Using Personal Genomic Data within Primary Care: A Bioinformatics Approach to Pharmacogenomics
Abstract
:1. Personalized Medicine and Pharmacogenomics
1.1. Personalized Medicine
1.2. Pharmacogenomics
1.3. Combining Family History and Genetics
2. The Data Gap
2.1. Providing Data to the Primary Care Professional
2.2. Contextual Knowledge
3. Research and Privacy
3.1. Direct Impact between Primary Care and Academic Research
3.2. Genomic Data, Privacy and Ethics
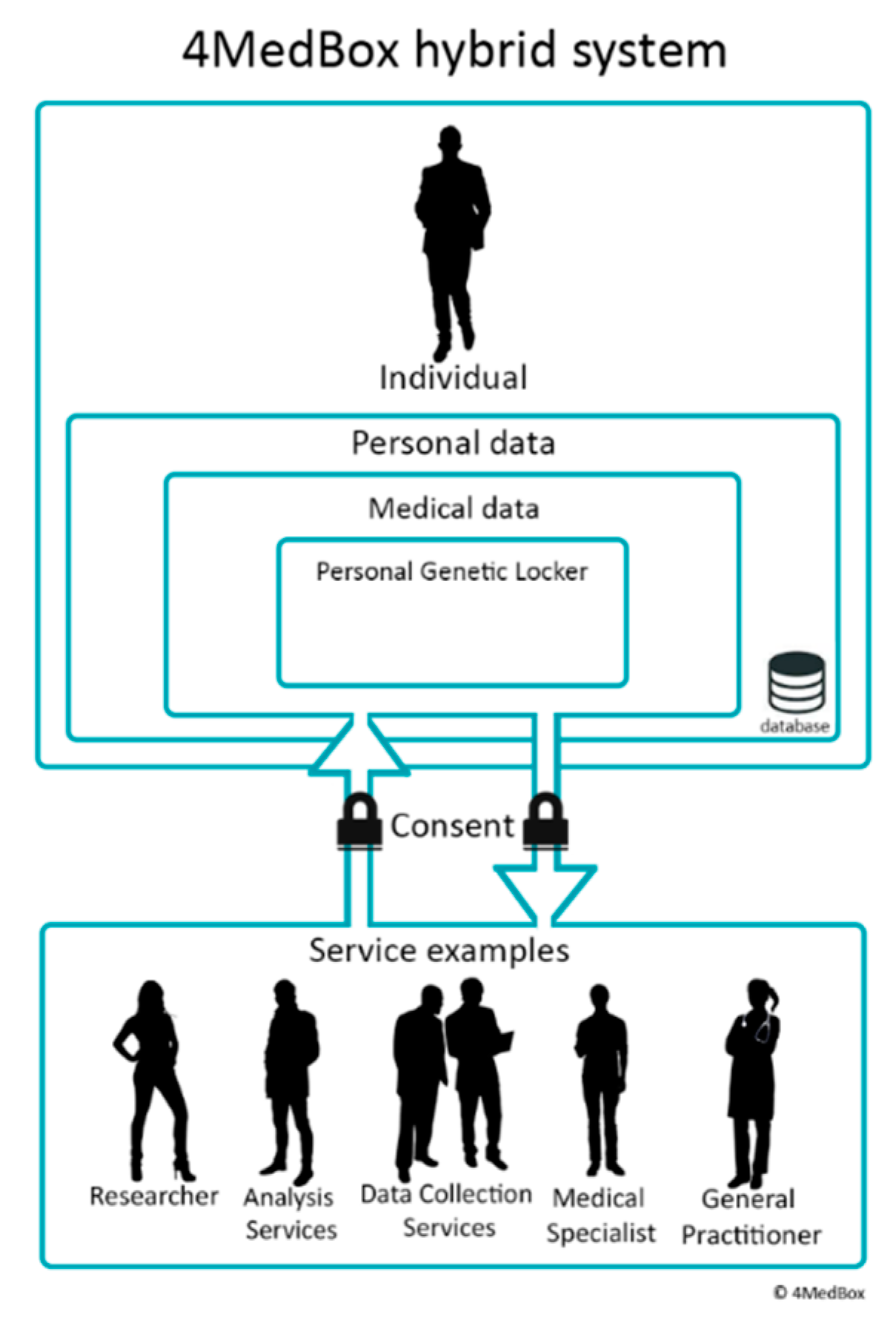
4. Personal Genetic Locker
4.1. First Use Case for Implementation
4.2. 4MedBox
5. Adoption
6. Implementation
6.1. Steps to Overcome Implementation Obstacles
- Language barriers for better adoption [36];
- Diversifying research populations [38];
- Inclusion of minors, parental consent and when rights are transferred from parent to child [39];
- Creating an ethical foundation for the system;
- Providing storage for personal genetic data;
- Provide data visiting capabilities based on FAIR principles;
- Providing primary care professionals the means to interpret the genetic data of their patients [40];
- Technological standards to share across primary care and research all over Europe;
- Architecture for such a system.
