Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review
Abstract
1. Introduction
1.1. Prostate Cancer
1.2. DL Applied to WSI
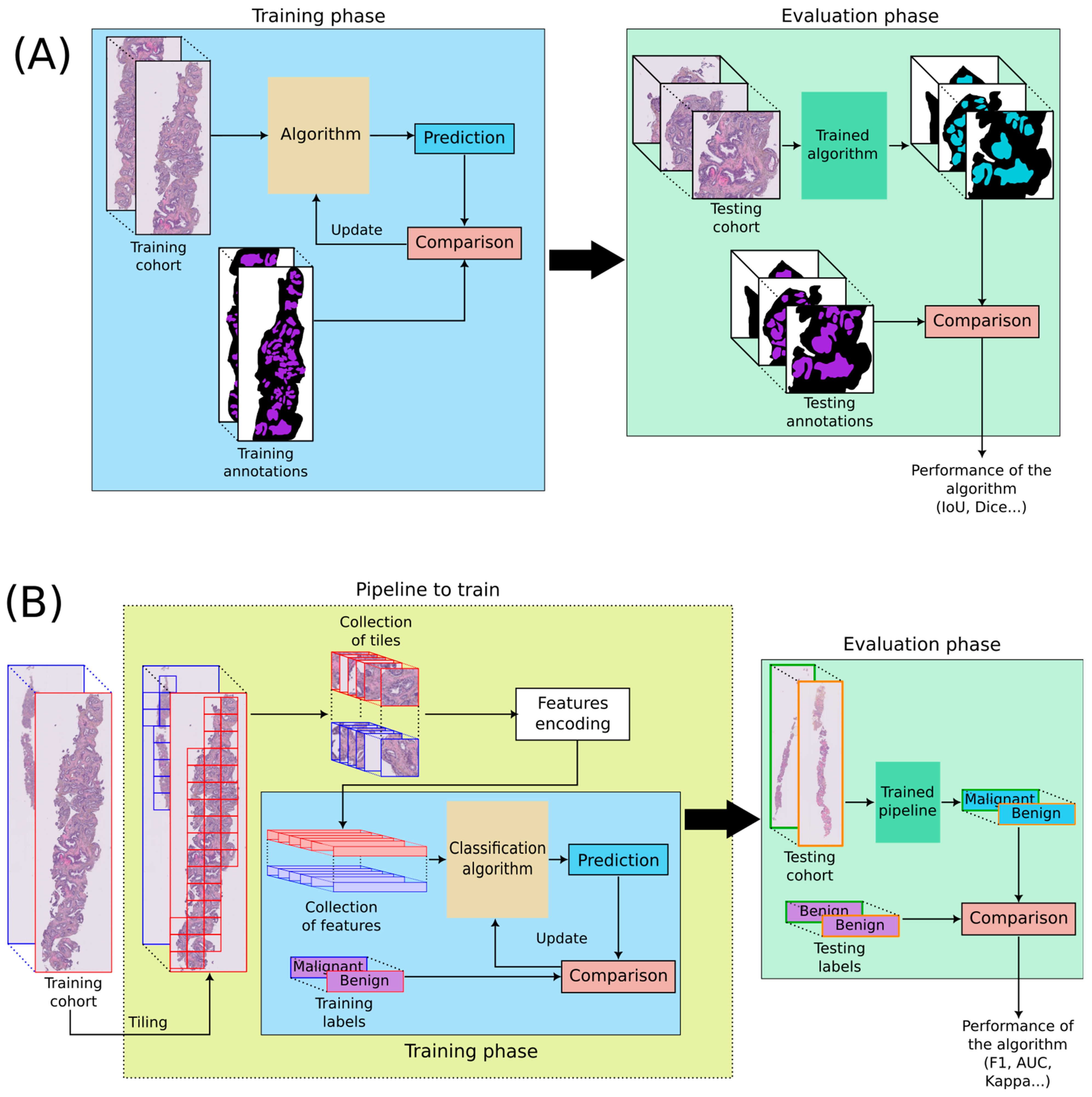
1.3. Applications and Evaluation of Algorithms
