LEAFY COTYLEDON 2: A Regulatory Factor of Plant Growth and Seed Development
Abstract
:1. Introduction
2. The Effect of LEC2 on Plant Early Embryo Morphogenesis
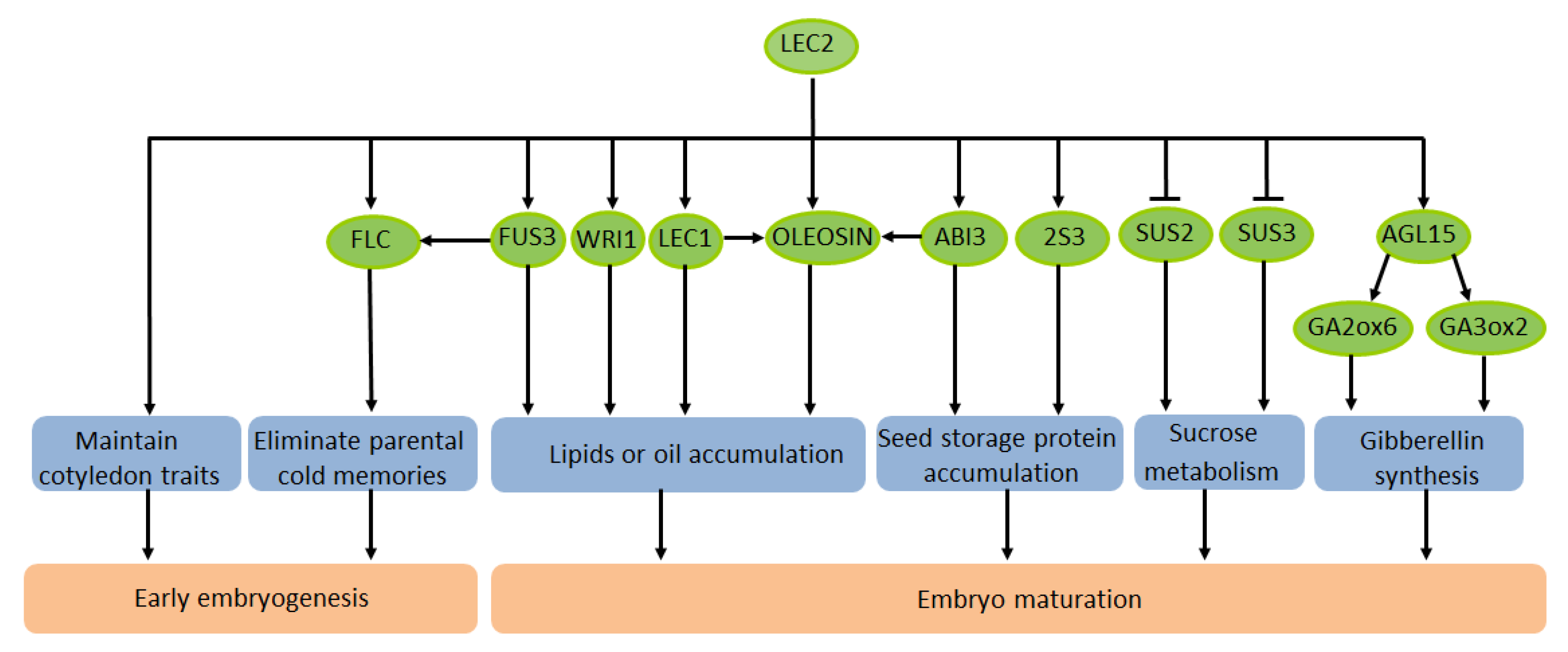
3. The Effect of LEC2 on the Maturation of Plant Seeds
4. The LEC2 Gene Plays a Crucial Role in Somatic Embryogenesis
5. The Function of LEC2 during Other Plant Developmental Stages
6. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LEC2 | LEAFY COTYLEDON 2 |
| ABI3 | ABA INSENSITIVE3 |
| FUS3 | FUSCA3 |
| LEC1 | LEAFY COTYLEDON1 |
| TAG | Triacylglycerol |
| SSP | Seed storage protein |
| AGL15 | AGAMOUS-LIKE15 |
| HAM | Hairy meristem |
| IAA30 | Indole acetic acid inducible 30 |
| SUS | Sucrose synthase |
| LEA | Late embryogenesis abundant |
| DOG1 | DELAY OF GERMINATION1 |
References
- Niu, D.; He, Y. LEAFY COTYLEDONs: Old genes with new roles beyond seed development. F1000Research 2019, 8, 2144. [Google Scholar] [CrossRef]
- Carbonero, P.; Iglesias-Fernandez, R.; Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. J. Exp. Bot. 2017, 68, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Cagliari, A.; Turchetto-Zolet, A.C.; Korbes, A.P.; Maraschin Fdos, S.; Margis, R.; Margis-Pinheiro, M. New insights on the evolution of leafy cotyledon1 (LEC1) type genes in vascular plants. Genomics 2014, 103, 380–387. [Google Scholar] [CrossRef] [Green Version]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [Green Version]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilioti, Z.; Ganopoulos, I.; Bossis, I.; Tsaftaris, A. LEC1-LIKE paralog transcription factor: How to survive extinction and fit in NF-Y protein complex. Gene 2014, 543, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; McCarty, D.R.; Suzuki, M. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 2013, 163, 1293–1305. [Google Scholar] [CrossRef] [Green Version]
- Vicente-Carbajosa, J.; Carbonero, P. Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005, 49, 645–651. [Google Scholar] [CrossRef]
- Braybrook, S.A.; Harada, J.J. LECs go crazy in embryo development. Trends Plant Sci. 2008, 13, 624–630. [Google Scholar] [CrossRef]
- Harada, J.J. Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J. Plant Physiol. 2001, 158, 405–409. [Google Scholar] [CrossRef]
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Li, X.; Jiang, C.; Wong, G.K.; Rothfels, C.J.; Rao, G. Evolutionary analysis of the LAFL genes involved in the land plant seed maturation program. Front. Plant Sci. 2017, 8, 439. [Google Scholar] [CrossRef] [Green Version]
- Braybrook, S.A.; Stone, S.L.; Park, S.; Bui, A.Q.; Le, B.H.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 3468–3473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Suzuki, M.; McCarty, D.R. Structural variation affecting DNA backbone interactions underlies adaptation of B3 DNA binding domains to constraints imposed by protein architecture. Nucleic Acids Res. 2021, 49, 4989–5002. [Google Scholar] [CrossRef] [PubMed]