6.2. First Study
7. Concluding Remarks and Future Directions
From Primary Care to Preventive Care
- Use the pharmacogenetics profile to get the best medication;
- Know about risks for hereditary disorders and treat them early where possible;
- Perform a genetic health-risk assessment for personalized follow-up and where possible reduce risks early by lifestyle adjustments;
- As soon as one wants children, use it to compare with the partner’s DNA to identify possible risks of hereditary disorders for the children.
Author Contributions
Funding
Conflicts of Interest
References
- Stanek, E.J.; Sanders, C.L.; Taber, K.A.; Khalid, M.; Patel, A.; Verbrugge, R.R.; Agatep, B.C.; Aubert, R.E.; Epstein, R.S.; Frueh, F.W. Adoption of pharmacogenomic testing by US physicians: Results of a nationwide survey. Clin. Pharmacol. Ther. 2012, 91, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Bank, P.C.; Swen, J.J.; Guchelaar, H.J. A nationwide survey of pharmacists’ perception of pharmacogenetics in the context of a clinical decision support system containing pharmacogenetics dosing recommendations. Pharmacogenomics 2017, 18, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Bank, P.C.D.; Caudle, K.E.; Swen, J.J.; Gammal, R.S.; Whirl-Carrillo, M.; Klein, T.E.; Relling, M.V.; Guchelaar, H.J. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 2018, 103, 599–618. [Google Scholar] [CrossRef] [PubMed]
- Bank, P.C.D.; Swen, J.J.; Guchelaar, H.J. Estimated nationwide impact of implementing a preemptive pharmacogenetic panel approach to guide drug prescribing in primary care in The Netherlands. BMC Med. 2019, 17, 110. [Google Scholar] [CrossRef] [Green Version]
- Petry, N.; Baye, J.; Aifaoui, A.; Wilke, R.A.; Lupu, R.A.; Savageau, J.; Gapp, B.; Massmann, A.; Hahn, D.; Hajek, C.; et al. Implementation of wide-scale pharmacogenetic testing in primary care. Pharmacogenomics 2019, 20, 903–913. [Google Scholar] [CrossRef]
- Houwink, E.J.F.; Hortensius, O.R.; van Boven, K.; Sollie, A.; Numans, M.E. Genetics in primary care: Validating a tool to pre-symptomatically assess common disease risk using an Australian questionnaire on family history. Clin. Transl. Med. 2019, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, P.; Scheuner, M.; Peterson-Oehlke, K.; Gwinn, M.; Faucett, A.; Khoury, M. Can family history be used as a tool for public health and preventive medicine? Genet. Med. 2002, 4, 304–310. [Google Scholar] [CrossRef]
- Acheson, L.; Wiesner, G.; Zyzanski, S.; Goodwin, M.; Stange, K. Family history-taking in community family practice: Implications for genetic screening. Genet. Med. 2000, 2, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Ruffin, M.T.; Nease, D.E.; Sen, A.; Pace, W.D.; Wang, C.; Acheson, L.S.; Rubinstein, W.S.; O’Neill, S.; Gramling, R.; For the Family History Impact Trial (fhitr) Group. Effect of preventive messages tailored to family history on health behaviors: The family healthware impact trial. Ann. Fam. Med. 2011, 9, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Houwink, E.J.F.; van Luijk, S.J.; Henneman, L.; van der Vleuten, C.; Dinant, G.J.; Cornel, M.C. Genetic educational needs and the role of genetics in primary care: A focus group study with multiple perspectives. BMC Fam. Pract. 2011, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Daelemans, S.; Vandevoorde, J.; Vansintejan, J.; Borgermans, L.; Devroey, D. The use of family history in primary health care: A qualitative study. Adv. Prev. Med. 2013, 2013, 695763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Esch, S.C.M.; Heideman, W.H.; Cleijne, W.; Cornel, M.C.; Snoek, F.J. Health care providers’ perspective on using family history in the prevention of type 2 diabetes: A qualitative study including different disciplines. BMC Fam. Pract. 2013, 14, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houwink, E.J.F.; Sollie, A.W.; Numans, M.E.; Cornel, M.C. Proposed roadmap to stepwise integration of genetics in family medicine and clinical research. Clin. Transl. Med. 2013, 2, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snoeck-Stroband, J.B. Towards clinical phenotyping of COPD: Effects of Inhaled Corticosteroide in the GLUCOLD Study. Ph.D. Thesis, Leiden University, Leiden, The Netherlands, 12 September 2012. [Google Scholar]