1.4. Aim of This Review
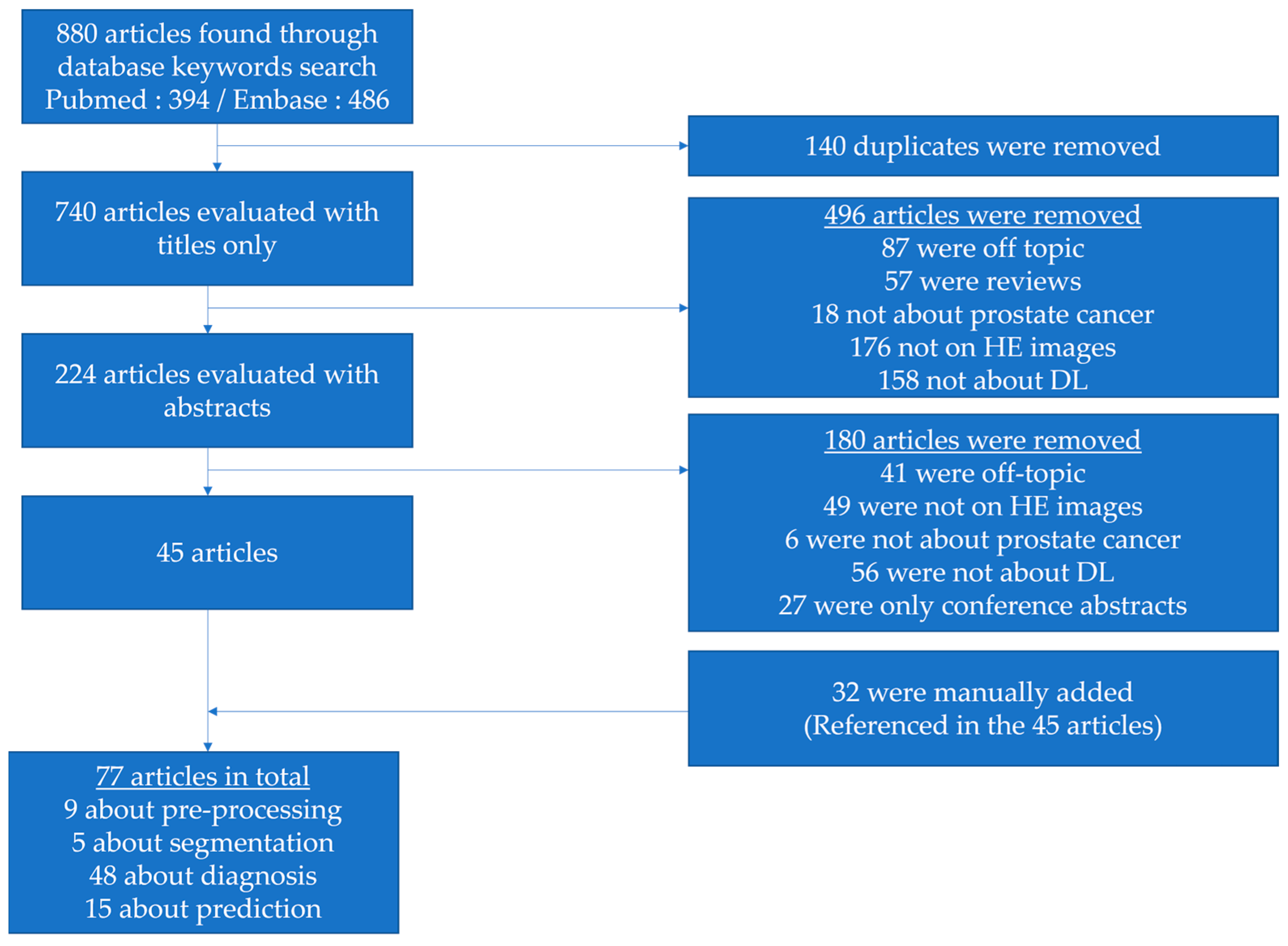
2. Materials and Methods
- be written in English,
- focus on prostate cancer,
- use pathology H&E-stained images,
- rely on deep learning.
