Evaluating AI-Based Mitosis Detection for Breast Carcinoma in Digital Pathology: A Clinical Study on Routine Practice Integration
Abstract
:1. Introduction
2. Related Works
- Providing evidence that AI assistance can reduce inter-observer variability in real-world clinical settings;
- Integrating a comprehensive deep learning pipeline—for IC-NST localization, mitosis detection, and hotspot identification on whole-slide images—into a user-friendly interface seamlessly embedded within the pathologist’s workflow, with no need for manual region-of-interest (ROI) selection or intervention;
- Conducting a clinical evaluation comparing pathologists’ performance with and without AI assistance;
- Performing a subgroup analysis to identify specific scenarios where AI assistance offers the most significant benefit.
3. Materials and Methods
3.1. Data Description
3.2. Pipeline Description
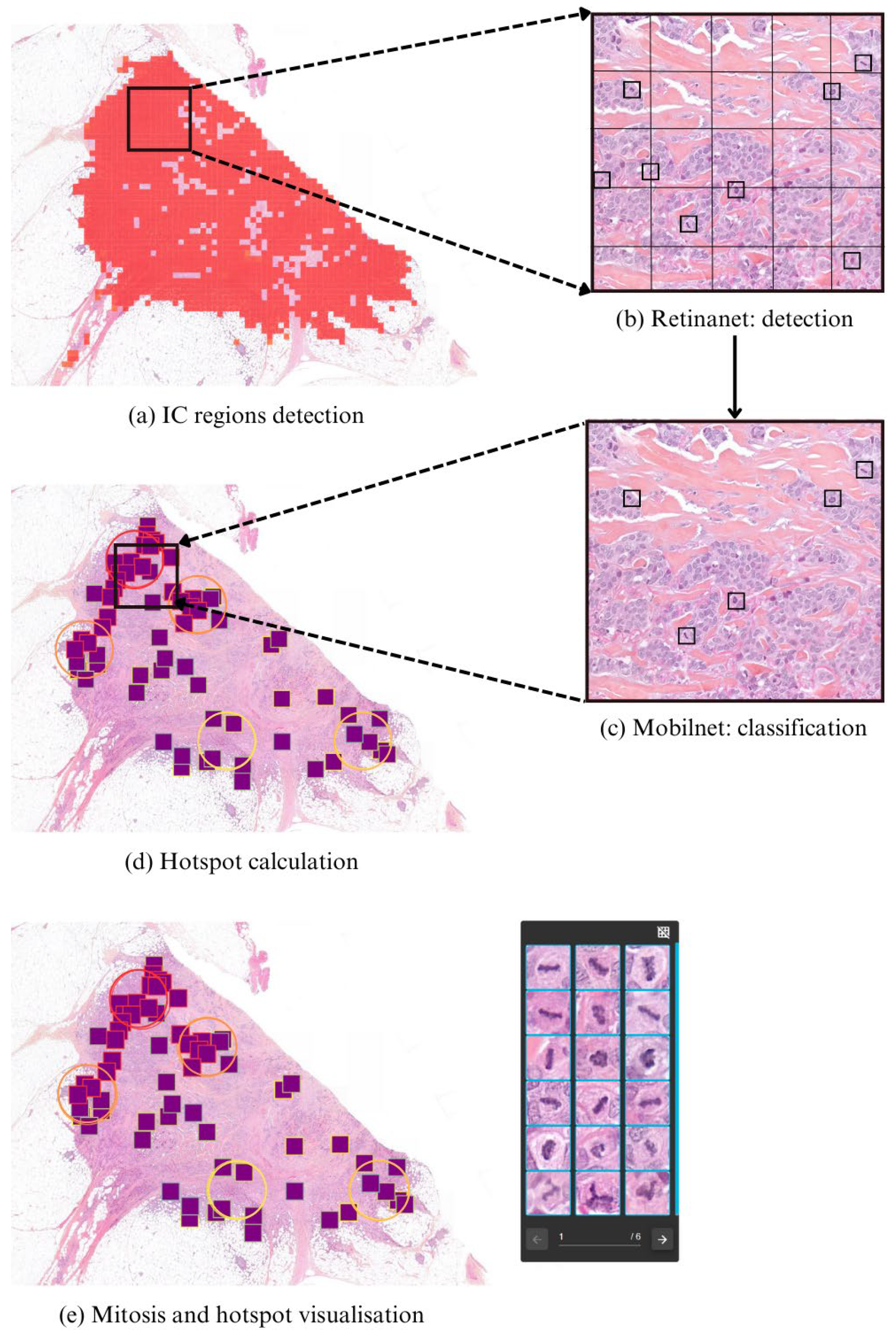
3.2.1. IC-NST Region Detection
3.2.2. Mitosis Candidate Localization
- -
- First, a RetinaNet-based object detector [36] analyzes 50 × 50 pixel crops to localize mitosis-like candidates.
- -
- These candidate regions are then evaluated by a Mo-bileNetV2-based classifier [37], which determines whether each candidate corresponds to a true mitotic figure.
- -
- Only predictions that exceed a predefined confidence threshold are retained.