- Bregendahl, M.E.; Orlando, L.A.; Palaniappan, L. Health Risk Assessments, Family Health History, and Predictive Genetic/Pharmacogenetic Testing. In Genomic and Precision Medicine, 3rd ed.; David, S.P., Ed.; Academic Press: Cambridge, MA, USA; An Imprint of Elsevier: Cambridge, MA, USA, 2017; pp. 75–88. [Google Scholar]
- Swen, J.J.; Nijenhuis, M.; van Rhenen, M.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.; van der Weide, J.; et al. Pharmacogenetic Information in Clinical Guidelines: The European Perspective. Clin. Pharmacol. Ther. 2018, 103, 795–801. [Google Scholar] [CrossRef]
- Houwink, E.J.F.; Kasteleyn, M.J.; Alpay, L.; Pearce, C.; Butler-Henderson, K.; Meijer, E.; van Kampen, S.; Versluis, A.; Bonten, T.N.; van Dalfsen, J.H.; et al. SERIES: EHealth in primary care. Part 3: EHealth and education in primary care. Eur. J. Gen. Pract. 2020, 26, 108–118. [Google Scholar] [CrossRef]
- Boers, S.N.; Jongsma, K.R.; Lucivero, F.; Aardoom, J.; Büchner, F.L.; de Vries, M.; Honkoop, P.; Houwink, E.J.F.; Kasteleyn, M.J.; Meijer, E.; et al. SERIES: EHealth in primary care. Part 2: Exploring the ethical implications of its application in primary care practice. Eur. J. Gen. Pract. 2020, 26, 26–32. [Google Scholar] [CrossRef] [Green Version]
- van der Kleij, R.M.J.J.; Kasteleyn, M.J.; Meijer, E.; Bonten, T.N.; Houwink, E.J.F.; Teichert, M.; van Luenen, S.; Vedanthan, R.; Evers, A.W.M.; Car, J.; et al. SERIES: EHealth in primary care. Part 1: Concepts, conditions and challenges. Eur. J. Gen. Pract. 2019, 25, 179–189. [Google Scholar] [CrossRef]
- Wilkinson, M.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Vayena, E.; Gasser, U. Between openness and privacy in genomics. PLoS Med. 2016, 13, e1001937. [Google Scholar] [CrossRef] [Green Version]
- Deist, T.M.; Dankers, F.J.W.M.; Ojha, P.; Marshall, M.S.; Janssen, T.; Faivre-Finn, C.; Masciocchi, C.; Valentini, V.; Wang, J.; Chen, J.; et al. Distributed learning on 20 000+ lung cancer patients—The Personal Health Train. Radiother. Oncol. 2020, 144, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Beyan, O.; Choudhury, A.; van Soest, J.; Kohlbacher, O.; Zimmermann, L.; Stenzhorn, H.; Karim, R.; Dumontier, M.; Decker, S.; da Silva Santos, L.O.B.; et al. Distributed Analytics on Sensitive Medical Data: The Personal Health Train. Data Intell. 2020, 2, 96–107. [Google Scholar] [CrossRef]
- Rigter, T.; Jansen, M.E.; de Groot, J.M.; Janssen, S.W.J.; Rodenburg, W.; Cornel, M.C. Implementation of Pharmacogenetics in Primary Care: A Multi-Stakeholder Perspective. Front Genet. 2020, 11, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Dunnen, J.T. The DNA Bank: High-Security bank accounts to protect and share your genetic identity. Hum. Mutat. 2015, 36, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Poikola, A.; Kuikkaniemi, K.; Kuittinen, O.; Honko, H.; Knuutila, A.; Lähteenoja, V. MyData—An Introduction to Human-Centric use of Personal Data, 3rd ed.; Ministery of Transport and Communications: Helsinki, Finland, 2020.
- Sambra, A.V.; Mansour, E.; Hawke, S.; Zereba, M.; Greco, N.; Ghanem, A.; Zagidulin, D.; Aboulnaga, A.; Berners-Lee, T. Solid: A Platform for Decentralized Social Applications Based on Linked Data; Technical Report; MIT CSAIL: Cambridge, MA, USA; Qatar Computing Research Institute: Ar-Rayyan, Qatar, 2016. [Google Scholar]
- MyData. Available online: https://web.archive.org/web/20201011172714/https://mydata.org/ (accessed on 11 October 2020).
