Evaluation of Single-Molecule Sequencing Technologies for Structural Variant Detection in Two Swedish Human Genomes
Abstract
1. Introduction
2. Materials and Methods
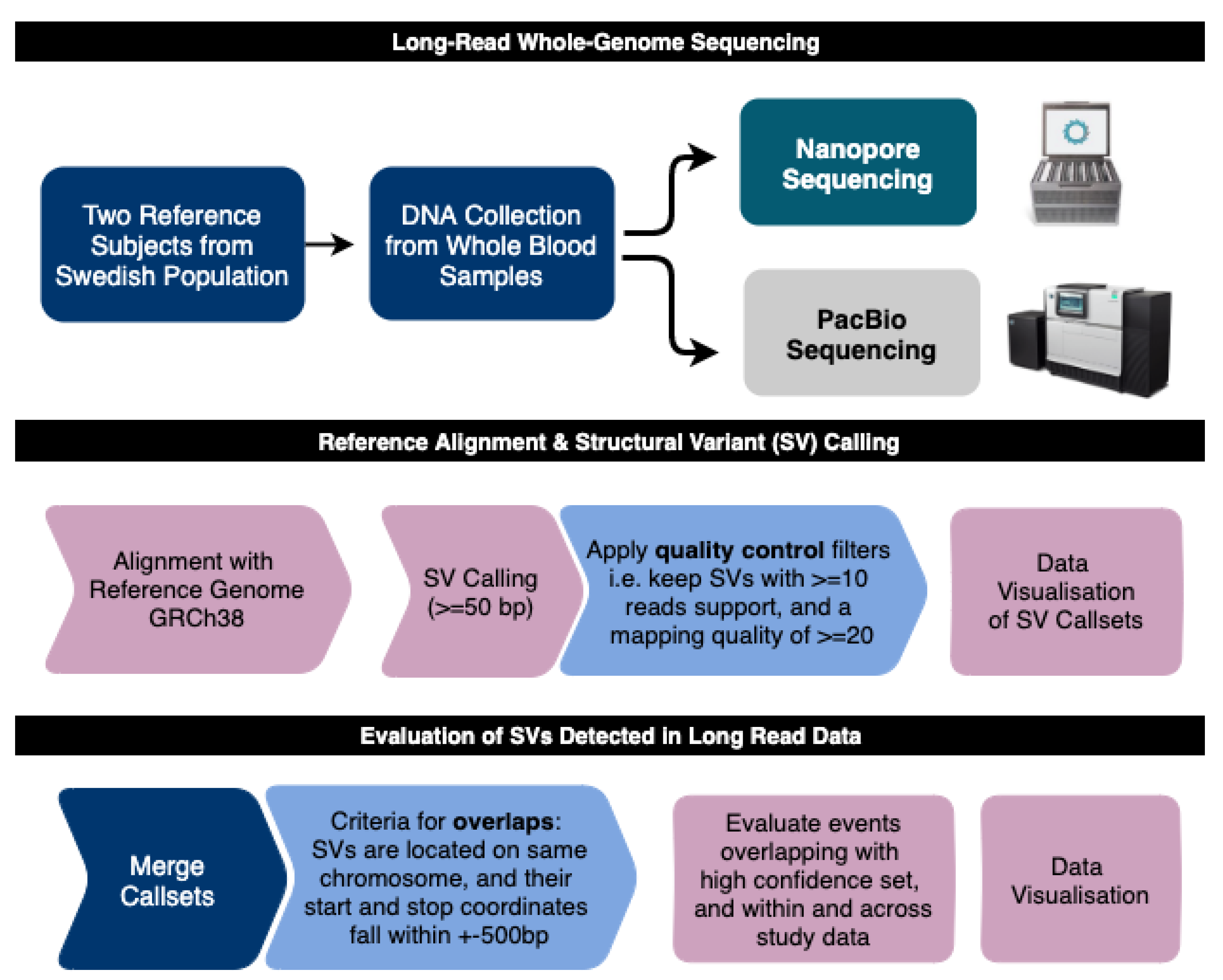
2.1. Sample Collection
2.2. Whole-Genome Sequencing of Two Swedish Genomes
2.3. Alignment and Reference Genome
Alignment of ONT and PacBio Reads
2.4. Structural Variant Calling and Analysis
2.5. Downsampling Analysis
3. Results
3.1. Overview of the Study
3.2. Mapping of Long-Read Data for two Swedish Individuals
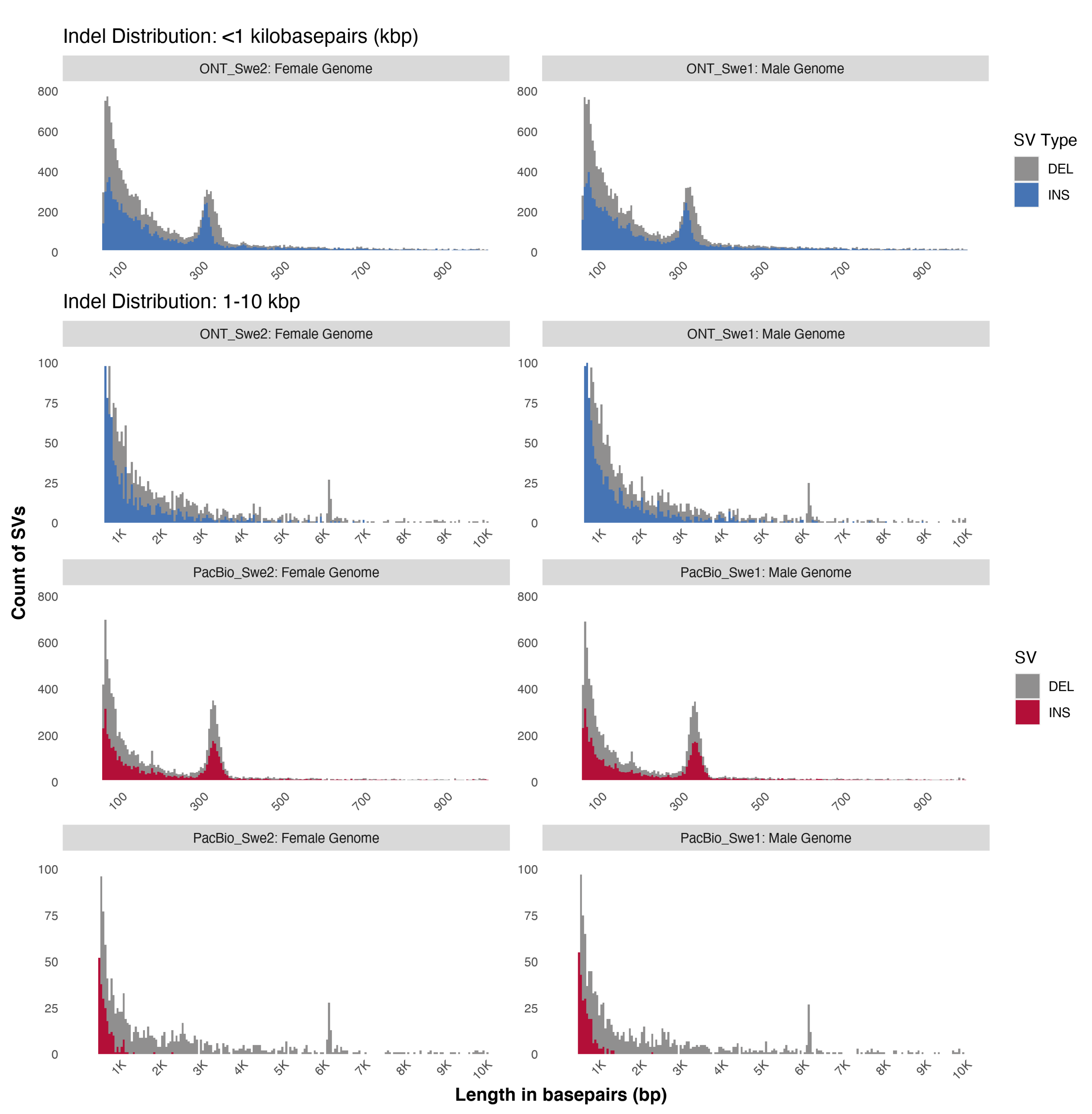
3.3. Characterisation of Structural Variation in ONT and PacBio Data
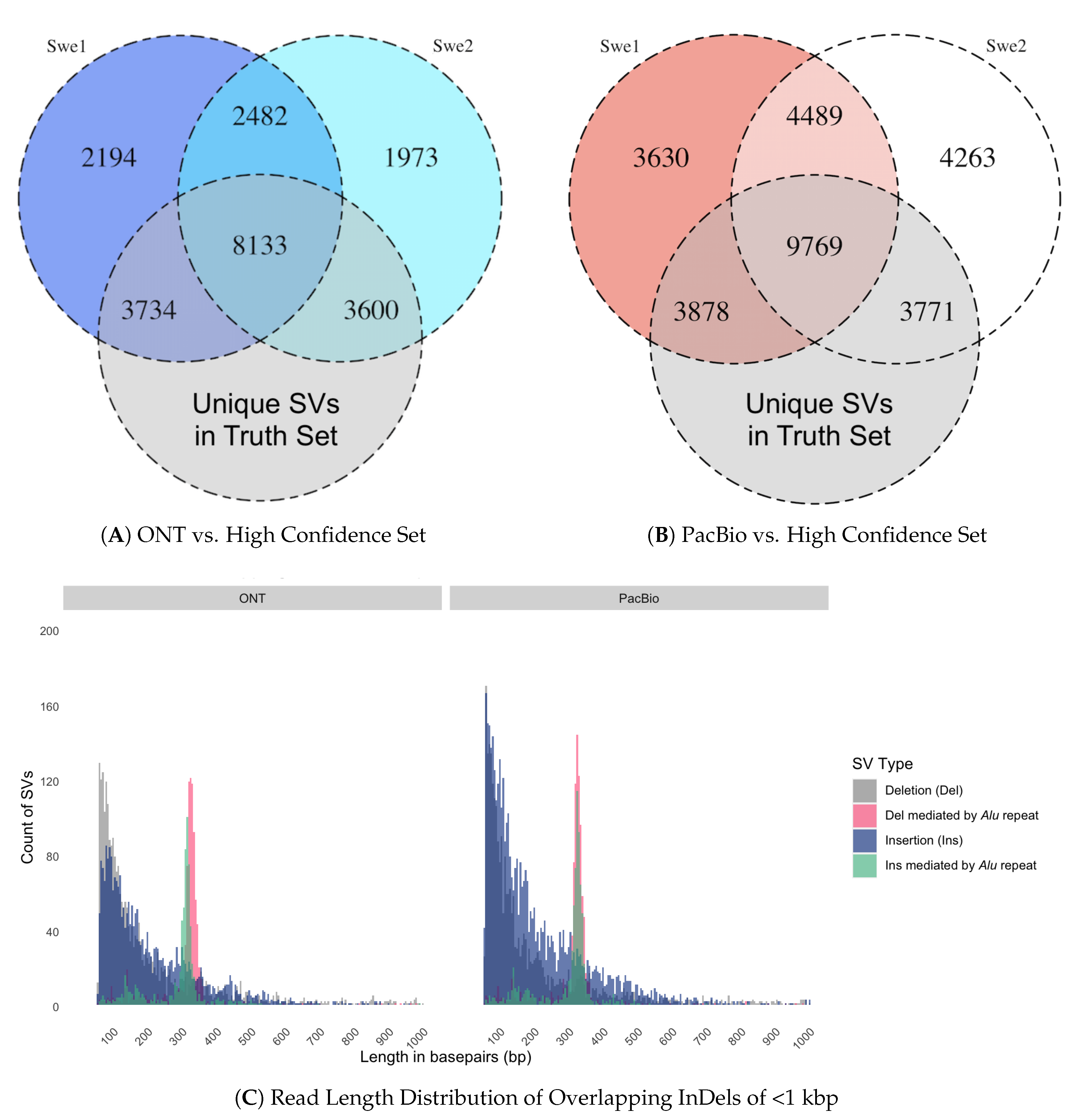
3.4. Comparing the Swedish SVs to a High Confidence Set of SVs
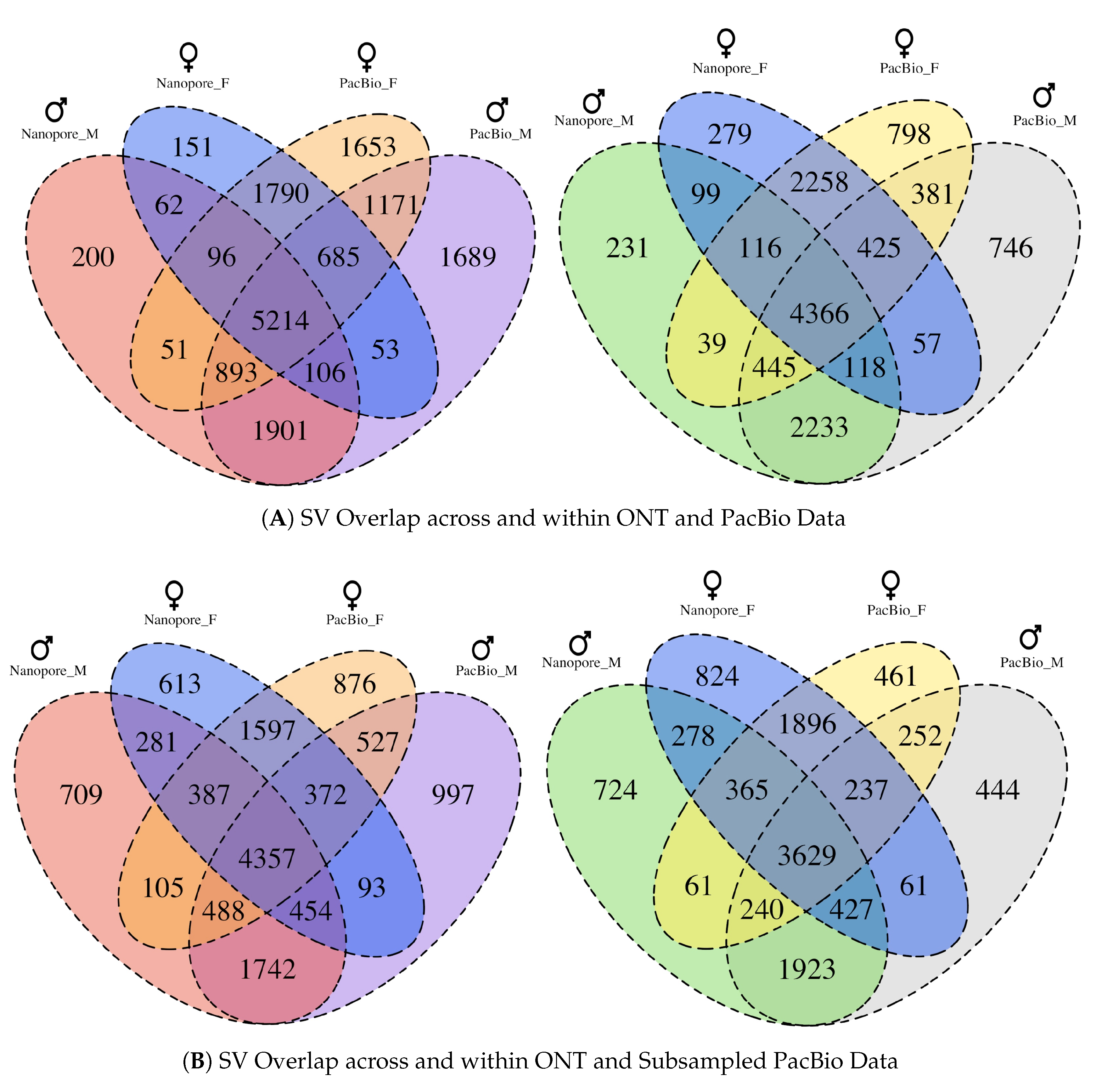
3.5. Comparison of SVs between Sequencing Technologies and Individuals
3.6. Comparison of SVs in ONT and Subsampled PacBio Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CLR | Continuous Long Read |
| ONT | Oxford Nanopore Technologies |
| PacBio | Pacific Biosciences |
| SV | Structural Variant |
| SweGen | Swedish Genetics Cohort |
| WGS | Whole-Genome Sequencing |
References
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Iafrate, A.J.; Feuk, L.; Rivera, M.N.; Listewnik, M.L.; Donahoe, P.K.; Qi, Y.; Scherer, S.W.; Lee, C. Detection of large-scale variation in the human genome. Nat. Genet. 