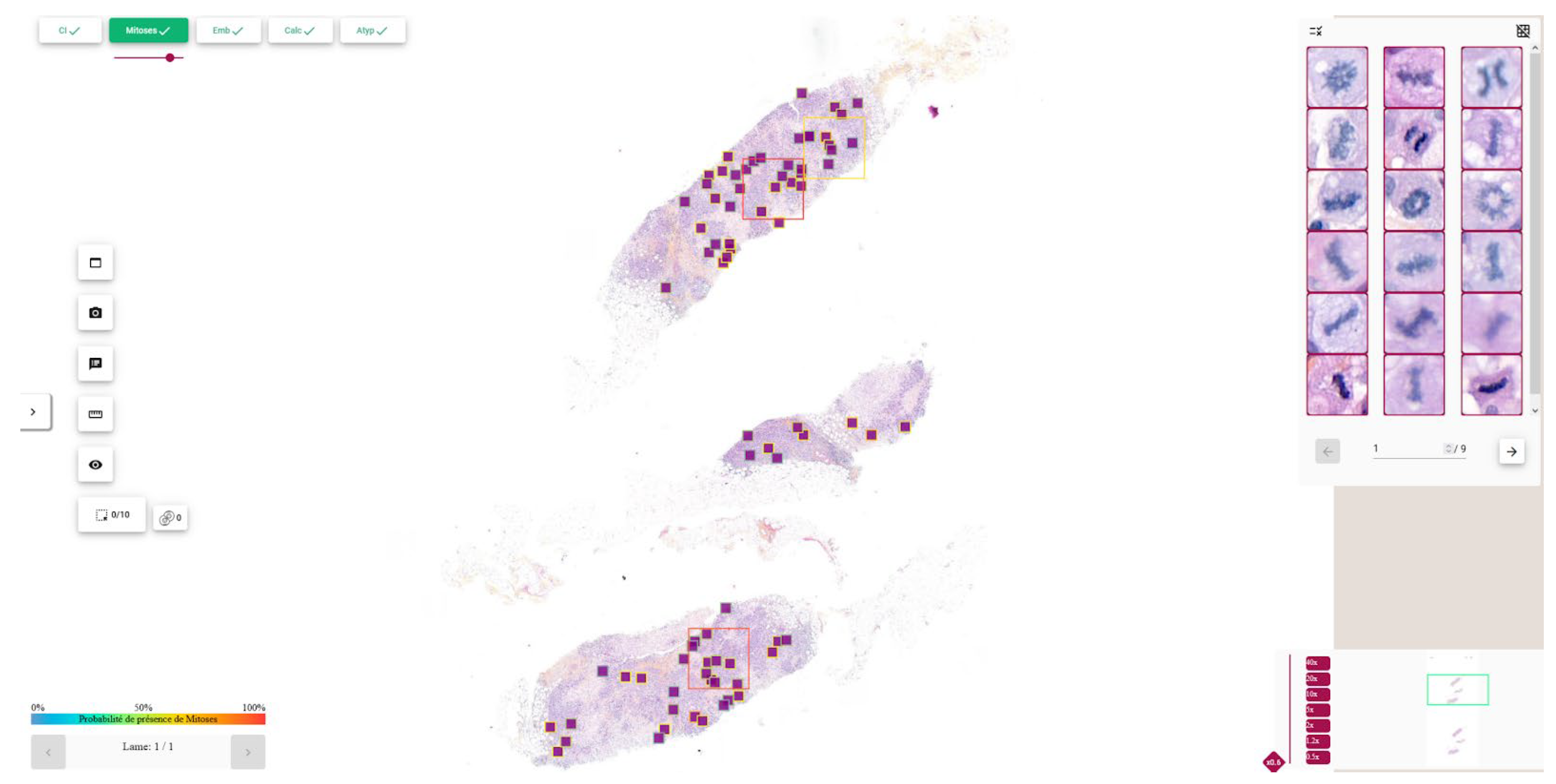
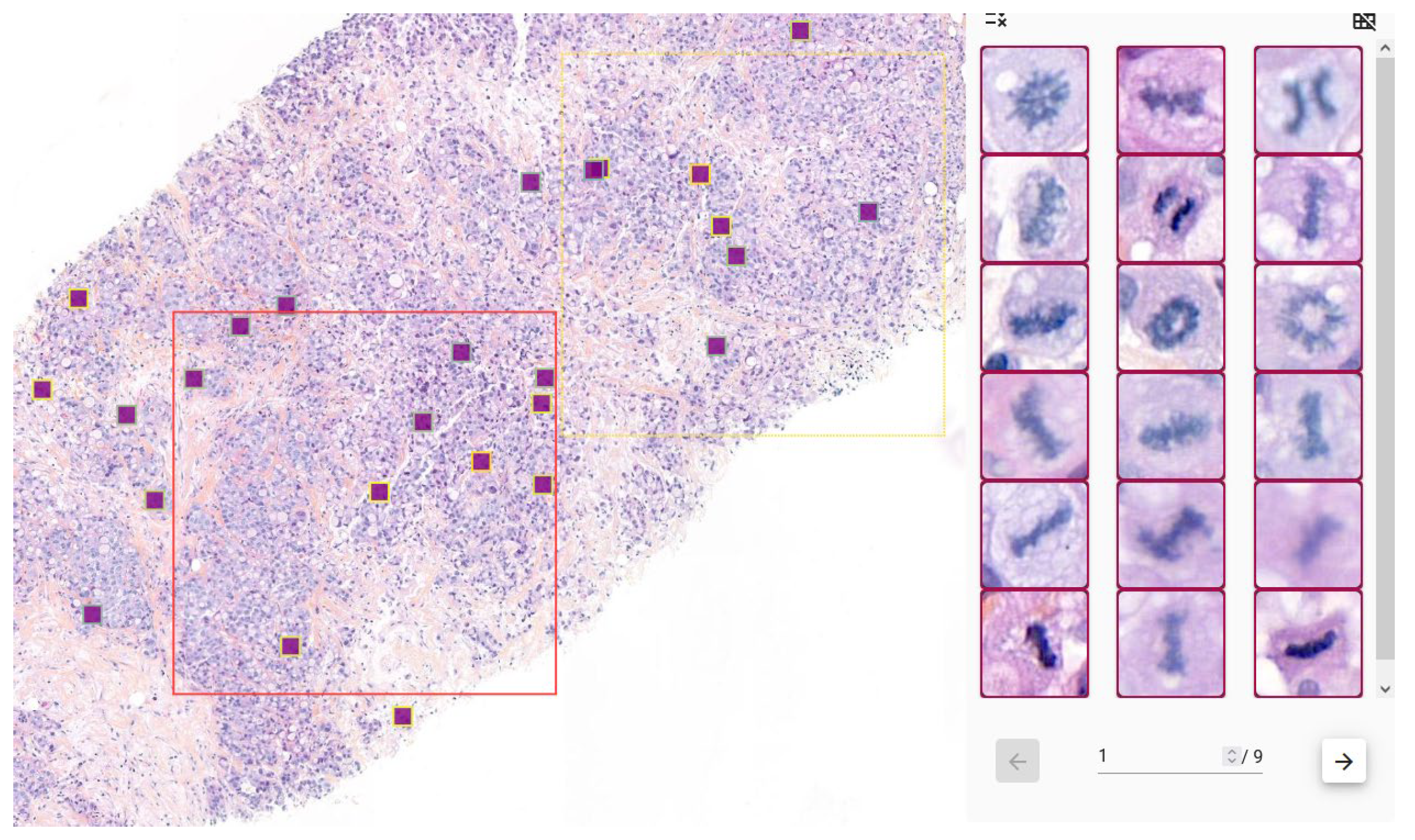
3.2.3. Hotspot Computation
- -
- is the hotspot score assigned to patch p.
- -
- is the number of mitoses within a core circular region _core (e.g., 1 mm2) centered on patch p.
- -
- is the number of mitoses within a broader circular context _context (e.g., 2 mm2), excluding the core region.
- -
- ε ∈ [0, 1] is a tunable weight controlling the influence of the surrounding mitotic activity.
- Input:
- -
- M = {(x1, y1), …, (xn, yn)} // mitoses
- -
- P = {(x1, y1), …, (xk, yk)} // patch centers
- -
- r1 = radius (1 mm2), r2 = radius (2 mm2)
- -
- ε = surrounding weight
- Function:
- Count(c, r, M):
- return |{ m ∈ M: dist(m, c) ≤ r }|
- Main:
- H ← {}
- for p ∈ P:
- n1 ← Count(p, r1, M)
- n2 ← Count(p, r2, M)
- H[p] ← n1 + ε·(n2 − n1)
- return sort_desc(H)
3.2.4. Visualization and Clinical Support
3.3. Data and Training
3.3.1. Datasets
- -
- For the training set: 2791 patches containing at least one mitosis.
- -
- For the testing set: 1341 patches without mitosis, and 146 patches with mitosis and 24,716 patches without mitosis.
- -
- Training set: 3106 mitoses and 8638 artifacts.
- -
- Testing set: 153 mitoses and 5081 artifacts.
3.3.2. Data Augmentation
3.3.3. Training Configuration
3.3.4. Analytical Validation of the Detection Pipeline
3.4. Design of the Clinical Study
3.4.1. Patients and Tissue Selection
3.4.2. Study Design
- -
- In the first session, each investigator reviewed half of the slides without access to the AI tool and the remaining half with AI assistance.
- -
- Then, after a washout period of several weeks (to minimize recall bias), the slide sets were switched: each investigator re-evaluated the cases, now using the opposite condition (i.e., slides previously reviewed with AI were now reviewed without, and vice versa).
