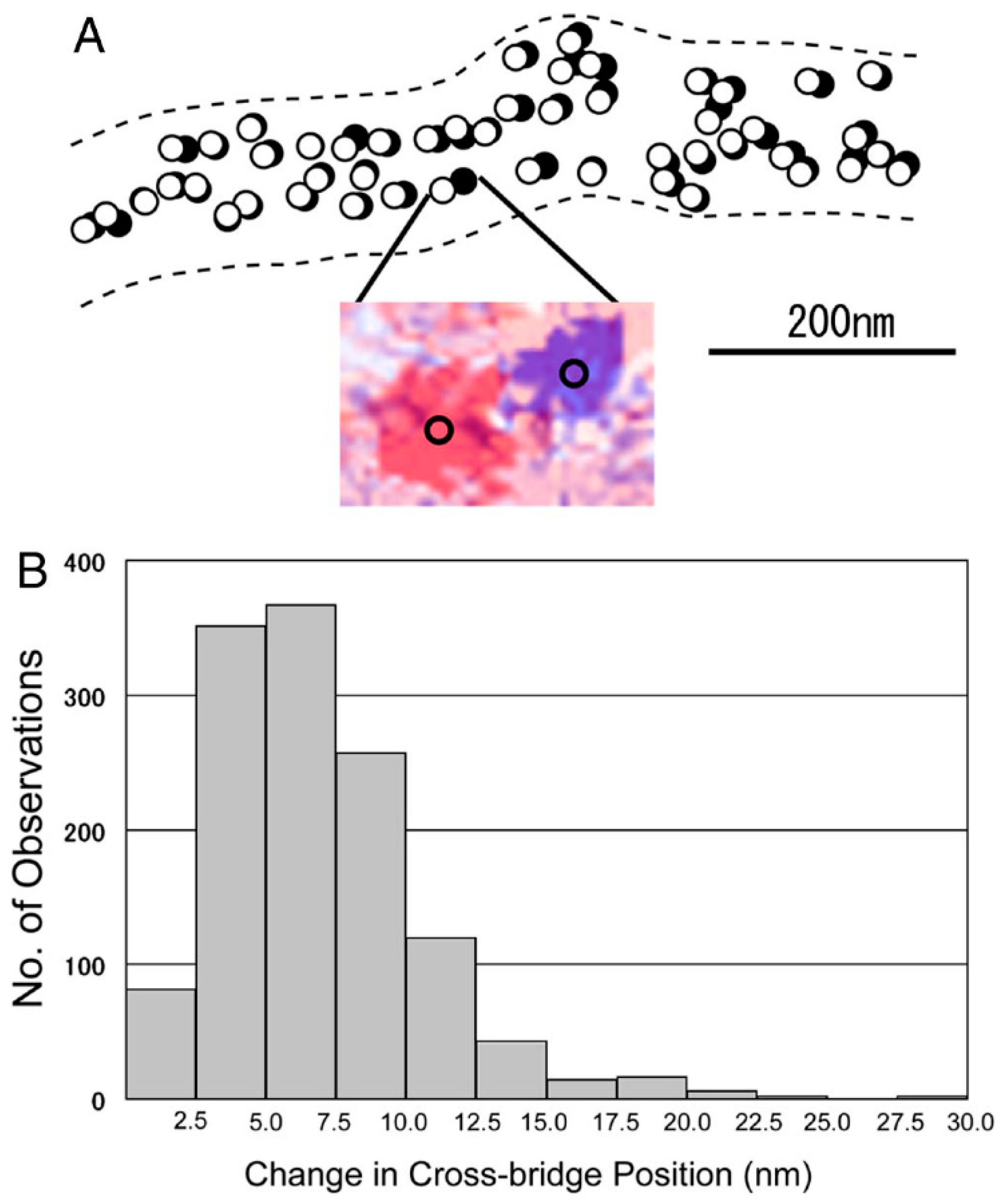
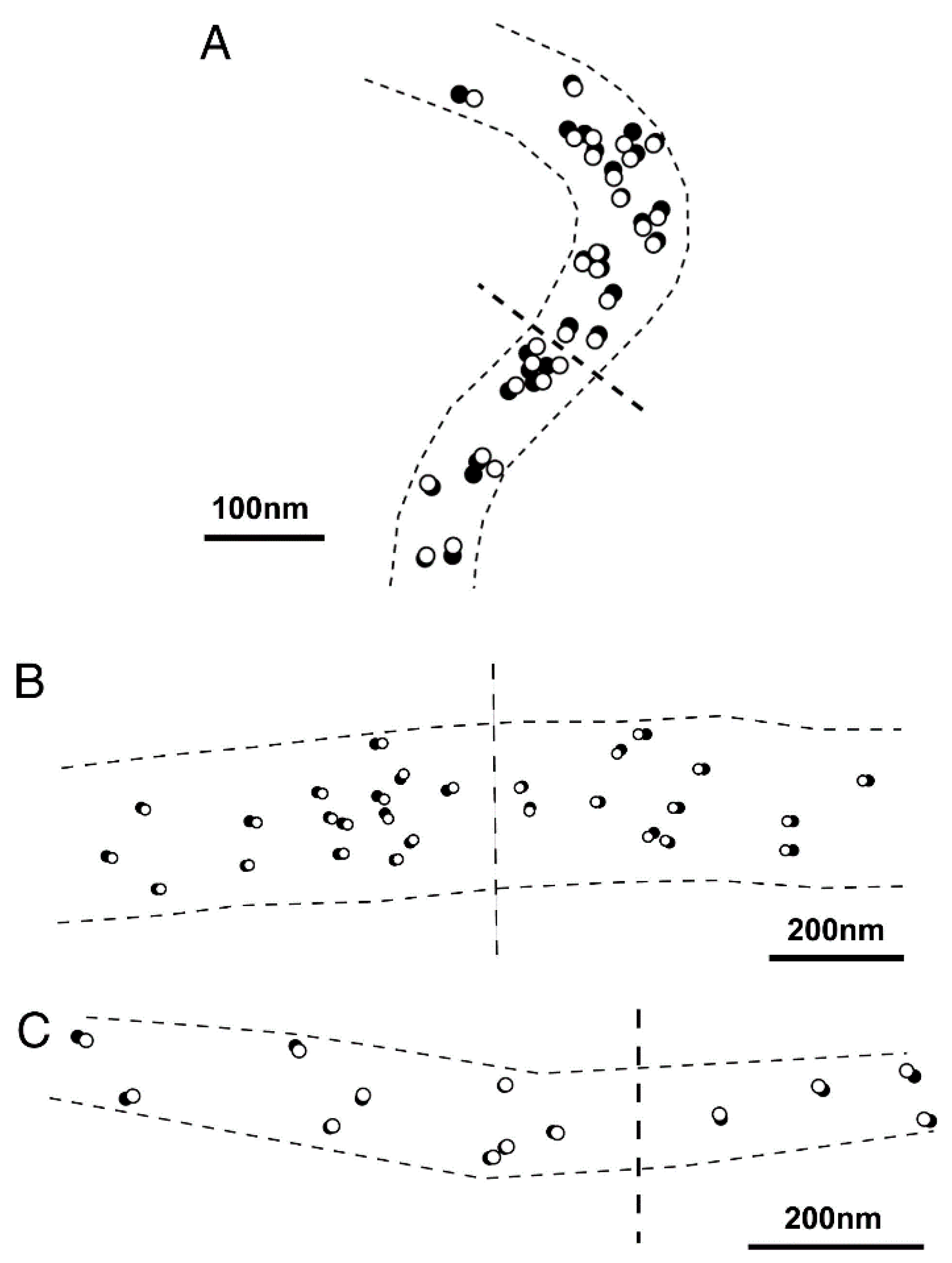
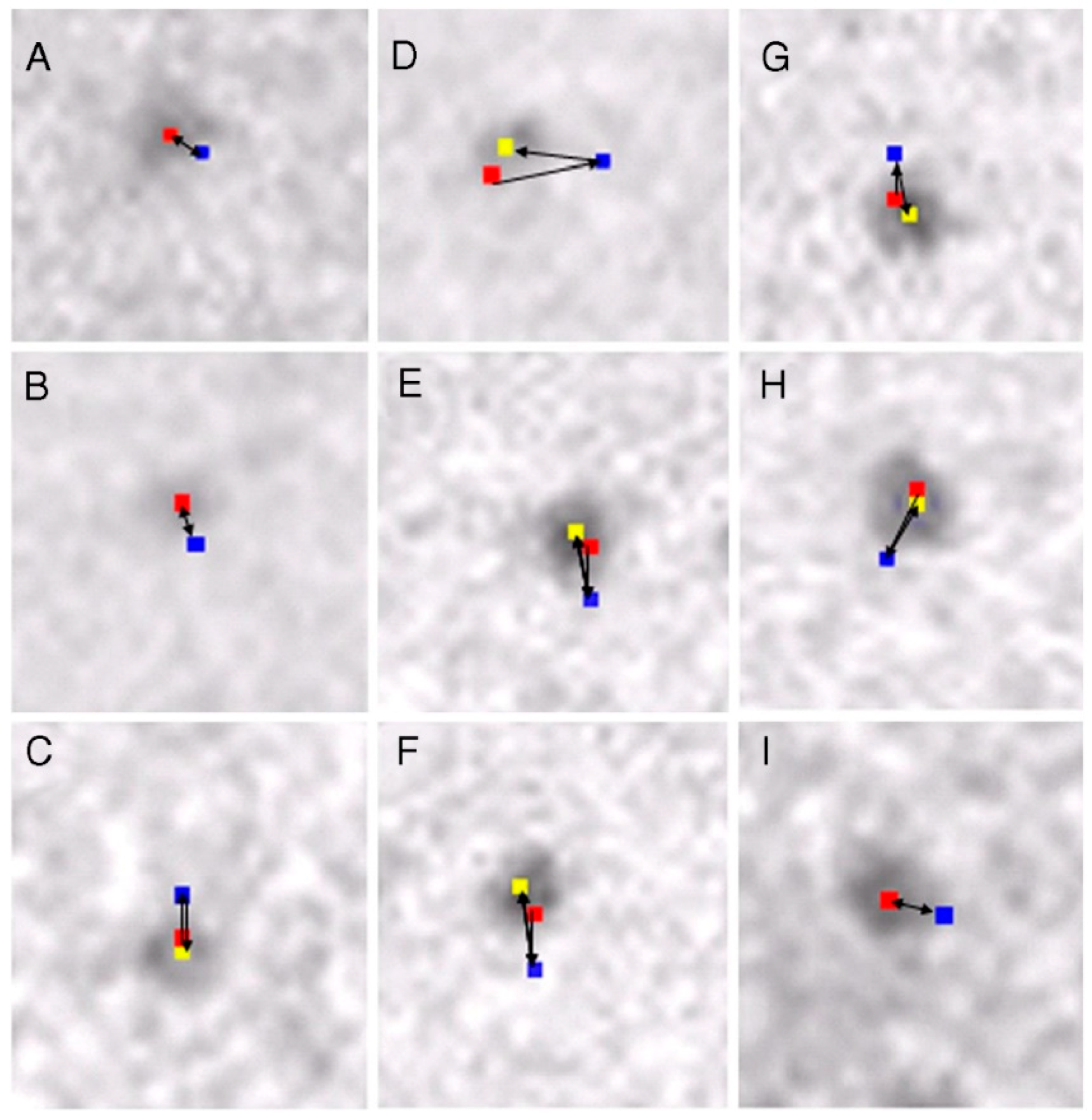
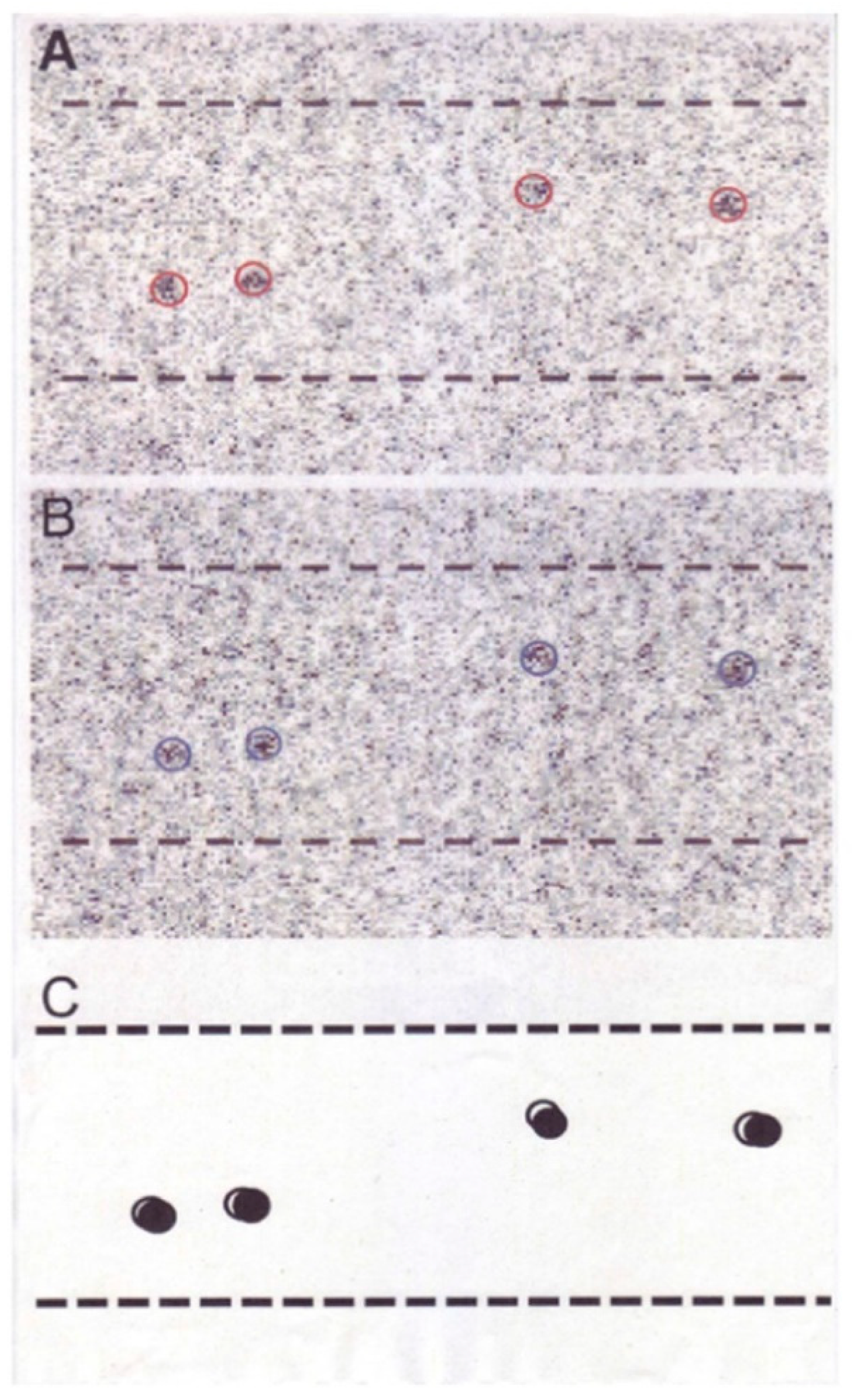
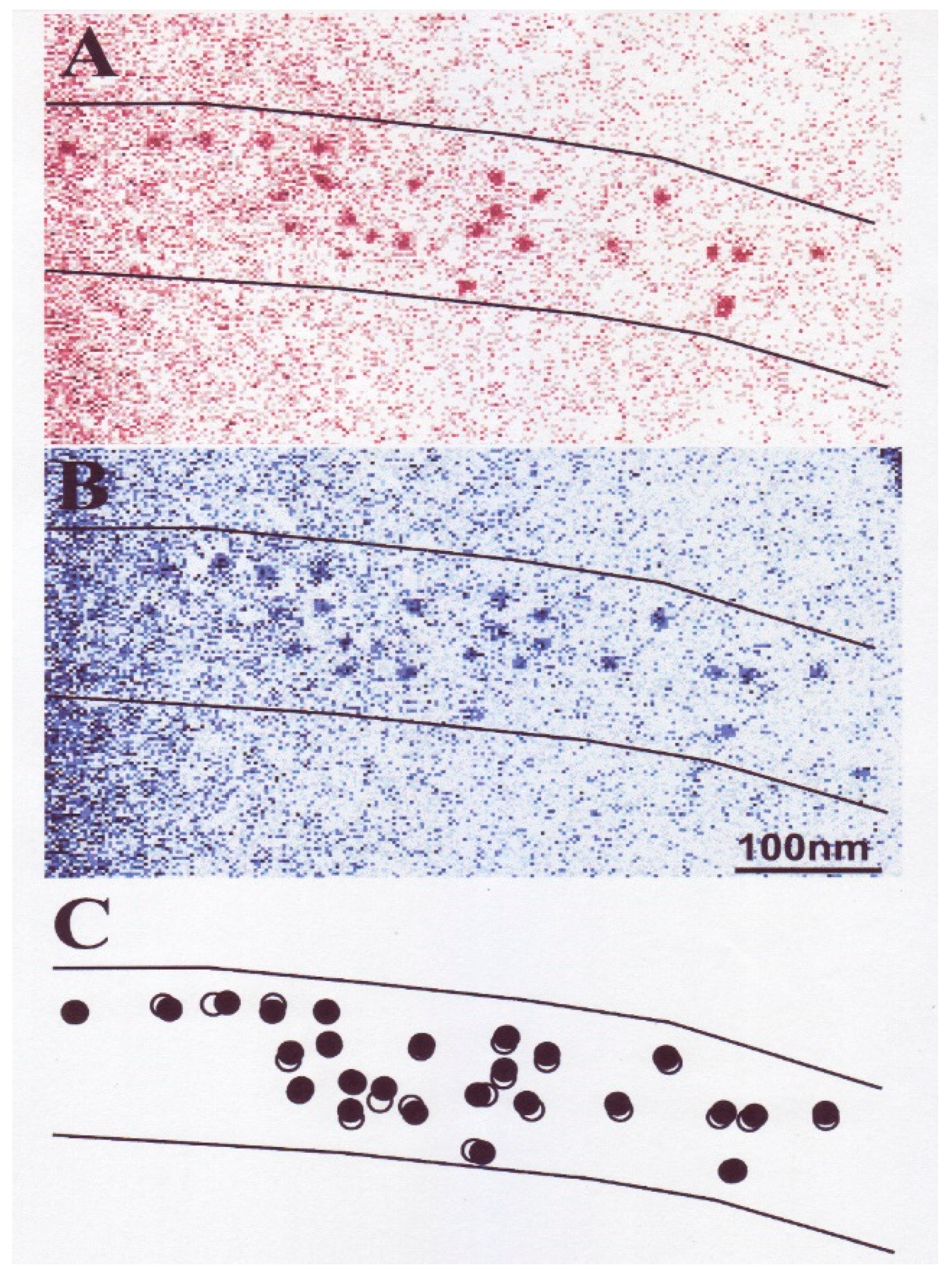
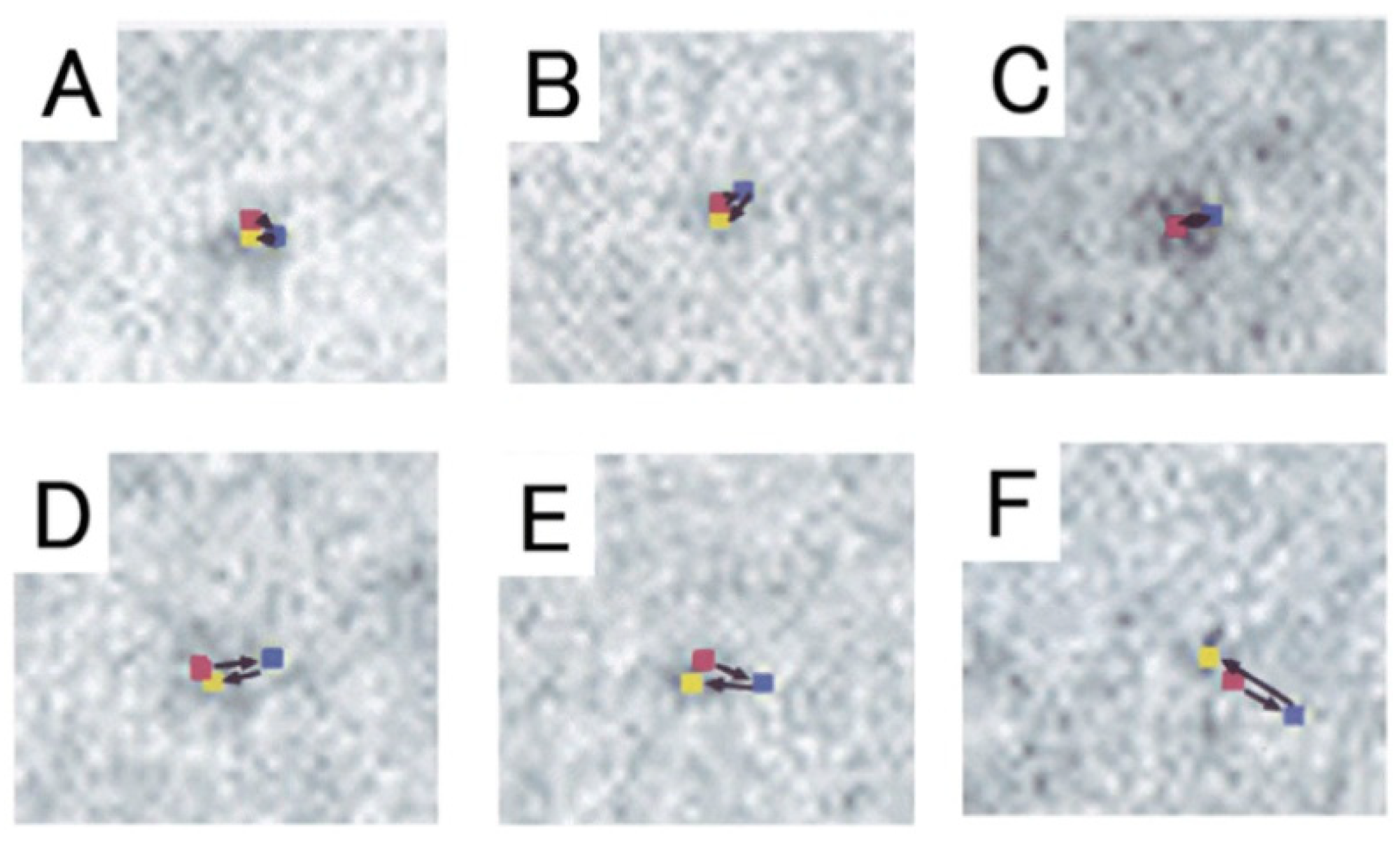
2.2. Position Marking of Individual Myosin Heads
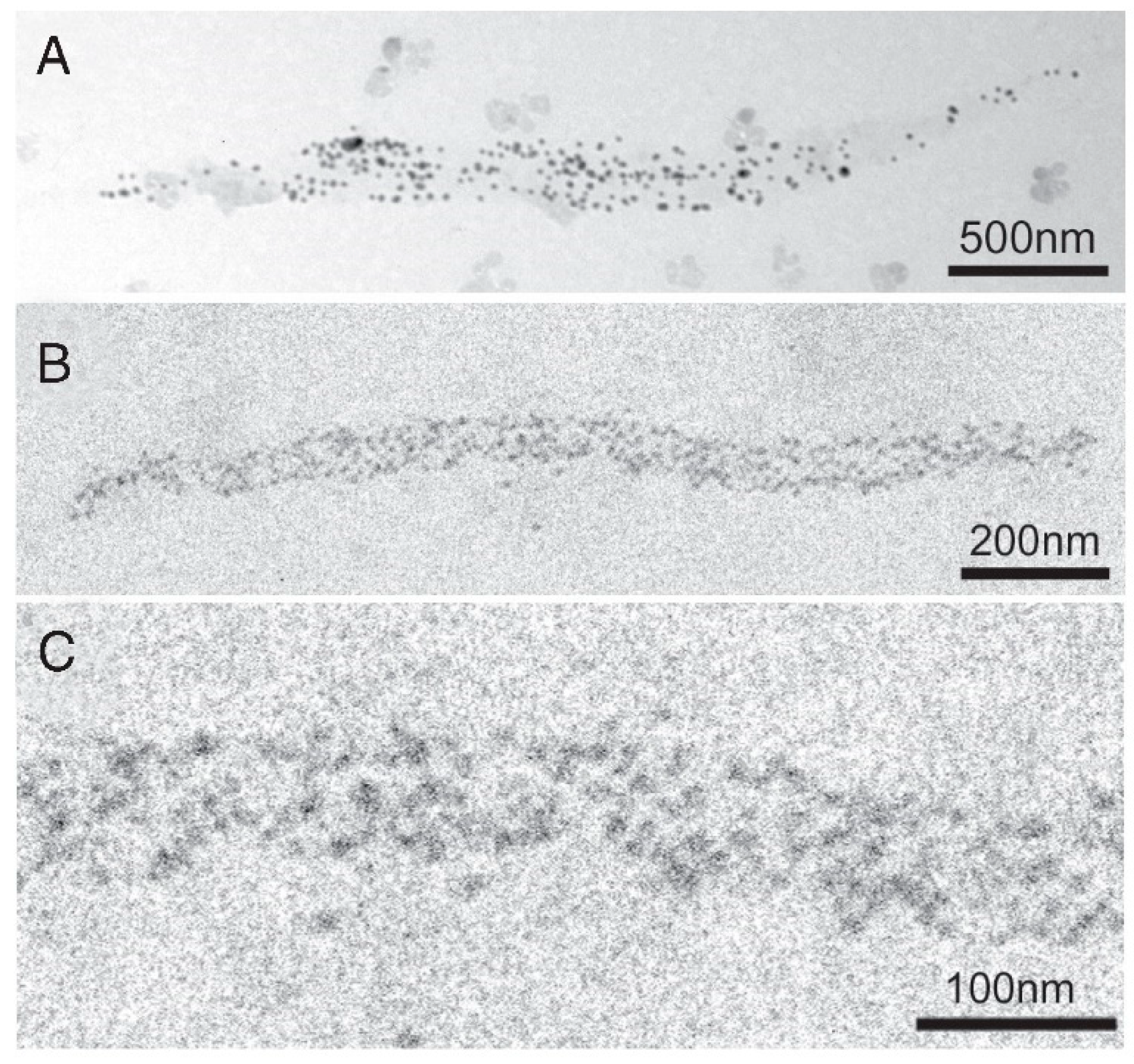
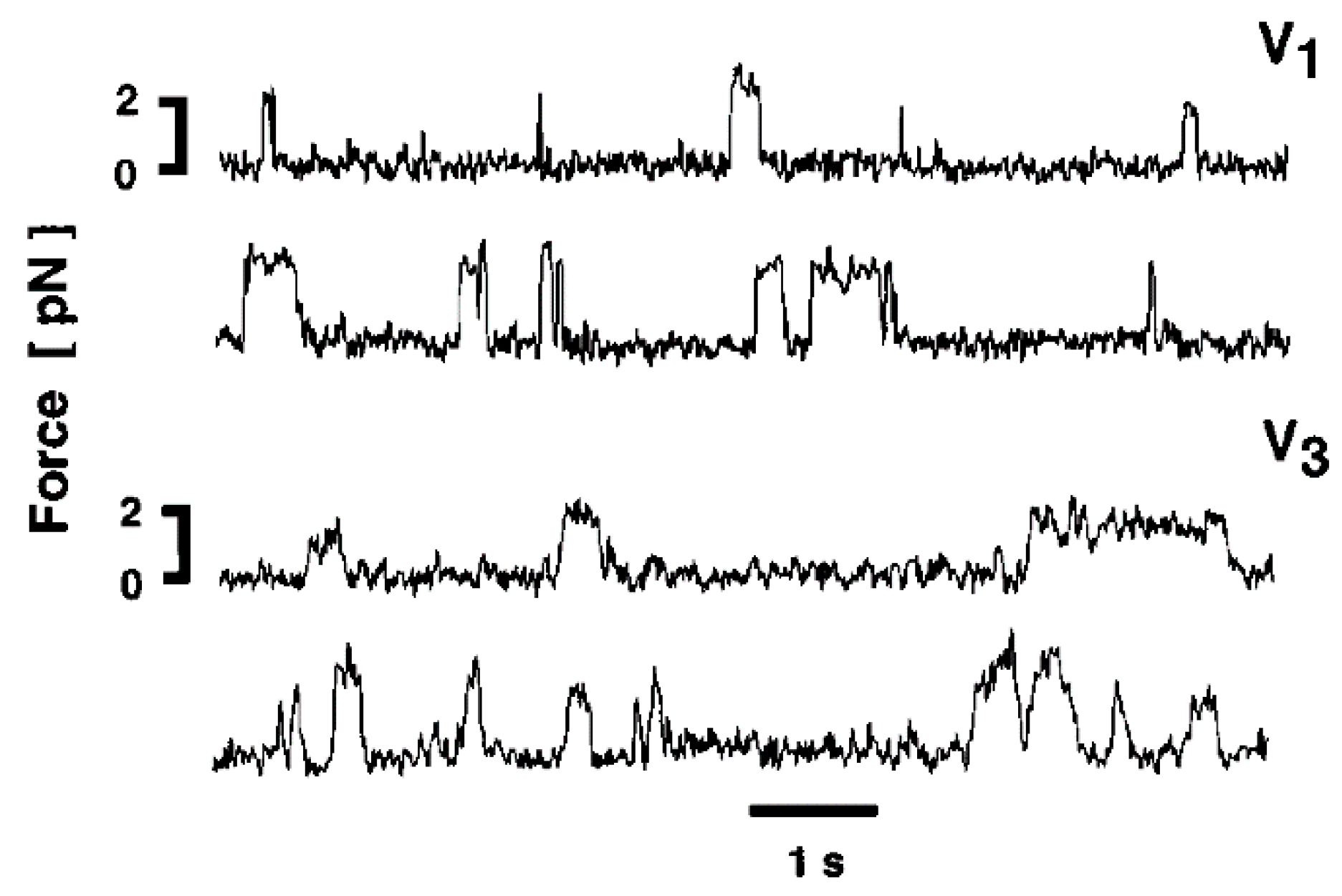
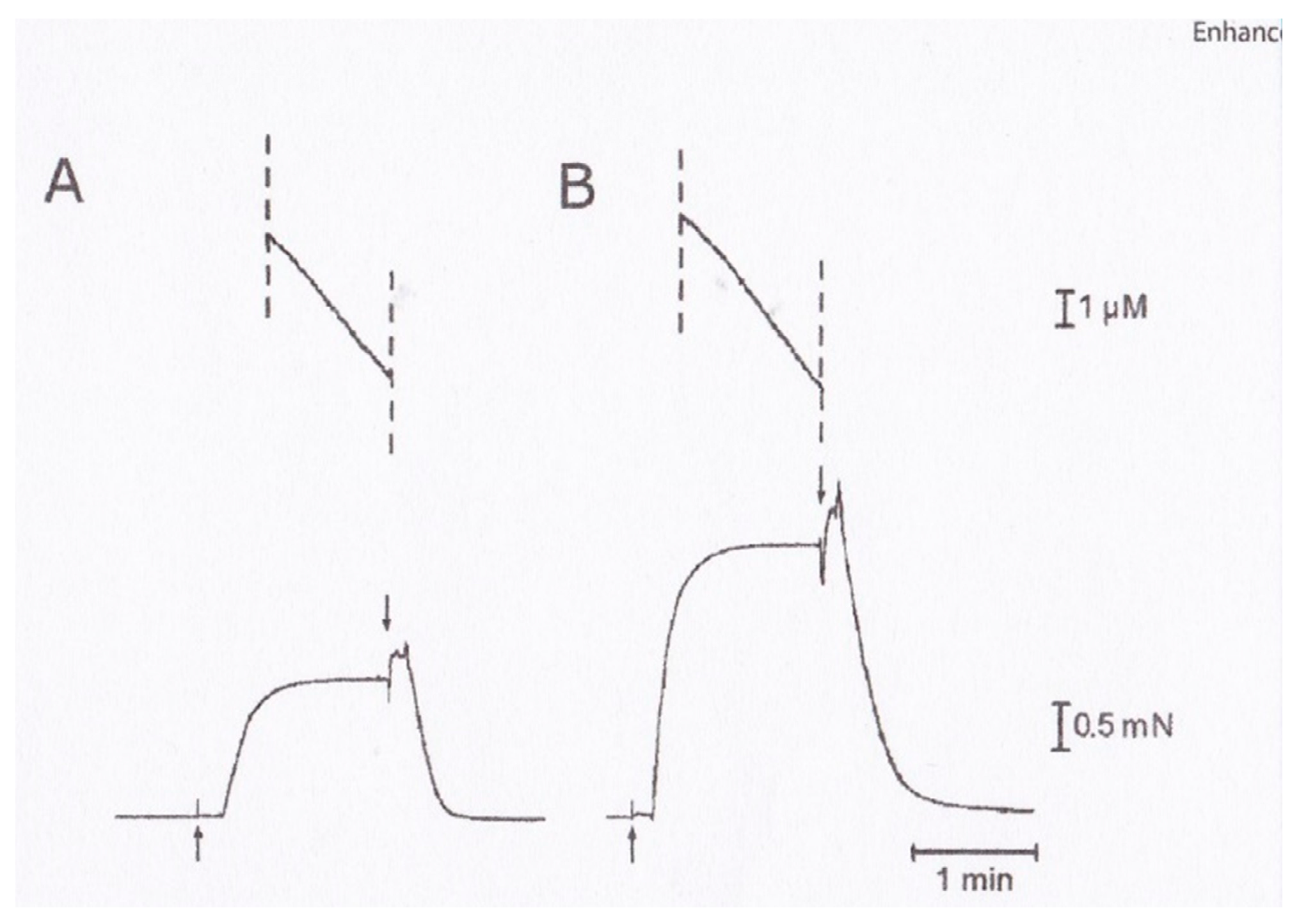
2.3. Recording and Analysis of the ATP-Induced Myosin Head Movement
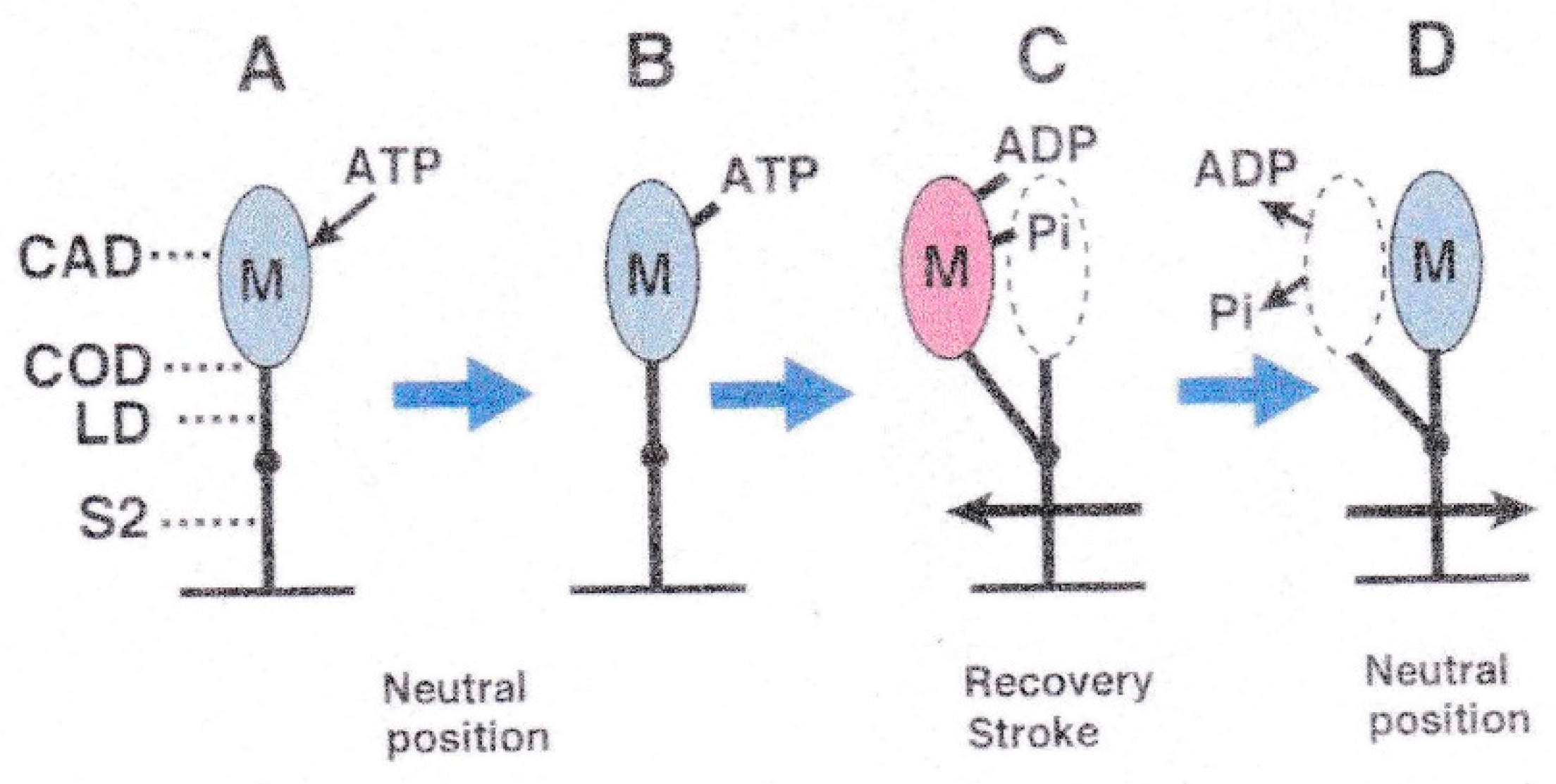
3. Myosin Head Movements in Response to ATP
3.1. Stability in the Position of Myosin Heads in the Absence of ATP
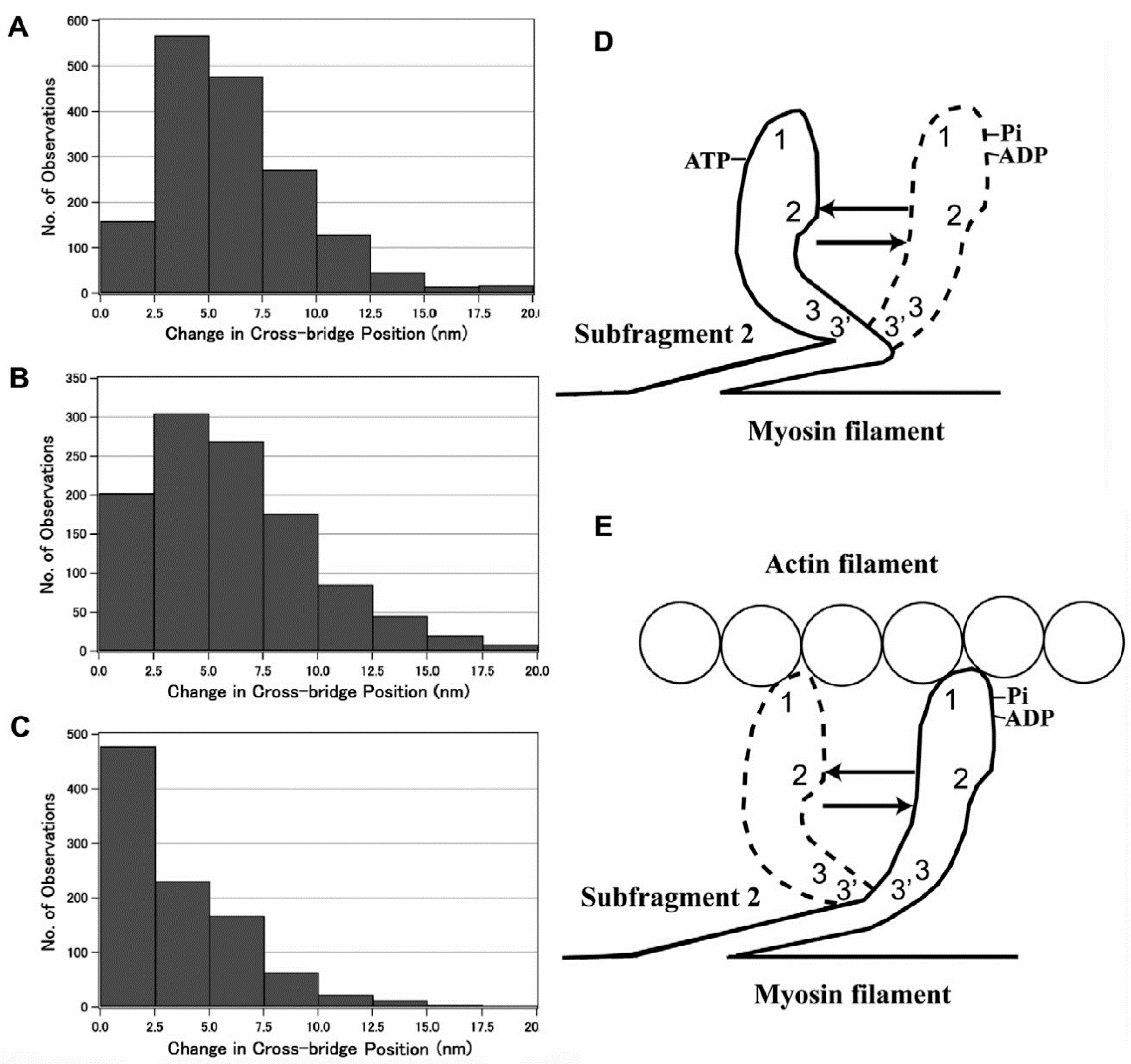
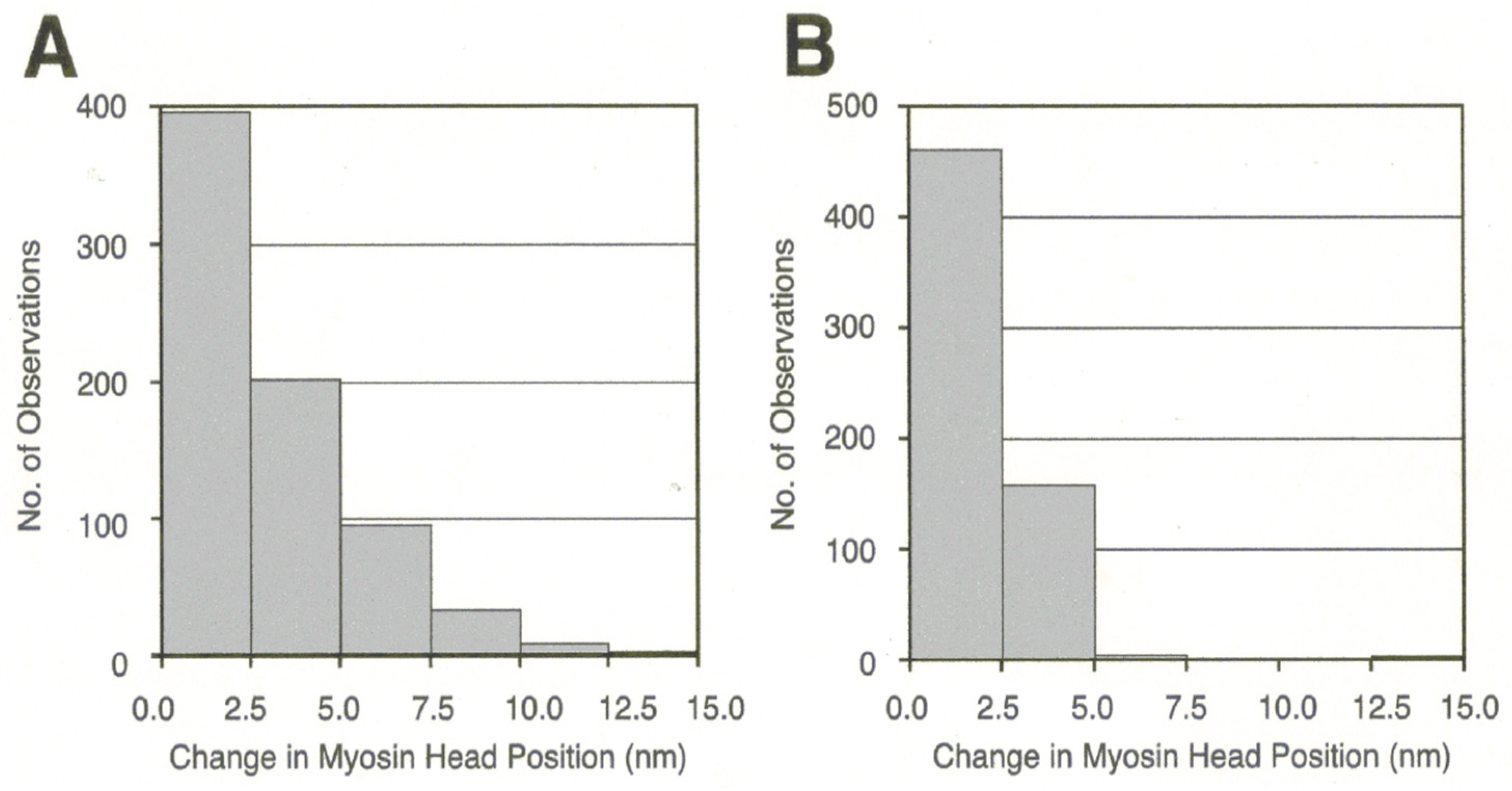
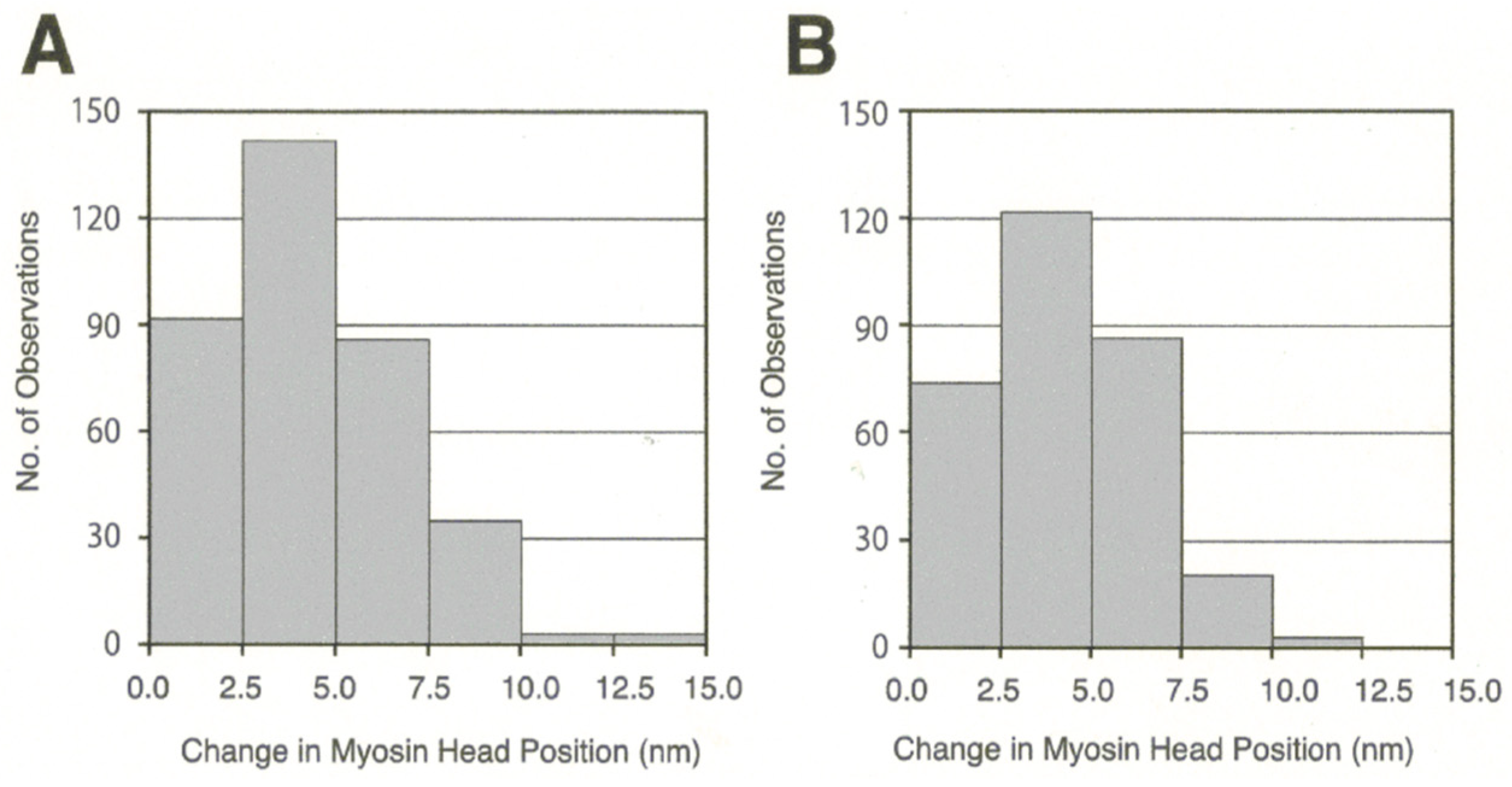
3.2. Amplitude and Direction of ATP-induced Myosin Head Movement in the Absence of Action Filament
3.3. Two Different Modes of Myosin Head Power Stroke
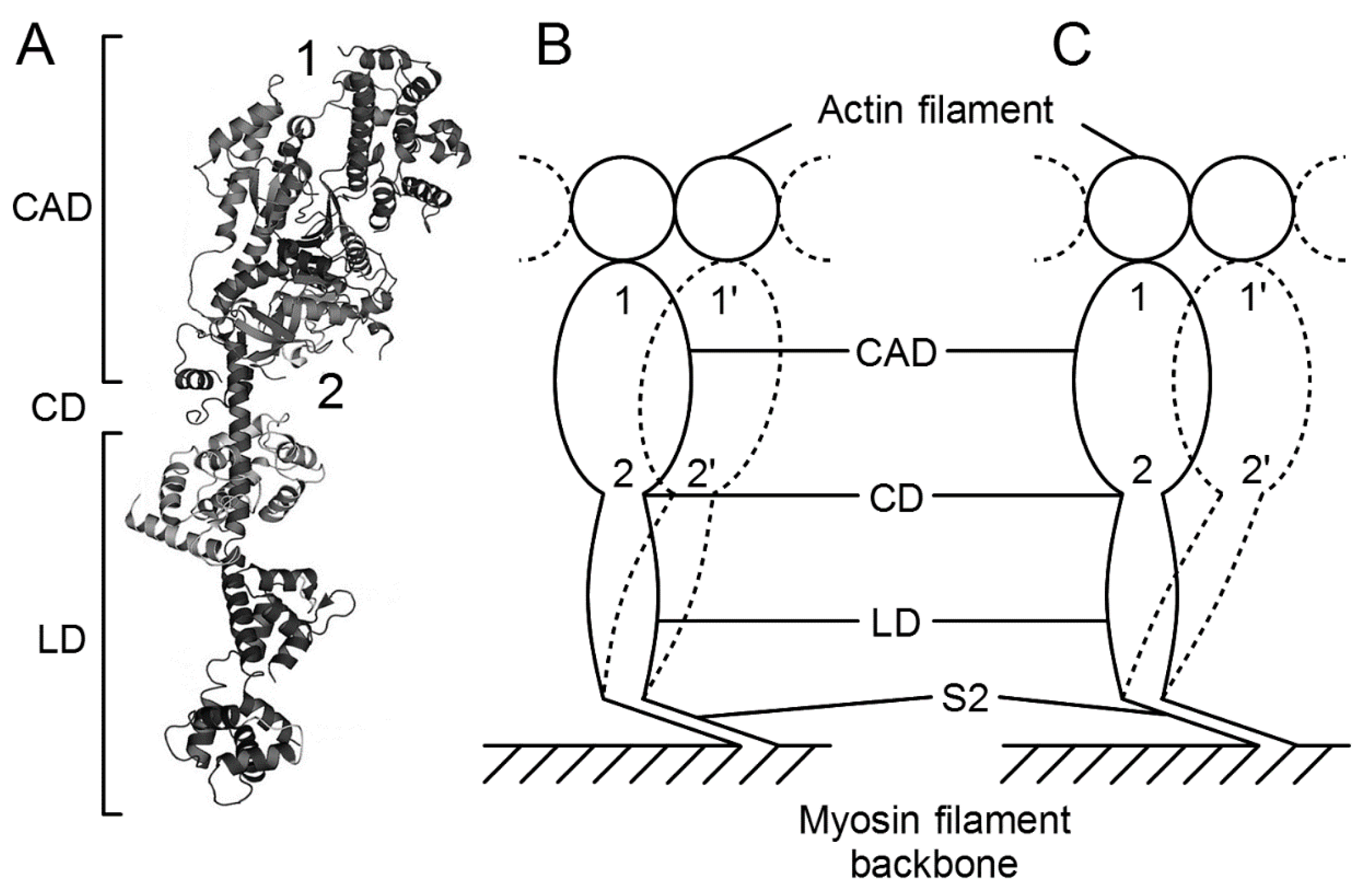
3.4. Fundamental Properties Incorporated in Myosin Heads as Revealed by EC Experiments and Freeze-Fracture Studies on Skinned Muscle Fibers
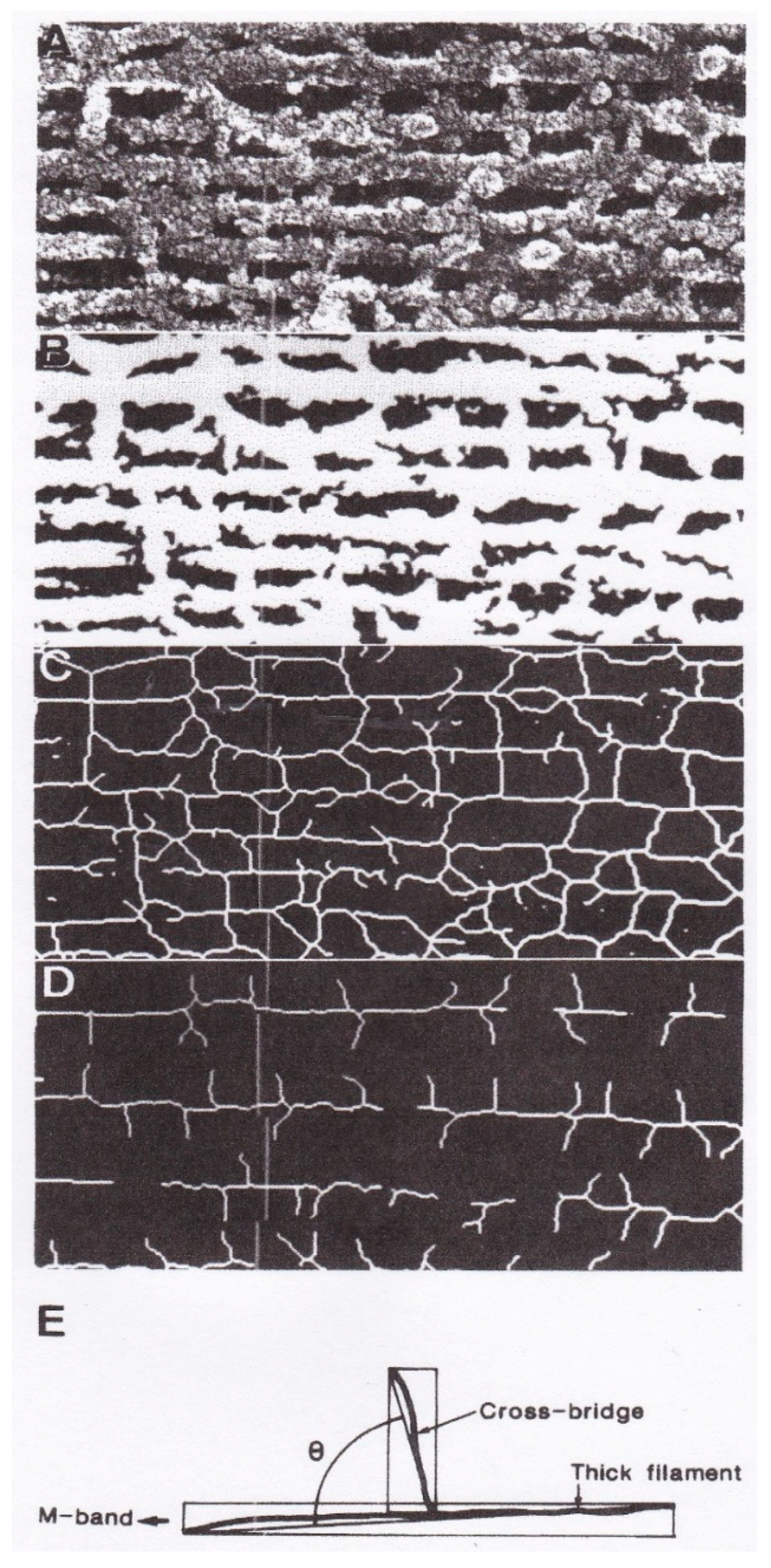
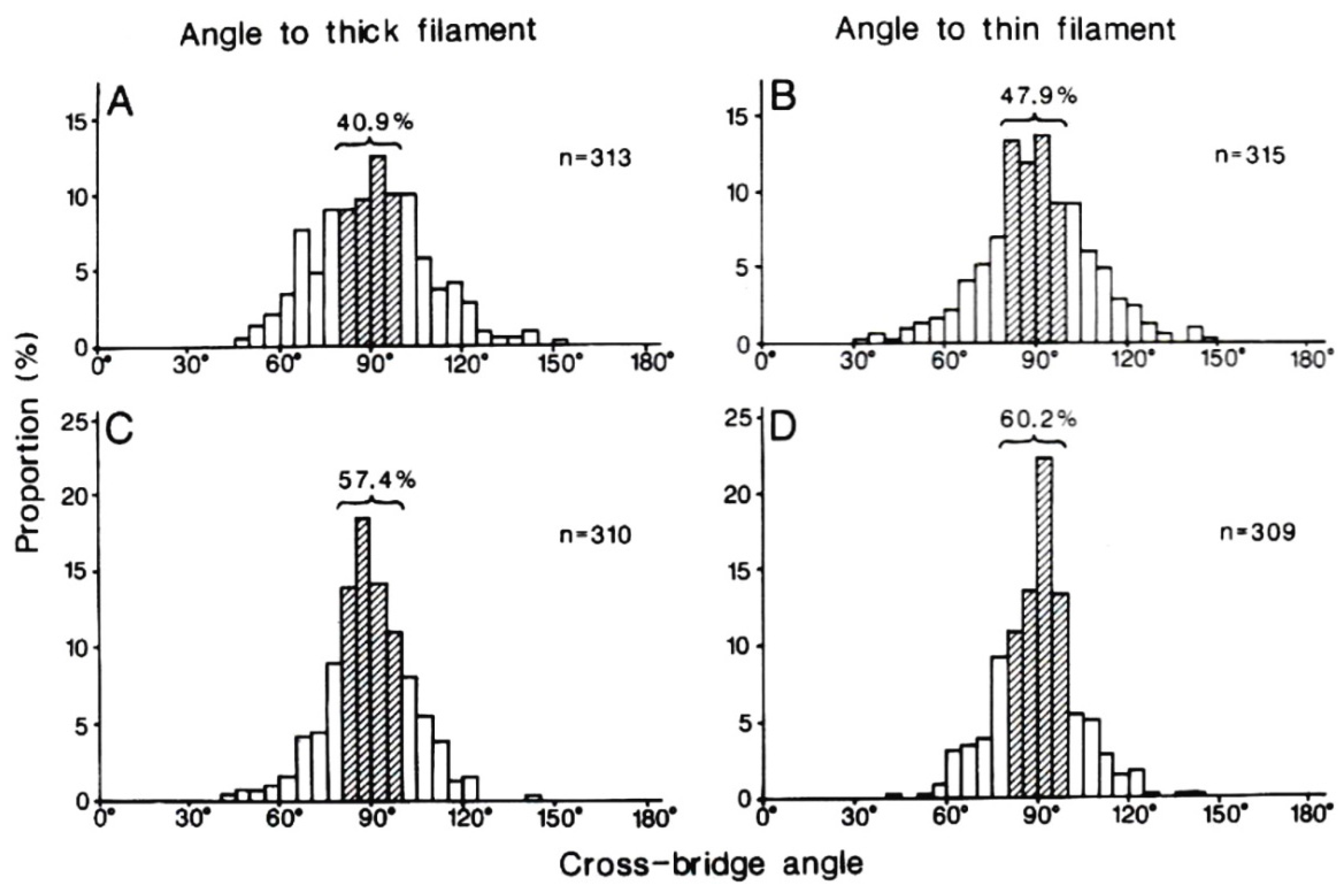