2004, 36, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Brkanac, Z.; Coe, B.P.; Baker, C.; Vives, L.; Vu, T.H.; Shafer, N.; Bernier, R.; Ferrero, G.B.; Silengo, M.; et al. Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes. PLoS Genet. 2011, 7, e1002334. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.L.; Workman, R.E.; Fan, Y.; Eshleman, J.R.; Timp, W. Nanopore sequencing detects structural variants in cancer. Cancer Biol. Ther. 2016, 17, 246–253. [Google Scholar] [CrossRef]
- Vu, T.; Davidson, S.L.; Borgesi, J.; Maksudul, M.; Jeon, T.J.; Shim, J. Piecing together the puzzle: Nanopore technology in detection and quantification of cancer biomarkers. RSC Adv. 2017, 7, 42653–42666. [Google Scholar] [CrossRef]
- Aganezov, S.; Goodwin, S.; Sherman, R.M.; Sedlazeck, F.J.; Arun, G.; Bhatia, S.; Lee, I.; Kirsche, M.; Wappel, R.; Kramer, M.; et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 2020, 30, 1258–1273. [Google Scholar] [CrossRef]
- Merker, J.D.; Wenger, A.M.; Sneddon, T.; Grove, M.; Zappala, Z.; Fresard, L.; Waggott, D.; Utiramerur, S.; Hou, Y.; Smith, K.S.; et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 2018, 20, 159–163. [Google Scholar] [CrossRef]
- Abel, H.J.; Larson, D.E.; Regier, A.A.; Chiang, C.; Das, I.; Kanchi, K.L.; Layer, R.M.; Neale, B.M.; Salerno, W.J.; Reeves, C.; et al. Mapping and characterization of structural variation in 17,795 human genomes. Nature 2020, 1–10. [Google Scholar] [CrossRef]