- Leviczky, T.; Molnar, E.; Papdi, C.; Oszi, E.; Horvath, G.V.; Vizler, C.; Nagy, V.; Pauk, J.; Bogre, L.; Magyar, Z. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 2019, 146, dev179333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.R.; Bian, S.M.; Tang, M.J.; Lu, Q.; Li, S.B.; Liu, X.G.; Tian, G.; Nguyen, V.; Tsang, E.W.T.; Wang, A.M.; et al. MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet. 2012, 8, e1003091. [Google Scholar] [CrossRef]
- Willmann, M.R.; Mehalick, A.J.; Packer, R.L.; Jenik, P.D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011, 155, 1871–1884. [Google Scholar] [CrossRef] [Green Version]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [Green Version]
- Gutzat, R.; Borghi, L.; Fuetterer, J.; Bischof, S.; Laizet, Y.; Hennig, L.; Feil, R.; Lunn, J.; Gruissem, W. RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 2011, 138, 2977–2986. [Google Scholar] [CrossRef] [Green Version]
- Meinke, D.W.; Franzmann, L.H.; Nickle, T.C.; Yeung, E.C. Leafy Cotyledon mutants of Arabidopsis. Plant Cell 1994, 6, 1049–1064. [Google Scholar] [CrossRef] [PubMed]
- Gaj, M.D.; Zhang, S.; Harada, J.J.; Lemaux, P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 2005, 222, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.S.; Dubreucq, B.; Miquel, M.; Caboche, M.; Lepiniec, L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005, 579, 4666–4670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef] [Green Version]
- Angeles-Nunez, J.G.; Tiessen, A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds, decreasing oil and protein content, while maintaining high levels of starch and sucrose in mature seeds. J. Plant Physiol. 2011, 168, 1891–1900. [Google Scholar] [CrossRef]
- Feeney, M.; Frigerio, L.; Cui, Y.; Menassa, R. Following vegetative to embryonic cellular changes in leaves of Arabidopsis overexpressing LEAFY COTYLEDON2. Plant Physiol. 2013, 162, 1881–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.U.; Jung, S.J.; Lee, K.R.; Kim, E.H.; Lee, S.M.; Roh, K.H.; Kim, J.B. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio 2013, 4, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Iwase, A.; Mita, K.; Nonaka, S.; Ikeuchi, M.; Koizuka, C.; Ohnuma, M.; Ezura, H.; Imamura, J.; Sugimoto, K. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 2015, 128, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.P.; Zhou, C.; Wang, S.S.; Yuan, J.; Zhang, X.S.; Su, Y.H. FUSCA 3 interacting with LEAFY COTYLEDON 2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 2017, 213, 1740–1754. [Google Scholar] [CrossRef]
- Tao, Z.; Hu, H.; Luo, X.; Jia, B.; Du, J.; He, Y. Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 2019, 5, 424–435. [Google Scholar] [CrossRef]
- Barreto, H.G.; Ságio, S.A.; Chalfun-Júnior, A.; Fevereiro, P.; Benedito, V.A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. Plant Cell Tissue Organ Cult. 2019, 139, 327–337. [Google Scholar] [CrossRef]