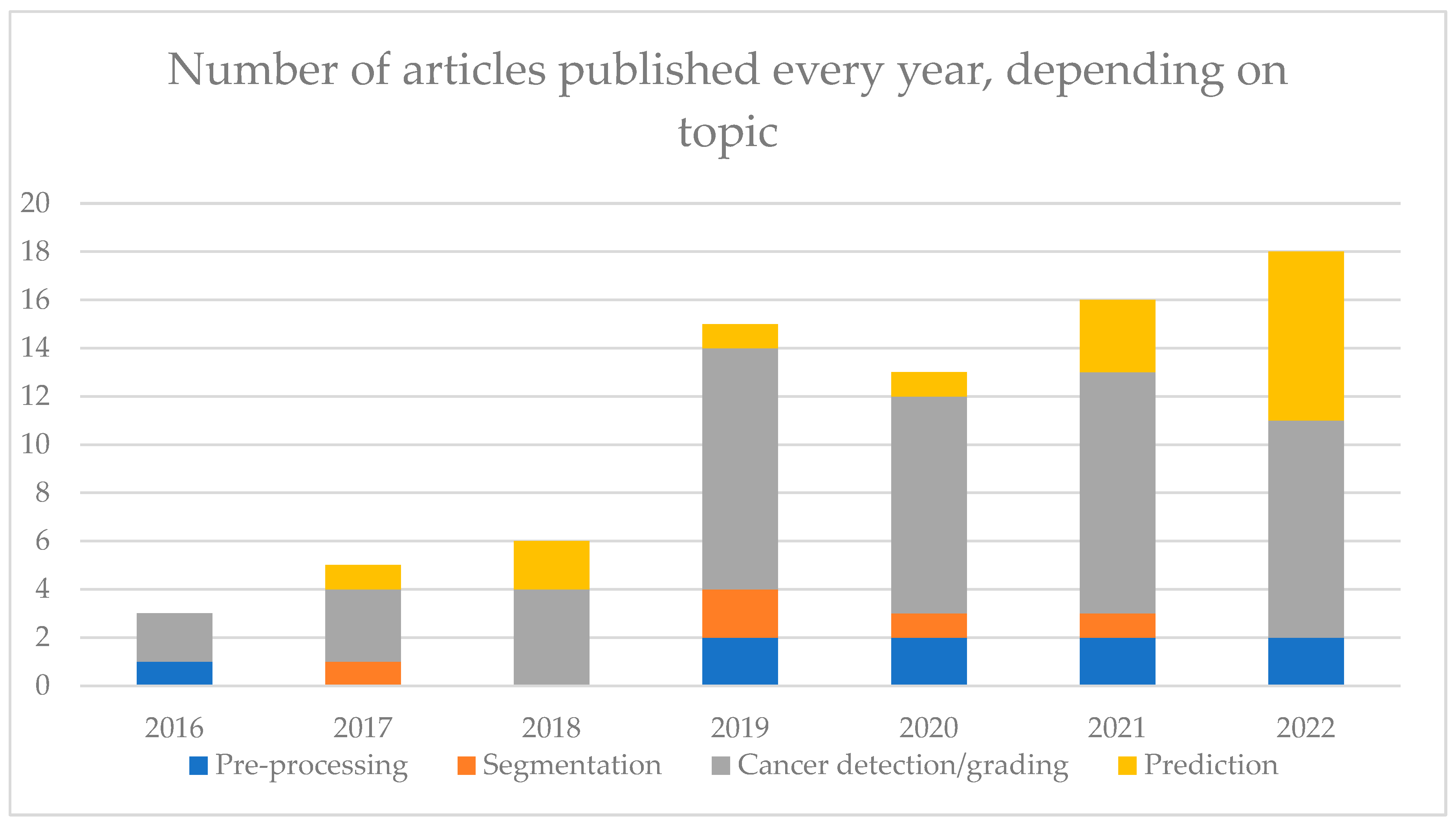
3. Results
3.1. Pre-Processing
3.1.1. Quality Assessment
3.1.2. Staining Normalization and Tile Selection
3.2. Diagnosis
3.2.1. Segmentation
3.2.2. Cancer Detection
3.2.3. Gleason Grading
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Källén, 2016 [63] | OverFeat | TCGA | 10-fold cross val | 213 WSIs | GP tile classification | ACC: 0.81 |
| Classify WSIs with a majority GP | ACC: 0.89 | |||||
| Jimenez Del Toro, 2017 [74] | GoogleNet | 141 WSIs | 47 WSIs | None | High vs. Low-grade classification | ACC:0.735 |
| Arvaniti, 2018 [21] | MobileNet + classifier | 641 TMAs | 245 TMAs | None | TMA grading | qKappa: 0.71/0.75 (0.71 pathologists) |
| Tile grading | qKappa: 0.55/0.53 (0.67 pathologists) | |||||
| Poojitha, 2019 [64] | CNNs | 80 samples | 20 samples | None | GP estimation at tile level (GP 2 to 5) | F1: 0.97 |
| Nagpal, 2019 [73] | InceptionV3 | 1159 WSIs | 331 WSIs | None | GG classification | ACC: 0.7 |
| High/low-grade classification (GG 2, 3 or 4 as threshold) | : 0.95 | |||||
| Survival analysis, according to Gleason | HR: 1.38 | |||||
| Silva-Rodriguez, 2020 [52] | Custom CNN | 182 WSIs | 5-fold cross-val | 703 tiles from 641 TMAs [21] | GP at the tile level | : 0.713 (IV) & 0.57 (EV) qKappa: 0.732 (IV) & 0.64 (EV) |
| GG at the WSI level | qKappa: 0.81 (0.77 with [21] method) | |||||
| Cribriform pattern detection at the tile level | AUC: 0.822 | |||||
| Otalora, 2021 [65] | MobileNet-based CNN | 641 TMAs 255 WSIs | 245 TMAs 46 WSIs | None | GG classification | wKappa: 0.52 |
| Hammouda, 2021 [78] | CNNs | 712 WSIs | 96 WSIs | None | GP at the tile level | : 0.76 |
| GG | : 0.6 | |||||
| Marini, 2021 [66] | Custom CNN | 641 TMAs 255 WSI | 245 TMAs 46 WSIs | None | GP at the tile level | qKappa: 0.66 |
| GS at TMA level | qKappa: 0.81 | |||||
| Marron-Esquivel, 2023 [77] | DenseNet121 | 15,020 patches | 2612 patches | None | Tile-level GP classification | qKappa: 0.826 |
| 102,324 patches (PANDA + fine-tuning) | qKappa: 0.746 | |||||
| Ryu, 2019 [72] | DeepDx Prostate | 1133 WSIs | 700 WSIs | None | GG classification | qKappa: 0.907 |
| Karimi, 2019 [61] | Custom CNN | 247 TMAs | 86 TMAs | None | Malignancy detection at the tile level | Sens: 0.86 Spec: 0.85 |
| GP 3 vs. 4/5 at tile level | Sens: 0.82 Spec: 0.82 | |||||
| Nagpal, 2020 [35] | Xception | 524 WSIs | 430 WSIs | 322 WSIs | Malignancy detection at the WSI level | AUC: 0.981 Agreement: 0.943 |
| GG1-2 vs. GG3-5 | AUC: 0.972 Agreement: 0.928 | |||||