4. Freeze-Fracture Studies on Myosin Head Configurations
5. Conclusions and Future Prospects
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Huxley, H.E.; Hanson, J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Lymn, R.W.; Taylor, E.W. Mechanism of adenosine triphosphat hydrolysis by actomyosin. Biochemistry 1971, 16, 4617–4624. [Google Scholar] [CrossRef]
- Sugi, H.; Tsuchiya, T. Muscle Mechanics I. Intact single fibers. In Current Methods in Muscle Physiology; Sugi, H., Ed.; Oxford University Press: Oxford, UK, 1998; pp. 3–31. [Google Scholar]
- Caorsi, V.; Ushakov, D.S.; West, T.G.; Setta-Kaffetsi, N.; Ferenczi, M.A. FRET characterization for cross-bridge dynamics in single-skinned rigor muscle fibres. Eur. Biophys. J. 2011, 40, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Squire, J.M.; Knupp, C. X-ray diffraction studies of muscle and the crossbridge cycle. Adv. Protein Chem. 2005, 71, 195–255. [Google Scholar] [PubMed]
- Sugi, H.; Minoda, H.; Inayoshi, Y.; Yumoto, F.; Miyakawa, T.; Miyauchi, Y.; Tanokura, M.; Akimoto, T.; Kobayashi, T.; Chae, S.; et al. Direct demonstration of the cross-bridge recovery stroke in muscle thick filaments in aqueous solution by using the hydration chamber. Proc. Natl. Acad. Sci. USA 2008, 45, 17396–17401. [Google Scholar] [CrossRef] [PubMed]