- Meinke, D.W. A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 1992, 258, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Qüesta, J.I.; Song, J.; Geraldo, N.; An, H.; Dean, C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 2016, 353, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Luo, X.; Li, Z.; Yang, W.; Wang, Y.; Liu, R.; Du, J.; He, Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016, 48, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- McCarty, D.R. Genetic control and integration of maturation and germination pathways in seed development. Annu. Rev. Plant Biol. 1995, 46, 71–93. [Google Scholar] [CrossRef]
- Goldberg, R.B.; Paiva, G.; Yadegari, R. Plant embryogenesis: Zygote to seed. Science 1994, 266, 605–614. [Google Scholar] [CrossRef]
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant. Biol. 2019, 61, 564–580. [Google Scholar] [CrossRef] [Green Version]
- Higgins, T. Synthesis and regulation of major proteins in seeds. Annu. Rev. Plant Phys. 1984, 35, 191–221. [Google Scholar] [CrossRef]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef] [Green Version]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; van Staden, J. New insights into plant somatic embryogenesis: An epigenetic view. Acta Physiol. Plant 2017, 39, 194. [Google Scholar] [CrossRef]
- Wang, H.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 2004, 16, 1206–1219. [Google Scholar] [CrossRef] [Green Version]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef] [Green Version]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoet, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Vaultier, M.N.; Rochat, C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 2004, 55, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Bieniawska, Z.; Paul Barratt, D.H.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef]
- Fallahi, H.; Scofield, G.N.; Badger, M.R.; Chow, W.S.; Furbank, R.T.; Ruan, Y.L. Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J. Exp. Bot. 2008, 59, 3283–3295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angeles-Nunez, J.G.; Kronenberger, J.; Wuilleme, S.; Lepiniec, L.; Rochat, C. Study of AtSUS2 localization in seeds reveals a strong association with plastids. Plant Cell Physiol. 2008, 49, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Nunez, J.G.; Tiessen, A. Regulation of AtSUS2 and AtSUS3 by glucose and the transcription factor LEC2 in different tissues and at different stages of Arabidopsis seed development. Plant Mol. Biol. 2012, 78, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.H. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, N.; Yang, Y.; Li, Y.; Wang, L.; Huang, P.; Gao, Y.; An, C. Efficient LEC2 activation of OLEOSIN expression requires two neighboring RY elements on its promoter. Sci. China C Life Sci. 2009, 52, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Kelemen, Z.; Thevenin, J.; Boulard, C.; Blanchet, S.; To, A.; Payre, M.; Berger, N.; Effroy-Cuzzi, D.; Franco-Zorrilla, J.M.; et al. Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol. 2016, 171, 1099–1112. [Google Scholar] [PubMed]