3.4.3. Statistical Analysis
4. Results
Study Outcomes
5. Discussion
6. Limitations and Further Works
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WSI | Whole Slide Images |
| IC-NST | Invasive Carcinoma of No Special Type |
| MS | Mitotic Score |
| MC | Mitotic Count |
| MH | Mitotic Hotspot |
| SFP | French Society of Pathology |
| CNN | Convolutional Neural Networks |
| HE | Hematoxylin Eosin |
| HES | Hematoxylin Eosin Safran |
| CI | Confidence Interval |
| ICC | Intraclass Correlation Coefficient |
| CK | Cohen’s Kappa |
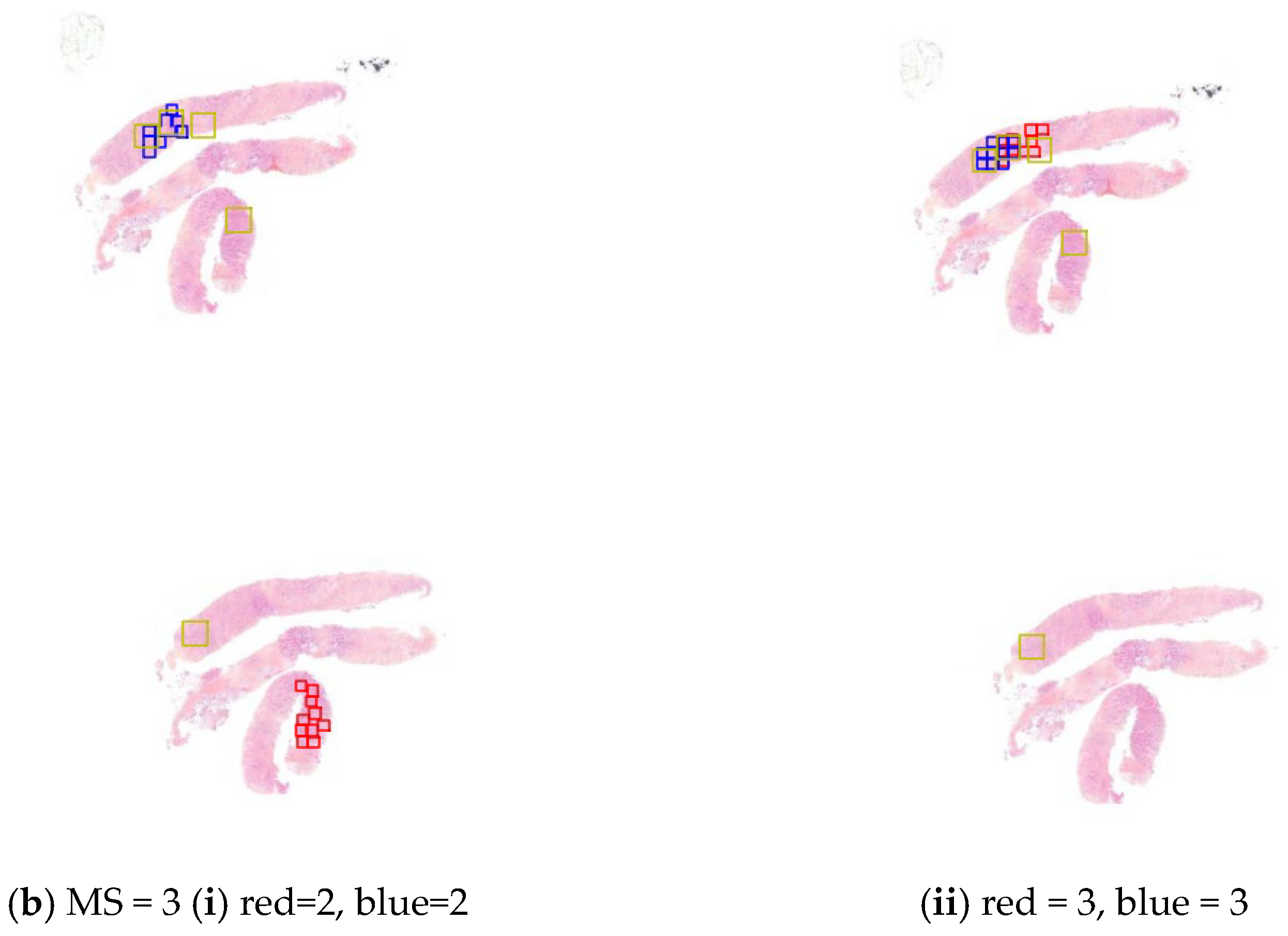
Appendix A. Analytical Validation of the Mitosis Detection Algorithm
Appendix A.1. Objective
- The number of mitoses counted by each pathologist.
- The mitotic score assigned (based on standard cut-offs).
- The agreement between mitoses annotated by pathologists and those detected by the algorithm.
Appendix A.2. Key Results
| Pathologist 1 | Pathologist 2 | Pathologist 3 | Pathologist 4 | ||
| Algorithm | ICC | 0.45 [0.10–0.68] | 0.67 [0.49–0.80] | 0.68 [0.50–0.80] | 0.77 [0.627–0.86] |
| CK | 0.42 | 0.58 | 0.62 | 0.69 | |
| Pathologist1 | ICC | 0.48 [0.12–0.70] | 0.78 [0.15–0.92] | 0.70 [0.17–0.88] | |
| CK | 0.34 | 0.47 | 0.47 | ||
| Pathologist2 | ICC | 0.77 [0.63–0.87] | 0.80 [0.67–0.89] | ||
| CK | 0.66 | 0.66 | |||
| Pathologist3 | ICC | 0.90 [0.83–0.94] | |||
| CK | 0.75 | ||||
| Pathologist4 | ICC | ||||
| CK |
Appendix A.3. Conclusions
Appendix B. Raw Confusion Matrices
| Expert consensus | Pathologist 1 without algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 27 | 2 | 0 | 29 | |
| 2 | 7 | 1 | 1 | 9 | |
| 3 | 4 | 5 | 3 | 12 | |
| Total | 38 | 8 | 4 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.378 (95% CI 0.179–0.577) without the use of the algorithm. | |||||
| Expert consensus | Pathologist 1 with algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 27 | 1 | 1 | 29 | |
| 2 | 4 | 4 | 1 | 9 | |
| 3 | 2 | 3 | 7 | 12 | |
| Total | 33 | 8 | 9 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.629 (95% CI 0.437–0.820) with the use of the algorithm. | |||||
| Expert consensus | Pathologist 2 without algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 28 | 1 | 0 | 29 | |
| 2 | 9 | 0 | 0 | 9 | |
| 3 | 2 | 6 | 4 | 12 | |
| Total | 39 | 7 | 4 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.457 (95% CI 0.267–0.647) without the use of the algorithm. | |||||
| Expert consensus | Pathologist 2 with algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 28 | 1 | 0 | 29 | |
| 2 | 7 | 2 | 0 | 9 | |
| 3 | 0 | 3 | 9 | 12 | |
| Total | 35 | 6 | 9 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.726 (95% CI 0.575–0.876) with the use of the algorithm. | |||||
| Pathologist 2 Without Algorithm | Pathologist 1 without algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 34 | 4 | 1 | 39 | |
| 2 | 4 | 2 | 1 | 7 | |
| 3 | 0 | 2 | 2 | 4 | |
| Total | 38 | 8 | 4 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.482 (95% CI 0.231–0.733) without the use of the algorithm. Intraclass correlation coefficient for mitotic count without algorithm: 0.591 (95% CI 0.375–0.746) | |||||
| Pathologist 2 with Algorithm | Pathologist 1 with algorithm | ||||
| Score | 1 | 2 | 3 | Total | |
| 1 | 30 | 4 | 1 | 35 | |
| 2 | 1 | 4 | 1 | 6 | |
| 3 | 2 | 0 | 7 | 9 | |
| Total | 33 | 8 | 9 | 50 | |
| Linearly weighted Cohen’s Kappa was 0.672 (95% CI 0.461–0.882) with the use of the algorithm. Intraclass correlation coefficient for mitotic count without algorithm: 0.883 (95% CI 0.803–0.932) | |||||
Appendix C
References
- Garcia, E.; Kundu, I.; Kelly, M.; Soles, R.; Mulder, L.; Talmon, G.A. The American Society for Clinical Pathology’s Job Satisfaction, Well-Being, and Burnout Survey of Pathologists. Am. J. Clin. Pathol. 2020, 153, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Roa, A.; Basavanhally, A.; González, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Automatic Detection of Invasive Ductal Carcinoma in Whole Slide Images with Convolutional Neural Networks. In Proceedings of the SPIE Proceedings; Gurcan, M.N., Madabhushi, A., Eds.; SPIE: Bellingham, WA, USA, 2014. [Google Scholar]