- Butler, E.P.; Hale, K.F. Dynamic Experiments in the Electron Microscope. Practical Methods in the Electron Microscope; North-Holland Pub. Co.: Amsterdam, The Netherlands, 1981; Volume 9, p. 457. [Google Scholar]
- Sugi, H.; Akimoto, T.; Sutoh, K.; Chaen, S.; Oishi, N.; Suzuki, S. Dynamic electron microscopy of ATP-induced myosin head movement in living muscle thick filaments. Proc. Natl. Acad. Sci. USA 1997, 94, 4378–4382. [Google Scholar] [CrossRef] [PubMed]
- Sugi, H.; Minoda, H.; Miyakawa, T.; Tanokura, M.; Chaen, S.; Kobayashi, T. The gas environmental chamber as a powerful tool to study structural changes of living muscle thick filaments coupled with ATP hydrolysis. In Current Basic and Pathological Approaches to the Functions of Muscle Cells and Tissues―From Molecules to Humans; Sugi, H., Ed.; InTech: Rijeka, Croatia, 2012; pp. 3–34. [Google Scholar]
- Sugi, H.; Chaen, S.; Akimoto, T.; Minoda, H.; Miyakawa, T.; Miyauchi, Y.; Tanokura, M.; Sugiura, S. Electron microscopic recording of myosin head power stroke in hydrated myosin filaments. Sci. Rep. 2015, 5, 15700. [Google Scholar] [CrossRef] [PubMed]