- Guo, F.; Liu, C.; Xia, H.; Bi, Y.; Zhao, C.; Zhao, S.; Hou, L.; Li, F.; Wang, X. Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 2013, 8, e71714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Wang, J.; Liu, C.; Li, C.; Qiu, J.; Zhao, C.; Xia, H.; Ma, C.; Wang, X.; Li, P. Expression of AtLEC2 and AtIPTs promotes embryogenic callus formation and shoot regeneration in tobacco. BMC Plant Biol. 2019, 19, 314. [Google Scholar] [CrossRef] [Green Version]
- Grimault, A.; Gendrot, G.; Chaignon, S.; Gilard, F.; Tcherkez, G.; Thevenin, J.; Dubreucq, B.; Depege-Fargeix, N.; Rogowsky, P.M. Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant Sci. 2015, 236, 116–125. [Google Scholar] [CrossRef]
- Manan, S.; Ahmad, M.Z.; Zhang, G.; Chen, B.; Haq, B.U.; Yang, J.; Zhao, J. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Feher, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. BBA-Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef]
- Salvo, S.A.; Hirsch, C.N.; Buell, C.R.; Kaeppler, S.M.; Kaeppler, H.F. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS ONE 2014, 9, e111407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaj, M.D. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47. [Google Scholar] [CrossRef]
- Wojcikowska, B.; Jaskola, K.; Gasiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Jha, P.; van Staden, J. LEAFY COTYLEDONs (LECs): Master regulators in plant embryo development. Plant Cell Tissue Organ Cult. 2020, 140, 475–487. [Google Scholar] [CrossRef]
- Chen, Y.F.; Etheridge, N.; Schaller, G.E. Ethylene signal transduction. Ann. Bot. 2005, 95, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Guo, H. Genetic basis of ethylene perception and signal transduction in Arabidopsis. J. Integr. Plant Biol. 2008, 50, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Wojcikowska, B.; Gaj, M.D. ERF022 impacts the induction of somatic embryogenesis in Arabidopsis through the ethylene-related pathway. Planta 2015, 241, 967–985. [Google Scholar] [CrossRef] [Green Version]
- Gliwicka, M.; Nowak, K.; Balazadeh, S.; Mueller-Roeber, B.; Gaj, M.D. Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e69261. [Google Scholar] [CrossRef] [Green Version]
- Belide, S.; Zhou, X.R.; Kennedy, Y.; Lester, G.; Shrestha, P.; Petrie, J.R.; Singh, S.P. Rapid expression and validation of seed-specific constructs in transgenic LEC2 induced somatic embryos of Brassica napus. Plant Cell Tissue Organ Cult. 2013, 113, 543–553. [Google Scholar] [CrossRef]
- Shires, M.E.; Florez, S.L.; Lai, T.S.; Curtis, W.R. Inducible somatic embryogenesis in Theobroma cacao achieved using the DEX-activatable transcription factor-glucocorticoid receptor fusion. Biotechnol. Lett. 2017, 39, 1747–1755. [Google Scholar] [CrossRef] [Green Version]
- Fister, A.S.; Landherr, L.; Perryman, M.; Zhang, Y.F.; Guiltinan, M.J.; Maximova, S.N. Glucocorticoid receptor-regulated TcLEC2 expression triggers somatic embryogenesis in Theobroma cacao leaf tissue. PLoS ONE 2018, 13, e0207666. [Google Scholar]
- Brand, A.; Quimbaya, M.; Tohme, J.; Chavarriaga-Aguirre, P. Arabidopsis LEC1 and LEC2 orthologous genes are key regulators of somatic embryogenesis in cassava. Front. Plant Sci. 2019, 10, 673. [Google Scholar] [CrossRef]