- Celik, Y.; Talo, M.; Yildirim, O.; Karabatak, M.; Acharya, U.R. Automated Invasive Ductal Carcinoma Detection Based Using Deep Transfer Learning with Whole-Slide Images. Pattern Recognit. Lett. 2020, 133, 232–239. [Google Scholar] [CrossRef]
- Peyret, R.; Pozin, N.; Sockeel, S.; Kammerer-Jacquet, S.-F.; Adam, J.; Bocciarelli, C.; Ditchi, Y.; Bontoux, C.; Depoilly, T.; Guichard, L.; et al. Multicenter Automatic Detection of Invasive Carcinoma on Breast Whole Slide Images. PLoS Digit. Health 2023, 2, e0000091. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; He, J.; Chao, X.; Chen, K.; Xu, Y.; Huang, Q.; Yun, J.; Li, M.; Luo, R.; Kuang, J.; et al. A Computational Tumor-Infiltrating Lymphocyte Assessment Method Comparable with Visual Reporting Guidelines for Triple-Negative Breast Cancer. EBioMedicine 2021, 70, 103492. [Google Scholar] [CrossRef]
- Elston, C.W.; Ellis, I.O. Pathological Prognostic Factors in Breast Cancer. I. The Value of Histological Grade in Breast Cancer: Experience from a Large Study with Long-Term Follow-up. Histopathology 2002, 41, 151. [Google Scholar] [CrossRef]
- Rakha, E.A.; Bennett, R.; Coleman, D.; Pinder, S.E.; Ellis, I.O. Review of the National External Quality Assessment (EQA) Scheme for Breast Pathology in the UK. J. Clin. Pathol. 2016, 70, 51–57. [Google Scholar] [CrossRef]
- Berbís, M.A.; McClintock, D.S.; Bychkov, A.; Van Der Laak, J.; Pantanowitz, L.; Lennerz, J.K.; Cheng, J.Y.; Delahunt, B.; Egevad, L.; Eloy, C.; et al. Computational Pathology in 2030: A Delphi Study Forecasting the Role of AI in Pathology within the next Decade. eBioMedicine 2023, 88, 104427. [Google Scholar] [CrossRef]
- Irshad, H. Automated Mitosis Detection in Histopathology Using Morphological and Multi-Channel Statistics Features. J. Pathol. Inform. 2013, 4, 10. [Google Scholar] [CrossRef]
- Tek, F.B. Mitosis Detection Using Generic Features and an Ensemble of Cascade Adaboosts. J. Pathol. Inform. 2013, 4, 12. [Google Scholar] [CrossRef]
- Mathew, T.; Kini, J.R.; Rajan, J. Computational Methods for Automated Mitosis Detection in Histopathology Images: A Review. Biocybern. Biomed. Eng. 2021, 41, 64–82. [Google Scholar] [CrossRef]
- Ibrahim, A.; Lashen, A.; Katayama, A.; Mihai, R.; Ball, G.; Toss, M.; Rakha, E. Defining the Area of Mitoses Counting in Invasive Breast Cancer Using Whole Slide Image. Mod. Pathol. 2021, 35, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Razavi, S.; Dambandkhameneh, F.; Androutsos, D.; Done, S.; Khademi, A. Cascade RCNN for MIDOG Challenge. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer International Publishing: Cham, Germany, 2021. [Google Scholar]
- Wilm, F.; Marzahl, C.; Breininger, K.; Aubreville, M. Domain Adversarial RetinaNet as a Reference Algorithm for the MItosis DOmain Generalization Challenge. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer International Publishing: Cham, Germany, 2021. [Google Scholar] [CrossRef]
- Yang, S.; Luo, F.; Zhang, J.; Wang, X. Sk-Unet Model with Fourier Domain for Mitosis Detection. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer International Publishing: Cham, Germany, 2021. [Google Scholar]
- Roy, G.; Dedieu, J.; Bertrand, C.; Moshayedi, A.; Mammadov, A.; Petit, S.; Hadj, S.B.; Fick, R.H.J. Robust Mitosis Detection Using a Cascade Mask-RCNN Approach With Domain-Specific Residual Cycle-GAN Data Augmentation. arXiv 2021, arXiv:2109.01878. [Google Scholar]
- Kausar, T.; Wang, M.; Ashraf, M.A.; Kausar, A. SmallMitosis: Small Size Mitotic Cells Detection in Breast Histopathology Images. IEEE Access 2021, 9, 905–922. [Google Scholar] [CrossRef]
- Sebai, M.; Wang, X.; Wang, T. MaskMitosis: A Deep Learning Framework for Fully Supervised, Weakly Supervised, and Unsupervised Mitosis Detection in Histopathology Images. Med. Biol. Eng. Comput. 2020, 58, 1603–1623. [Google Scholar] [CrossRef]