- European Joint Programme Rare Diseases. Available online: https://web.archive.org/web/20200921092310/https://www.ejprarediseases.org/ (accessed on 21 September 2020).
- Digital House of Europe. Available online: https://www.digitalhouseofeurope.eu (accessed on 28 October 2020).
- Bates, B.R.; Lynch, J.A.; Bevan, J.L.; Condit, C.M. Warranted concerns, warranted outlooks: A focus group study of public understandings of genetic research. Soc. Sci. Med. 2005, 60, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Lemke, A.A.; Wolf, W.A.; Hebert-Beirne, J.; Smith, M.E. Public and biobank participant attitudes toward genetic research participation and data sharing. Public Health Genom. 2010, 13, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Callier, S.L.; Abudu, R.; Mehlman, M.J.; Singer, M.E.; Neuhauser, D.; Caga-Anan, C.; Wiesner, G.L. Ethical, legal, and social implications of personalized genomic medicine research: Current literature and suggestions for the future. Bioethics 2016, 30, 698–705. [Google Scholar] [CrossRef]
- Millward, L.J. Focus groups. In Research Methods in Psychology, 4th ed.; Breakwell, G.M., Smith, J.A., Wright, D.B., Eds.; SAGE Publications Inc.: London, UK, 2012; pp. 411–437. [Google Scholar]
- Krueger, R.A.; Casey, M.A. Focus Groups: A Practical Guide for Applied Research, 5th ed.; Sage: London, UK, 2014; pp. 2–6. [Google Scholar]
- Garrido, T.; Kanter, M.; Meng, D.; Turley, M.; Wang, J.; Sue, V.; Scott, L. Race/ethnicity, personal health record access, and quality of care. Am. J. Manag. Care 2015, 21, e103–e113. [Google Scholar]
- Sartain, S.A.; Stressing, S.; Prieto, J. Patients’ views on the effectiveness of patient-held records: A systematic review and thematic synthesis of qualitative studies. Health Expect. 2015, 18, 2666–2677. [Google Scholar] [CrossRef] [Green Version]
- Hindorff, L.A.; Bonham, V.L.; Ohno-Machado, L. Enhancing diversity to reduce health information disparities and build an evidence base for genomic medicine. Pers. Med. 2018, 15, 403–412. [Google Scholar] [CrossRef] [Green Version]
- McGowan, M.L.; Prows, C.A.; DeJonckheere, M.; Brinkman, W.B.; Vaughn, L.; Myers, M.F. Adolescent and Parental Attitudes About Return of Genomic Research Results: Focus Group Findings Regarding Decisional Preferences. J. Empir. Res. Hum. Res. Ethics 2018, 13, 371–382. [Google Scholar] [CrossRef]
- Emery, J.; Hayflick, S. The challenge of integrating genetic medicine into primary care. BMJ 2001, 322, 1027–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overkleeft, R.; Tommel, J.; Evers, A.W.M.; den Dunnen, J.T.; Roos, M.; Hoefmans, M.-J.; Schrader, W.E.; Swen, J.J.; Numans, M.E.; Houwink, E.J.F. Using Personal Genomic Data within Primary Care: A Bioinformatics Approach to Pharmacogenomics. Genes 2020, 11, 1443. https://doi.org/10.3390/genes11121443
Overkleeft R, Tommel J, Evers AWM, den Dunnen JT, Roos M, Hoefmans M-J, Schrader WE, Swen JJ, Numans ME, Houwink EJF. Using Personal Genomic Data within Primary Care: A Bioinformatics Approach to Pharmacogenomics. Genes. 2020; 11(12):1443. https://doi.org/10.3390/genes11121443
Chicago/Turabian StyleOverkleeft, Rick, Judith Tommel, Andrea W. M. Evers, Johan T. den Dunnen, Marco Roos, Marie-José Hoefmans, Walter E. Schrader, Jesse J. Swen, Mattijs E. Numans, and Elisa J. F. Houwink. 2020. "Using Personal Genomic Data within Primary Care: A Bioinformatics Approach to Pharmacogenomics" Genes 11, no. 12: 1443. https://doi.org/10.3390/genes11121443
APA StyleOverkleeft, R., Tommel, J., Evers, A. W. M., den Dunnen, J. T., Roos, M., Hoefmans, M.-J., Schrader, W. E., Swen, J. J., Numans, M. E., & Houwink, E. J. F. (2020). Using Personal Genomic Data within Primary Care: A Bioinformatics Approach to Pharmacogenomics. Genes, 11(12), 1443. https://doi.org/10.3390/genes11121443