- Santos, D.; Fevereiro, P. Loss of DNA methylation affects somatic embryogenesis in Medicago truncatula. Plant Cell Tissue Organ Cult. 2002, 70, 155–161. [Google Scholar] [CrossRef]
- Duque, A.S.; Pires, A.S.; dos Santos, D.M.; Fevereiro, P. Efficient somatic embryogenesis and plant regeneration from long-term cell suspension cultures of Medicago truncatula cv. Jemalong. In Vitro Cell. Dev. Biol.-Plant 2006, 42, 270–273. [Google Scholar]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Lee, K.R.; Jung, S.J.; Shin, H.A.; Go, Y.S.; Suh, M.C.; Kim, J.B. Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 2015, 13, 1346–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rikiishi, K.; Maekawa, M. Seed maturation regulators are related to the control of seed dormancy in wheat (Triticum aestivum L.). PLoS ONE 2014, 9, e107618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentsink, L.; Jowett, J.; Hanhart, C.J.; Koornneef, M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 17042–17047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reidt, W.; Wohlfarth, T.; Ellerstrom, M.; Czihal, A.; Tewes, A.; Ezcurra, I.; Rask, L.; Baumlein, H. Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000, 21, 401–408. [Google Scholar] [CrossRef]
- Wu, L.; Joshi, C.P.; Chiang, V.L. A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J. 2000, 22, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Nookaraju, A.; Pandey, S.K.; Fujino, T.; Kim, J.Y.; Suh, M.C.; Joshi, C.P. Enhanced accumulation of fatty acids and triacylglycerols in transgenic tobacco stems for enhanced bioenergy production. Plant Cell Rep. 2014, 33, 1041–1052. [Google Scholar] [CrossRef]
| Biological Function | Reference |
|---|---|
| Induces somatic embryos and embryonic development in vegetative cells | [4] |
| Initiates somatic embryo development | [12] |
| Regulates the expression of storage protein genes | [11] |
| Induces somatic embryogenesis | [22] |
| Triggers the stockpile of oil and seed-specific mRNAs | [23] |
| Induces maturation traits and auxin activity | [24] |
| Affects the contents of oil and protein, starch, and sucrose | [25] |
| Changes the shape and anatomy of leaves | [26] |
| Triggers the expression of genes encoding seed maturation and oil body protein regulators in trophic organization | [27] |
| Promotes embryogenic callus formation in roots | [28] |
| Controls the formation of lateral roots | [29] |
| Involved in early embryogenesis Participates in the development of somatic embryos | [30] [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Sun, G.; Liu, C.; Liu, S. LEAFY COTYLEDON 2: A Regulatory Factor of Plant Growth and Seed Development. Genes 2021, 12, 1896. https://doi.org/10.3390/genes12121896
Liu B, Sun G, Liu C, Liu S. LEAFY COTYLEDON 2: A Regulatory Factor of Plant Growth and Seed Development. Genes. 2021; 12(12):1896. https://doi.org/10.3390/genes12121896
Chicago/Turabian StyleLiu, Boling, Ge Sun, Changju Liu, and Shijuan Liu. 2021. "LEAFY COTYLEDON 2: A Regulatory Factor of Plant Growth and Seed Development" Genes 12, no. 12: 1896. https://doi.org/10.3390/genes12121896