- Sugi, H.; Chaen, S.; Akimoto, T.; Tanokura, M.; Miyakawa, T.; Minoda, H. Application of liquid cell microscopy to study function of muscle proteins. In Liquid Cell Electron Microscopy; Ross, F.M., Ed.; Cambridge University Press: Cambridge, UK, 2017; pp. 371–390. [Google Scholar]
- Sugi, H.; Akimoto, T.; Chaen, S.; Miyakawa, T.; Tanokura, M.; Minoda, H. Electron microscopic visualization and recording of ATP-induced myosin head power stroke producing muscle contraction using the gas environmental chamber. In Muscle Contraction and Cell Motility: Fundamentals and Developments; Sugi, H., Ed.; Pan Stanford Publishing: Singapore, 2017; pp. 3–34. [Google Scholar]
- Sutoh, K.; Tokunaga, M.; Wakabayashi, T. Electron microscopic mapping of myosin head with site-directed antibodies. J. Mol. Biol. 1989, 206, 357–363. [Google Scholar] [CrossRef]
- Huxley, A.F. Muscle structure and theories of contraction. Prog. Biophys. Biophys. Chem. 1957, 7, 255–318. [Google Scholar] [PubMed]
- Oiwa, K.; Chaen, S.; Sugi, H. Measurement of work done by ATP-induced sliding between rabbit muscle myosin and algal cell actin cables in vitro. J. Physiol. 