- Collins, R.L.; Brand, H.; Karczewski, K.J.; Zhao, X.; Alföldi, J.; Francioli, L.C.; Khera, A.V.; Lowther, C.; Gauthier, L.D.; Wang, H.; et al. A structural variation reference for medical and population genetics. Nature 2020, 581, 444–451. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Baudino, T.A. Targeted Cancer Therapy: The Next Generation of Cancer Treatment. Curr. Drug Discov. Technol. 2015, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Zhou, J.; Yang, Q.; Liang, F.; Wang, D.; Ma, N.; Gao, B.; Du, J.; Lin, G.; Wang, K.; et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 2018, 155, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Regier, A.A.; Farjoun, Y.; Larson, D.E.; Krasheninina, O.; Kang, H.M.; Howrigan, D.P.; Chen, B.J.; Kher, M.; Banks, E.; Ames, D.C.; et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018, 9, 4038. [Google Scholar] [CrossRef] [PubMed]
- Fakhro, K.A.; Staudt, M.R.; Ramstetter, M.D.; Robay, A.; Malek, J.A.; Badii, R.; Al-Marri, A.A.N.; Khalil, C.A.; Al-Shakaki, A.; Chidiac, O.; et al. The Qatar genome: A population-specific tool for precision medicine in the Middle East. Hum. Genome Var. 2016, 3, 1–7. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High Throughput Sequencing: An Overview of Sequencing Chemistry. Indian