| Pantanowitz, 2020 [36] | IBEX | 549 WSIs | 2501 WSIs | 1627 WSIs | Cancer detection at the WSI level | AUC: 0.997 (IV) & 0.991 (EV) |
| Low vs. high grade (GS 6 vs. GS 7–10) | AUC: 0.941 (EV) | |||||
| GP3/4 vs. GP5 | AUC: 0.971 (EV) | |||||
| Perineural invasion detection | AUC: 0.957 (EV) | |||||
| Ström, 2020 [37] | InceptionV3 | 6935 WSIs | 1631 WSIs | 330 WSIs | Malignancy detection at the WSI level | AUC: 0.997 (IV) 0.986 (EV) |
| GG classification | Kappa: 0.62 | |||||
| Li, 2021 [39] | Weakly supervised VGG11bn | 13,115 WSIs | 7114 WSIs | 79 WSIs | Malignancy of slides | AUC: 0.982 (IV) & 0.994 (EV) |
| Low vs. high grade at the WSI level | Kappa: 0.818 Acc: 0.927 | |||||
| Kott, 2021 [62] | ResNet | 85 WSIs | 5-fold cross-val | None | Malignancy detection at the tile level | AUC: 0.83 ACC: 0.85 for fine-tuned detection |
| GP classification at the tile level | Sens: 0.83 Spec: 0.94 | |||||
| Marginean, 2021 [79] | CNN | 698 WSIs | 37 WSIs | None | Cancer area detection | Sens: 1 Spec: 0.68 |
| GG classification | : 0.6 | |||||
| Jung, 2022 [75] | DeepDx Prostate | Pre-trained | Pre-trained | 593 WSIs | Correlation with reference pathologist (pathology report comparison) | Kappa: 0.654 (0.576) qKappa: 0.904 (0.858) |
| Silva-Rodriguez, 2022 [40] | VGG16 | 252 WSIs | 98 WSIs | None | Cancer detection at the tile level | AUC: 0.979 |
| GS at the tile level | AUC: 0.899 | |||||
| GP at the tile level | : 0.65 (0.75 prev. Paper) qKappa: 0.655 | |||||
| Bulten, 2022 [76] | Evaluation of multiple algorithms (PANDA challenge) | 10,616 WSIs | 545 WSIs | 741 patients (EV1) 330 patients (EV2) | GG classification | qKappa: 0.868 (EV2) qKappa: 0.862 (EV1) |
| Li, 2018 [68] | Multi-scale U-Net | 187 tiles from 17 patients | 37 tiles from 3 patients | None | Segment Stroma, benign and malignant gland segmentation | IoU: 0.755 (0.750 classic U-Net) |
| Stroma, benign and GP 3/4 gland segmentation | IoU: 0.658 (0.644 classic U-Net) | |||||
| Li, 2019 * [29] | R-CNN | 513 WSIs | 5 fold cross val | None | Stroma, benign, low- and high-grade gland segmentation | : 0.79 (mean amongst classes) |
| Bulten, 2019 * [28] | U-Net | 62 WSIs | 40 WSIs | 20 WSIs | Benign vs. GP | IoU: 0.811 (IV) & 0.735 (EV) F1: 0.893 (IV) & 0.835 (EV) |
| Lokhande, 2020 [69] | FCN8 based on ResNet50 | 172 TMAs | 72 TMAs | None | Benign, grade 3/4/5 segmentation | Dice: 0.74 (average amongst all classes) |
| Li, 2018 [71] | Multi-Scale U-Net-based CNN | 50 patients | 20 patients | None | Contribution of EM for multi-scale U-Net improvement | : 0.35 (U-Net) : 0.49 (EM-adaptative 30%) |