1991, 437, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Minoda, H.; Okabe, T.; Inayoshi, Y.; Miyakawa, T.; Miyauchi, Y.; Tanokura, M.; Katayama, E.; Wakabayashi, T.; Akimoto, T.; Sugi, H. Electron microscopic evidence for the myosin head lever arm mechanism in hydrated myosin filaments using the gas environmental chamber. Biochem. Biophys. Res. Commun. 2011, 405, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Nishizuka, T.; Seo, R.; Tadakuma, H.; Kinosita, K.; Ishiwata, H. Characterization of single actomyosin rigor onds: Load dependence of lifetime and mechanical properties. Biophys. J. 2000, 79, 962–974. [Google Scholar] [CrossRef]
- Finer, J.T.; Simmons, R.M.; Spudich, J.A. Single myosin molecule mechanics: Piconewton forces and nanometer steps. Nature 1994, 368, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, S.; Kobayakawa, N.; Fujita, H.; Yamashita, H.; Momomura, S.; Chaen, S.; Omata, M.; Sugi, H. Comparison of unitary displacements and forces between 2 cardiac myosin isoforms by the optical trap technique: Molecular basis for cardiac adaptation. Circ. Res. 1998, 82, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Sugi, H.; Kobayashi, T.; Chaen, S.; Ohnuki, Y.; Saeki, Y.; Sugiura, S. Enhancement