J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Ebbert, M.T.W.; Jensen, T.D.; Jansen-West, K.; Sens, J.P.; Reddy, J.S.; Ridge, P.G.; Kauwe, J.S.K.; Belzil, V.; Pregent, L.; Carrasquillo, M.M.; et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Shafin, K.; Pesout, T.; Lorig-Roach, R.; Haukness, M.; Olsen, H.E.; Bosworth, C.; Armstrong, J.; Tigyi, K.; Maurer, N.; Koren, S.; et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- De Roeck, A.; De Coster, W.; Bossaerts, L.; Cacace, R.; De Pooter, T.; Van Dongen, J.; D’Hert, S.; De Rijk, P.; Strazisar, M.; Van Broeckhoven, C.; et al. NanoSatellite: Accurate characterization of expanded tandem repeat length and sequence through whole genome long-read sequencing on PromethION. Genome Biol. 2019, 20, 239. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Rijk, P.D.; Roeck, A.D.; Pooter, T.D.; D’Hert, S.; Strazisar, M.; Sleegers, K.; Broeckhoven, C.V. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019, 29, 1178–1187. [Google Scholar] [CrossRef]
- Beyter, D.; Ingimundardottir, H.; Eggertsson, H.P.; Bjornsson, E.; Kristmundsdottir, S.; Mehringer, S.; Jonsson, H.; Hardarson, M.T.; Magnusdottir, D.N.; Kristjansson, R.P.; et al. Long read sequencing of 1817 Icelanders provides insight into the role of structural variants in human disease. bioRxiv 2019, 848366. [Google Scholar] [CrossRef]
- Steinberg, K.M.; Schneider, V.A.; Graves-Lindsay, T.A.; Fulton, R.S.; Agarwala, R.; Huddleston, J.; Shiryev, S.A.; Morgulis, A.; Surti, U.; Warren, W.C.; et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014, 24, 2066–2076. [Google Scholar] [CrossRef]