| Bulten, 2020 [41] | Extended Unet | 5209 biopsies from 1033 patients | 550 biopsies from 210 patients | 886 cores | Malignancy detection at the WSI level | AUC: 0.99 (IV) & 0.98 (EV) |
| GG > 2 detection | AUC: 0.978 (IV) & 0.871 (EV) | |||||
| 100 biopsies | None | GG classification | qKappa: 0.819 (general pathologists) & 0.854 (DL on IV) & 0.71 (EV) | |||
| Hassan, 2022 [70] | ResNet50 | 18,264 WSIs | 3251 WSIs | None | Tissue segmentation for GG presence | : 0.48 : 0.375 |
| Lucas, 2019 [67] | Inception V3 | 72 WSIs | 24 WSIs | None | Malignancy detection at the pixel level | Sens: 0.90 Spec: 0.93 |
| GP3 & GP4 segmentation at pixel level | Sens: 0.77 Spec: 0.94 |
3.3. Prediction
3.3.1. Clinical Outcome Prediction
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Kumar, 2017 [81] | CNNs | 160 TMAs | 60 TMAs | None | Nucleus detection for tile selection | ACC: 0.89 |
| Recurrence prediction | AUC: 0.81(DL) & 0.59 (clinical data) | |||||
| Ren, 2018 [83] | AlexNet + LSTM | 271 patients | 68 patients | None | Recurrence-free survival prediction | HR: 5.73 |
| Ren, 2019 [88] | CNN + LSTM | 268 patients | 67 patients | None | Survival model | HR: 7.10 when using image features |
| Leo, 2021 [87] | Segmentation-based CNNs | 70 patients | NA | 679 patients | Cribriform pattern recognition | Pixel TPV: 0.94 Pixel TNV: 0.79 |
| Prognosis classification using cribriform area measurements | Univariable HR: 1.31 Multivariable HR: 1.66 | |||||
| Wessels, 2021 [84] | xse_ResNext34 | 118 patients | 110 patients | None | LNM prediction based on initial RP slides | AUC: 0.69 |
| Esteva, 2022 [85] | ResNet | 4524 patients | 1130 patients | None | Distant metastasis at five years (5Y) and ten years (10Y) | AUC: 0.837 (5Y) AUC: 0.781 (10Y) |
| Prostate cancer-specific survival | AUC: 0.765 | |||||
| Overall survival at ten years | AUC: 0.652 | |||||
| Pinckaers, 2022 [82] | ResNet50 | 503 patients | 182 patients | 204 patients | Univariate analysis for DL predicted biomarker evaluation | OR: 3.32 (IV) HR: 4.79 (EV) |
| Liu, 2022 [86] | 10-CNN ensemble model | 9192 benign biopsies from 1211 patients | 2851 benign biopsies from 297 patients | None | Cancer detection at the patient level | AUC: 0.727 |
| Cancer detection at patient level from benign WSIs | AUC: 0.739 | |||||
| Huang, 2022 [80] | NA | 243 patients | None | 173 patients | Recurrence prediction at three years | AUC: 0.78 |