of force generated by individual myosin heads in skinned rabit psoas muscle fibers at low ionic strength. PLoS ONE 2013, 8, e63658. [Google Scholar] [CrossRef] [PubMed]
- Suda, H.; Ishikawa, A.; Fukami, A. Evaluation of the critical electron dose on the contractile ability of hydrated muscle fibers in the film-sealed environmental cell. J. Electron Microsc. 1992, 41, 223–229. [Google Scholar]
- Sugi, H.; Yamaguchi, M.; Ohno, T.; Kobayashi, T.; Chaen, S.; Okuyama, H. Tension recovery following ramp-shaped release in high-Ca and low-Ca rigor muscle fibers: Evidence for the dynamic state of AMADP myosin heads in the absence of ATP. PLoS ONE 2016, 11, e0162003. [Google Scholar] [CrossRef] [PubMed]
- Radocaj, A.; Weiss, T.; Helsby, W.I.; Brenner, B.; Kraft, T. Force-generating cross-bridges during ramp-shaped releases: Evidence for a new structural state. Biophys. J. 2009, 96, 1430–1446. [Google Scholar] [CrossRef] [PubMed]
- Sugi, H.; Chaen, S.; Kobayashi, T.; Abe, T.; Kimura, K.; Saeki, Y.; Ohnuki, Y.; Miyakawa, T.; Tanokura, M.; Sugiura, S. Definite differences between in vitro actin-myosin sliding and muscle contraction as revealed using antibodies to myosin head. PLoS ONE 2014, 9, e93272. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A.; Holmes, K.C. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005, 71, 161–193. [Google Scholar] [PubMed]
- Hauser, J.E.; Salpeter, S.R. Organization of acetylcholine receptors in quick-frozen, deep-etched, and rotary-replicated “torpedo” postsynaptic membrane. J. Cell Biol. 1979, 82, 150–173. [Google Scholar] [CrossRef]
- Hauser, J.E.; Cooke, R. Actin-myosin interactions visualized by the quick-freeze, deep-etch replica technique. J. Mol. Biol. 1983, 169, 97–122. [Google Scholar] [CrossRef]
- Suzuki, S.; Oshimi, Y.; Sugi, H. Freeze-fracture studies on the cross-bridge angle distribution at various states and the tin filament stiffness in single skinned frog muscle fibers. J. Electron Microsc. 1993, 42, 107–116. [Google Scholar]
- Morris, C.; Homsher, E. The use of caged compounds. In Current Methods in Muscle Physiology: Advantages, Problems and Limitations; Sugi, H., Ed.; Oxford University Press: Oxford, UK, 1998; pp. 71–89. [Google Scholar]
- Campbell, G.H.; McKeoun, J.T.; Santala, M.K. Time-resolved electron microscopy for in situ experiments. Appl. Phys. Rev. 2014, 1, 41101. [Google Scholar] [CrossRef]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugi, H.; Chaen, S.; Akimoto, T. Electron Microscopic Recording of the Power and Recovery Strokes of Individual Myosin Heads Coupled with ATP Hydrolysis: Facts and Implications. Int. J. Mol. Sci. 2018, 19, 1368. https://doi.org/10.3390/ijms19051368
Sugi H, Chaen S, Akimoto T. Electron Microscopic Recording of the Power and Recovery Strokes of Individual Myosin Heads Coupled with ATP Hydrolysis: Facts and Implications. International Journal of Molecular Sciences. 2018; 19(5):1368. https://doi.org/10.3390/ijms19051368
Chicago/Turabian StyleSugi, Haruo, Shigeru Chaen, and Tsuyoshi Akimoto. 2018. "Electron Microscopic Recording of the Power and Recovery Strokes of Individual Myosin Heads Coupled with ATP Hydrolysis: Facts and Implications" International Journal of Molecular Sciences 19, no. 5: 1368. https://doi.org/10.3390/ijms19051368
APA StyleSugi, H., Chaen, S., & Akimoto, T. (2018). Electron Microscopic Recording of the Power and Recovery Strokes of Individual Myosin Heads Coupled with ATP Hydrolysis: Facts and Implications. International Journal of Molecular Sciences, 19(5), 1368. https://doi.org/10.3390/ijms19051368