- Huddleston, J.; Chaisson, M.J.P.; Steinberg, K.M.; Warren, W.; Hoekzema, K.; Gordon, D.; Graves-Lindsay, T.A.; Munson, K.M.; Kronenberg, Z.N.; Vives, L.; et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017, 27, 677–685. [Google Scholar] [CrossRef]
- Ameur, A.; Che, H.; Martin, M.; Bunikis, I.; Dahlberg, J.; Höijer, I.; Häggqvist, S.; Vezzi, F.; Nordlund, J.; Olason, P.; et al. De Novo Assembly Two Swedish Genomes Reveals Missing Segments Human GRCh38 Reference Improves Variant Calling Population-Scale Sequencing Data. Genes 2018, 9, 486. [Google Scholar] [CrossRef]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Francioli, L.C.; Hormozdiari, F.; Marschall, T.; Hehir-Kwa, J.Y.; Abdellaoui, A.; Lameijer, E.W.; Moed, M.H.; Koval, V.; Renkens, I.; et al. Characteristics of de novo structural changes in the human genome. Genome Res. 2015, 25, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 1–18. [Google Scholar] [CrossRef]
- Chaisson, M.J.P.; Sanders, A.D.; Zhao, X.; Malhotra, A.; Porubsky, D.; Rausch, T.; Gardner, E.J.; Rodriguez, O.L.; Guo, L.; Collins, R.L.; et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, Y.; Jiang, Y.; Li, J.; Gao, Y.; Cui, Z.; Liu, Y.; Liu, B.; Wang, Y. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020, 21, 189. [Google Scholar] [CrossRef] [PubMed]