| Sandeman, 2022 [42] | Custom CNN (AIForIA) | 331 patients | 391 patients | 126 patients | Malignant vs. benign | AUC: 0.997 |
| Grade grouping | ACC: 0.67 wKappa: 0.77 | |||||
| Outcome prediction | HR: 5.91 |
3.3.2. Genomic Signatures Prediction
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Schaumberg, 2018 [92] | ResNet50 | 177 patients | None | 152 patients | SPOP mutation prediction | AUC: 0.74 (IV) AUC: 0.86 (EV) |
| Schmauch, 2020 [89] | HE2RNA | 8725 patients (Pan-cancer) | 5-fold cross val | None | Prediction of gene signatures specific to prostate cancer | PC: 0.18 (TP63) 0.12 (KRT8 & KRT18) |
| Chelebian, 2021 [91] | CNN from [37] fine-tuned | Pre-trained ([37]) | 7 WSIs | None | Correlation between clusters identified with AI and spatial transcriptomics | No global metric |
| Dadhania, 2022 [93] | MobileNetV2 | 261 patients | 131 patients | None | ERG gene rearrangement status prediction | AUC: 0.82 to 0.85 (depending on resolution) |
| Weitz, 2022 [90] | NA | 278 patients | 92 patients | None | CCP gene expression prediction | PC: 0.527 |
| BRICD5 expression prediction | PC: 0.749 | |||||
| SPOPL expression prediction | PC: 0.526 |
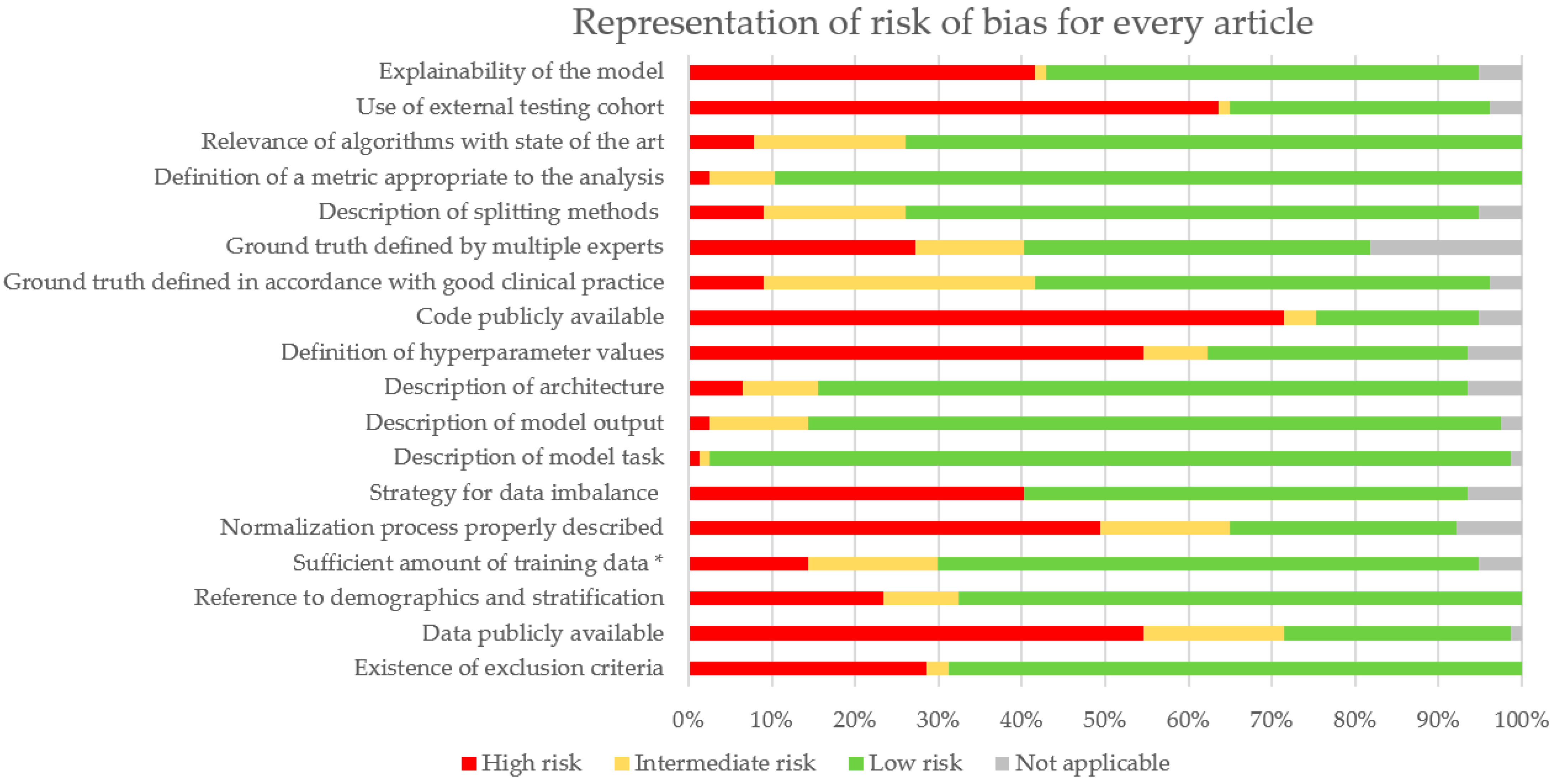
3.4. Risk of Bias Analysis
4. Discussion
4.1. Summary of Results
4.2. Biases
4.3. Limits of AI
4.4. Impact of AI in Routine Activity
4.5. Multimodal Approach for Predictive Algorithms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Descotes, J.-L. Diagnosis of Prostate Cancer. Asian J. Urol. 2019, 6, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.I.; Allsbrook, W.C.; Amin, M.B.; Egevad, L.L.; ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2005, 29, 1228–1242. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A.; The Grading Committee. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244. [Google Scholar] [CrossRef] [PubMed]
- Williams, I.S.; McVey, A.; Perera, S.; O’Brien, J.S.; Kostos, L.; Chen, K.; Siva, S.; Azad, A.A.; Murphy, D.G.; Kasivisvanathan, V.; et al. Modern Paradigms for Prostate Cancer Detection and Management. Med. J. Aust. 2022, 217, 424–433. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Syst. Rev. 2021, 10, 89. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Sounderajah, V.; Ashrafian, H.; Rose, S.; Shah, N.H.; Ghassemi, M.; Golub, R.; Kahn, C.E.; Esteva, A.; Karthikesalingam, A.; Mateen, B.; et al. A Quality Assessment Tool for Artificial Intelligence-Centered Diagnostic Test Accuracy Studies: QUADAS-AI. Nat. Med. 2021, 27, 1663–1665. [Google Scholar] [CrossRef]