- Aubreville, M.; Bertram, C.; Marzahl, C.; Maier, A.; Klopfleisch, R. A Large-Scale Dataset for Mitotic Figure Assessment on Whole Slide Images of Canine Cutaneous Mast Cell Tumor. Sci. Data 2019, 6, 274. [Google Scholar] [CrossRef]
- Mitosis Detection in Breast Cancer Histological Images (MITOS Dataset). Available online: http://ludo17.free.fr/mitos_2012/dataset.html (accessed on 24 February 2025).
- Mitos Atypia 14 Contest. Available online: https://mitos-atypia-14.grand-challenge.org/ (accessed on 24 February 2025).
- Aubreville, M. Mitosis Domain Generalization in Histopathology Images—The MIDOG Challenge. Med. Image Anal. 2023, 84, 102699. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Wu, W.; Lyu, H.; Tang, X.; Zhou, C.; Xu, F.; Luo, B.; Jiang, Y.; Liu, X.; et al. A Novel Dilated Contextual Attention Module for Breast Cancer Mitosis Cell Detection. Front. Physiol. 2024, 15, 1337554. [Google Scholar] [CrossRef]
- Subramanian, R.; Rubi, R.D.; Tapadia, R.; Karthik, K.; Ahmed, M.F.; Manudeep, A. Web Based Mitosis Detection on Breast Cancer Whole Slide Images Using Faster R-CNN and YOLOv5. Int. J. Adv. Comput. Sci. Appl. 2022, 13, 560–565. [Google Scholar] [CrossRef]
- Jahanifar, M.; Shephard, A.; Zamanitajeddin, N.; Graham, S.; Raza, S.E.A.; Minhas, F.; Rajpoot, N. Mitosis Detection, Fast and Slow: Robust and Efficient Detection of Mitotic Figures. Med. Image Anal. 2024, 94, 103132. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Z.; Pan, X.; Yu, K.; Lan, R.; Guan, J.; Li, B. A Novel Dataset and a Two-Stage Deep Learning Method for Breast Cancer Mitosis Nuclei Identification. Digit. Signal Process. 2025, 158, 104978. [Google Scholar] [CrossRef]
- Pantanowitz, L.X.; Hartman, D.J.; Qi, Y.; Cho, E.Y.; Suh, B.; Paeng, K.; Dhir, R.; Michelow, P.M.; Hazelhurst, S.; Song, S.Y.; et al. Accuracy and Efficiency of an Artificial Intelligence Tool When Counting Breast Mitoses. Diagn. Pathol. 2020, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- van Bergeijk, S.A.; Stathonikos, N.; Hoeve, N.D.; Lafarge, M.W.; Nguyen, T.Q.; van Diest, P.J.; Veta, M. Deep Learning Supported Mitoses Counting on Whole Slide Images: A Pilot Study for Validating Breast Cancer Grading in the Clinical Workflow. J. Pathol. Inform. 2023, 14, 100316. [Google Scholar] [CrossRef] [PubMed]
- Balkenhol, M.C.A.; Tellez, D.; Vreuls, W.; Clahsen, P.C.; Pinckaers, H.; Ciompi, F.; Bult, P.; van der Laak, J.A.W.M. Deep Learning Assisted Mitotic Counting for Breast Cancer. Lab. Investig. 2019, 99, 1596–1606. [Google Scholar] [CrossRef]
- Ibrahim, A.; Lashen, A.; Toss, M.; Mihai, R.; Rakha, E. Assessment of Mitotic Activity in Breast Cancer: Revisited in the Digital Pathology Era. J. Clin. Pathol. 2022, 75, 365–372. [Google Scholar] [CrossRef]
- Williams, B.; Hanby, A.; Millican-Slater, R.; Verghese, E.; Nijhawan, A.; Wilson, I.; Besusparis, J.; Clark, D.; Snead, D.; Rakha, E.; et al. Digital Pathology for Primary Diagnosis of Screen-Detected Breast Lesions—Experimental Data, Validation and Experience from Four Centres. Histopathology 2020, 76, 968–975. [Google Scholar] [CrossRef]
- Shaker, O.G.; Kamel, L.H.; Morad, M.A.; Shalaby, S.M. Reproducibility of Mitosis Counting in Breast Cancer between Whole Slide Images and Glass Slides. Pathol. Res. Pract. 2020, 216, 152993. [Google Scholar] [CrossRef]
- Ginter, P.S.; Lee, Y.J.; Suresh, A.; Acs, G.; Yan, S.; Reisenbichler, E.S. Mitotic Count Assessment on Whole Slide Images of Breast Cancer: A Comparative Study with Conventional Light Microscopy. Am. J. Surg. Pathol. 2021, 45, 1656–1664. [Google Scholar] [CrossRef]
- Rakha, E.A.; Toss, M.S.; Al-Khawaja, D.; Mudaliar, K.; Gosney, J.R.; Ellis, I.O.; Dalton, L.W. Impact of Whole Slide Imaging on Mitotic Count and Grading of Breast Cancer: A Multi-Institutional Concordance Study. J. Clin. Pathol. 2018, 71, 680–686. [Google Scholar] [CrossRef]
- Cytomine. Available online: https://cytomine.com/ (accessed on 24 February 2025).