- Ameur, A.; Dahlberg, J.; Olason, P.; Vezzi, F.; Karlsson, R.; Martin, M.; Viklund, J.; Kähäri, A.K.; Lundin, P.; Che, H.; et al. SweGen: A whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur. J. Hum. Genet. 2017, 25, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Heller, D.; Vingron, M. SVIM: Structural variant identification using mapped long reads. Bioinformatics 2019, 35, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Jolly, C.; Hoti, M.; Speed, D.; Shaw, L.; Rallis, C.; Balloux, F.; Dessimoz, C.; Bähler, J.; Sedlazeck, F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef]
- Audano, P.A.; Sulovari, A.; Graves-Lindsay, T.A.; Cantsilieris, S.; Sorensen, M.; Welch, A.E.; Dougherty, M.L.; Nelson, B.J.; Shah, A.; Dutcher, S.K.; et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell 2019, 176, 663–675.e19. [Google Scholar] [CrossRef]
- Bowden, R.; Davies, R.W.; Heger, A.; Pagnamenta, A.T.; de Cesare, M.; Oikkonen, L.E.; Parkes, D.; Freeman, C.; Dhalla, F.; Patel, S.Y.; et al. Sequencing of human genomes with nanopore technology. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Luan, M.W.; Zhang, X.M.; Zhu, Z.B.; Chen, Y.; Xie, S.Q. Evaluating Structural Variation Detection Tools for Long-Read Sequencing Datasets in Saccharomyces Cerevisiae. Front. Genet. 2020, 11, 159. [Google Scholar] [CrossRef]
| Sample | Total Sequence Data, Gb | Mean Mapping Coverage | GC Content, % |
|---|---|---|---|
| ONT_Swe1 | 104.5 | 31.69× | 40.72 |