- Cabitza, F.; Campagner, A. The IJMEDI Checklist for Assessment of Medical AI. Int. J. Med. Inform. 2021, 153. [Google Scholar] [CrossRef]
- Schömig-Markiefka, B.; Pryalukhin, A.; Hulla, W.; Bychkov, A.; Fukuoka, J.; Madabhushi, A.; Achter, V.; Nieroda, L.; Büttner, R.; Quaas, A.; et al. Quality Control Stress Test for Deep Learning-Based Diagnostic Model in Digital Pathology. Mod. Pathol. 2021, 34, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, M.; Browning, L.; Sirinukunwattana, K.; Malacrino, S.; Khalid Alham, N.; Colling, R.; Cui, Y.; Rakha, E.; Hamdy, F.C.; Verrill, C.; et al. Automated Quality Assessment of Large Digitised Histology Cohorts by Artificial Intelligence. Sci. Rep. 2022, 12, 5002. [Google Scholar] [CrossRef] [PubMed]
- Brendel, M.; Getseva, V.; Assaad, M.A.; Sigouros, M.; Sigaras, A.; Kane, T.; Khosravi, P.; Mosquera, J.M.; Elemento, O.; Hajirasouliha, I. Weakly-Supervised Tumor Purity Prediction from Frozen H&E Stained Slides. eBioMedicine 2022, 80, 104067. [Google Scholar] [CrossRef] [PubMed]
- Anghel, A.; Stanisavljevic, M.; Andani, S.; Papandreou, N.; Rüschoff, J.H.; Wild, P.; Gabrani, M.; Pozidis, H. A High-Performance System for Robust Stain Normalization of Whole-Slide Images in Histopathology. Front. Med. 2019, 6, 193. [Google Scholar] [CrossRef]
- Otálora, S.; Atzori, M.; Andrearczyk, V.; Khan, A.; Müller, H. Staining Invariant Features for Improving Generalization of Deep Convolutional Neural Networks in Computational Pathology. Front. Bioeng. Biotechnol. 2019, 7, 198. [Google Scholar] [CrossRef]
- Arvaniti, E.; Fricker, K.S.; Moret, M.; Rupp, N.; Hermanns, T.; Fankhauser, C.; Wey, N.; Wild, P.J.; Rüschoff, J.H.; Claassen, M. Automated Gleason Grading of Prostate Cancer Tissue Microarrays via Deep Learning. Sci. Rep. 2018, 8, 12054. [Google Scholar] [CrossRef]
- Rana, A.; Lowe, A.; Lithgow, M.; Horback, K.; Janovitz, T.; Da Silva, A.; Tsai, H.; Shanmugam, V.; Bayat, A.; Shah, P. Use of Deep Learning to Develop and Analyze Computational Hematoxylin and Eosin Staining of Prostate Core Biopsy Images for Tumor Diagnosis. JAMA Netw. Open 2020, 3, e205111. [Google Scholar] [CrossRef]
- Sethi, A.; Sha, L.; Vahadane, A.R.; Deaton, R.J.; Kumar, N.; Macias, V.; Gann, P.H. Empirical Comparison of Color Normalization Methods for Epithelial-Stromal Classification in H and E Images. J. Pathol. Inform. 2016, 7, 17. [Google Scholar] [CrossRef]