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Guichard, L. Evaluation du Score Mitotique des Carcinomes Mammaires Infiltrants: Développement et Apport d’un Algorithme de Détection de Mitoses. PhD Thesis, Université Paris-Saclay, Gif-sur-Yvette, France, 2022. [Google Scholar]
- Aubreville, M.; Stathonikos, N.; Donovan, T.A.; Klopfleisch, R.; Ammeling, J.; Ganz, J.; Wilm, F.; Veta, M.; Jabari, S.; Eckstein, M.; et al. Domain Generalization across Tumor Types, Laboratories, and Species—Insights from the 2022 Edition of the Mitosis Domain Generalization Challenge. Med. Image Anal. 2024, 94, 103155. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- McHugh, M.L. Interrater Reliability: The Kappa Statistic. Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Bertram, C.A.; Aubreville, M.; Donovan, T.A.; Bartel, A.; Wilm, F.; Marzahl, C.; Assenmacher, C.-A.; Becker, K.; Bennett, M.; Corner, S.; et al. Computer-Assisted Mitotic Count Using a Deep Learning–Based Algorithm Improves Interobserver Reproducibility and Accuracy. Vet. Pathol. 2022, 59, 211–226. [Google Scholar] [CrossRef]
- Gu, H.; Onstott, E.; Yan, W.; Xu, T.; Wang, R.; Wu, Z.; Chen, X.A.; Haeri, M. Z-Stack Scanning Can Improve AI Detection of Mitosis: A Case Study of Meningiomas. arXiv 2025, arXiv:2501.15743. [Google Scholar]
- Nerrienet, N.; Peyret, R.; Sockeel, M.; Sockeel, S. Standardized CycleGAN Training for Unsupervised Stain Adaptation in Invasive Carcinoma Classification for Breast Histopathology. J. Med. Imaging 2023, 10, 067502. [Google Scholar] [CrossRef]



| Slides | Training | 12 (Bicêtre) and 150 (MIDOG21) | |||
| Testing | 17 (Bicêtre) | ||||
| Patches | Detection | Classification | |||
| Size | 256 × 256 pixel | 50 × 50 pixel | |||
| Magnification | ×20 | ×20 | |||
| Class | Mitotic | Not mitotic | Mitosis | Artifacts | |
| number in training | 2791 | 1341 | 3106 | 8638 | |
| number in testing | 146 | 24,716 | 153 | 5081 | |
| Model | Loss Function | Optimizer | Learning Rate Strategy | Hyperparameters |
|---|---|---|---|---|
| RetinaNet | L1 + Focal loss | SDG | Piecewise constant decay | LR = 0.01, momentum = 0.9, weight decay = 1 × 10−4 |
| MobileNetV2 | BCE | Adam | Constant | LR = 0.0001 |
| Dataset | Recall | Precision |
|---|---|---|
| Private | 43.8% | 27.6% |
| MIDOG 2022 | 33.1% | 37.2% |
| MITOS-ATYPIA [19] | 39.6% | 28.6% |
| Cohort (n = 50) | |
|---|---|
| Number of Cases | |
| Gender | |
| Female | 50 (100%) |
| Male | 0 (0%) |
| Age | |
| ≥50 years | 42 (84%) |
| <50 years | 8 (16%) |
| Pathological tumor stage (for breast resection only—25 cases) | |
| pT1 | 18 (72%) |
| pT2 | 4 (16%) |
| pT3 | 1 (4%) |
| pT4 | 2 (8%) |
| Pathological lymph node stage (for breast resection only—25 cases) | |
| N0 (including isolated tumor cells) | 12 (48%) |
| N1 | 9 (36%) |
| N2 | 0 (0%) |
| N3 | 1 (4%) |
| Nx | 3 (12%) |
| Histologic subtype | |
| Invasive carcinoma of no special type | 39 (78%) |
| with neuroendocrine differentiation | 2 (4%) |
| Mixed invasive carcinoma of no special type | |
| with mucinous carcinoma | 1 (2%) |
| with invasive micropapillary carcinoma | 1 (2%) |
| Invasive lobular carcinoma | 6 (12%) |
| Pure invasive micropapillary carcinoma | 1 (2%) |
| Tumor ER/PR and HER2 status | |
| ER+/PR+/HER2- | 39 (78%) |
| ER+/PR-/HER2- | 6 (12%) |
| ER-/PR-/HER2- | 2 (4%) |
| ER-/PR-/HER2+ | 3 (6%) |
| Lymphovascular invasion | |
| Negative | 47 (94%) |
| Positive | 3 (6%) |
| In situ carcinoma associated | |
| Yes | 18 (36%) |
| No | 32 (64%) |
| Mitotic score | |
| 1 | 29 (55%) |
| 2 | 12 (24%) |
| 3 | 9 (18%) |