| ONT_Swe2 | 104.4 | 31.49× | 40.41 |
| PacBio_Swe1 | 235.7 | 66.54× | 40.41 |
| PacBio_Swe2 | 232.9 | 65.56× | 40.46 |
| SV Type | ONT Swe1 | ONT Swe2 | PacBio Swe1 | PacBio Swe2 |
|---|---|---|---|---|
| Deletions | 7769 | 7820 | 9052 | 9101 |
| Insertions | 8746 | 8369 | 12,441 | 12,218 |
| Duplications | 185 | 147 | 331 | 291 |
| Inversions | 133 | 128 | 196 | 193 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, N.; Petri, A.; Gyllensten, U.; Feuk, L.; Ameur, A. Evaluation of Single-Molecule Sequencing Technologies for Structural Variant Detection in Two Swedish Human Genomes. Genes 2020, 11, 1444. https://doi.org/10.3390/genes11121444
Fatima N, Petri A, Gyllensten U, Feuk L, Ameur A. Evaluation of Single-Molecule Sequencing Technologies for Structural Variant Detection in Two Swedish Human Genomes. Genes. 2020; 11(12):1444. https://doi.org/10.3390/genes11121444
Chicago/Turabian StyleFatima, Nazeefa, Anna Petri, Ulf Gyllensten, Lars Feuk, and Adam Ameur. 2020. "Evaluation of Single-Molecule Sequencing Technologies for Structural Variant Detection in Two Swedish Human Genomes" Genes 11, no. 12: 1444. https://doi.org/10.3390/genes11121444
APA StyleFatima, N., Petri, A., Gyllensten, U., Feuk, L., & Ameur, A. (2020). Evaluation of Single-Molecule Sequencing Technologies for Structural Variant Detection in Two Swedish Human Genomes. Genes, 11(12), 1444. https://doi.org/10.3390/genes11121444