| Score 1 (n = 29) | Score 2 (n = 9) | Score 3 (n = 12) | Biopsies (n = 25) | Specimens (n = 25) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AI assistance | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes |
| Score accuracy (%) | 94.83 | 94.83 | 5.56 | 33.33 | 29.17 | 66.67 | 60.00 | 72.00 | 66.00 | 82.00 |
| Linear weighted CK | 0.47 | / | 0 | 0.31 | 031 | 0.47 | 0.17 | 0.53 | 0.55 | 0.73 |
| % of slides where readers’ counting zone intersect | 48.3 | 48.3 | 44.4 | 55.6 | 33.3 | 66.7 | 32.0 | 60.0 | 56.0 | 60.0 |
| % of slides where AI hotspot intersect | ||||||||||
| Reader1’s counting zone Reader2’s counting zone | 37.9 58.6 | 79.3 89.7 | 66.7 77.8 | 77.8 88.9 | 50.0 68.7 | 83.3 91.7 | 40.0 60.0 | 84.0 100.0 | 48.0 68.0 | 76.0 88.0 |
| Paper | Study Design | Automatic ROI Selection | Main Results |
|---|---|---|---|
| Balkenhol et al. (2019), [29] | Pathologists assessed semi automatically pre-extracted high-power fields (HPFs) MC with microscope vs. digital slides with AI assistance. | Partial | They demonstrate a +0.13 improvement in Cohen’s Kappa for MS agreement, and a +0.02 increase in ICC for MC agreement |
| Pantanowitz et al. (2020), [27] | Pathologists assessed pre-extracted high-power fields (HPFs) MS with and without AI assistance. | No | AI assistance led to an 11.82% increase in MS accuracy. The study focused on accuracy improvement when using AI in selected high-power fields. |
| van Bergeijk et al. (2023), [28] | Slide reading in a clinical setup, comparing microscope-based reading vs. WSI with and without AI assistance. Pathologists assessed mitotic count with and without AI. | Yes | AI-assisted mitotic count was found to be possibly non-inferior to conventional microscopic evaluation. The study suggests AI assistance could be integrated into clinical workflows. |
| Ours | Clinical study evaluating AI-assisted mitotic counting in a real-world setup. Pathologists analyzed WSI with and without AI assistance, and results were compared against ground truth. AI automatically selected hotspots. | Yes | Our study demonstrated a +14% increase in MS accuracy, a +0.19 improvement in Cohen’s Kappa for MS agreement, a +0.29 increase in ICC for MC agreement, and a +16% improvement in hotspot agreement, highlighting the benefits of AI assistance in mitotic counting. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simmat, C.; Guichard, L.; Sockeel, S.; Pozin, N.; Peyret, R.; Lacroix-Triki, M.; Miquel, C.; Gauthier, A.; Sockeel, M.; Prévot, S. Evaluating AI-Based Mitosis Detection for Breast Carcinoma in Digital Pathology: A Clinical Study on Routine Practice Integration. Diagnostics 2025, 15, 1127. https://doi.org/10.3390/diagnostics15091127
Simmat C, Guichard L, Sockeel S, Pozin N, Peyret R, Lacroix-Triki M, Miquel C, Gauthier A, Sockeel M, Prévot S. Evaluating AI-Based Mitosis Detection for Breast Carcinoma in Digital Pathology: A Clinical Study on Routine Practice Integration. Diagnostics. 2025; 15(9):1127. https://doi.org/10.3390/diagnostics15091127
Chicago/Turabian StyleSimmat, Clara, Loris Guichard, Stéphane Sockeel, Nicolas Pozin, Rémy Peyret, Magali Lacroix-Triki, Catherine Miquel, Arnaud Gauthier, Marie Sockeel, and Sophie Prévot. 2025. "Evaluating AI-Based Mitosis Detection for Breast Carcinoma in Digital Pathology: A Clinical Study on Routine Practice Integration" Diagnostics 15, no. 9: 1127. https://doi.org/10.3390/diagnostics15091127
APA StyleSimmat, C., Guichard, L., Sockeel, S., Pozin, N., Peyret, R., Lacroix-Triki, M., Miquel, C., Gauthier, A., Sockeel, M., & Prévot, S. (2025). Evaluating AI-Based Mitosis Detection for Breast Carcinoma in Digital Pathology: A Clinical Study on Routine Practice Integration. Diagnostics, 15(9), 1127. https://doi.org/10.3390/diagnostics15091127