- Swiderska-Chadaj, Z.; de Bel, T.; Blanchet, L.; Baidoshvili, A.; Vossen, D.; van der Laak, J.; Litjens, G. Impact of Rescanning and Normalization on Convolutional Neural Network Performance in Multi-Center, Whole-Slide Classification of Prostate Cancer. Sci. Rep. 2020, 10, 14398. [Google Scholar] [CrossRef]
- Salvi, M.; Molinari, F.; Acharya, U.R.; Molinaro, L.; Meiburger, K.M. Impact of Stain Normalization and Patch Selection on the Performance of Convolutional Neural Networks in Histological Breast and Prostate Cancer Classification. Comput. Methods Programs Biomed. Update 2021, 1, 100004. [Google Scholar] [CrossRef]
- Ren, J.; Sadimin, E.; Foran, D.J.; Qi, X. Computer Aided Analysis of Prostate Histopathology Images to Support a Refined Gleason Grading System. In Proceedings of the Medical Imaging 2017: Image Processing, Orlando, FL, USA, 24 February 2017; Volume 10133, pp. 532–539. [Google Scholar]
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A Hybrid Deep Learning Approach for Gland Segmentation in Prostate Histopathological Images. Artif. Intell. Med. 2021, 115, 102076. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Bándi, P.; Hoven, J.; van de Loo, R.; Lotz, J.; Weiss, N.; van der Laak, J.; van Ginneken, B.; Hulsbergen-van de Kaa, C.; Litjens, G. Epithelium Segmentation Using Deep Learning in H&E-Stained Prostate Specimens with Immunohistochemistry as Reference Standard. Sci. Rep. 2019, 9, 864. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, J.; Sarma, K.V.; Ho, K.C.; Shen, S.; Knudsen, B.S.; Gertych, A.; Arnold, C.W. Path R-CNN for Prostate Cancer Diagnosis and Gleason Grading of Histological Images. IEEE Trans. Med. Imaging 2019, 38, 945–954. [Google Scholar] [CrossRef]
- Bukowy, J.D.; Foss, H.; McGarry, S.D.; Lowman, A.K.; Hurrell, S.L.; Iczkowski, K.A.; Banerjee, A.; Bobholz, S.A.; Barrington, A.; Dayton, A.; et al. Accurate Segmentation of Prostate Cancer Histomorphometric Features Using a Weakly Supervised Convolutional Neural Network. J. Med. Imaging 2020, 7, 057501. [Google Scholar] [CrossRef]
- Duong, Q.D.; Vu, D.Q.; Lee, D.; Hewitt, S.M.; Kim, K.; Kwak, J.T. Scale Embedding Shared Neural Networks for Multiscale Histological Analysis of Prostate Cancer. In Proceedings of the Medical Imaging 2019: Digital Pathology, San Diego, CA, USA, 18 March 2019; Volume 10956, pp. 15–20. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-Grade Computational Pathology Using Weakly Supervised Deep Learning on Whole Slide Images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Pinckaers, H.; Bulten, W.; van der Laak, J.; Litjens, G. Detection of Prostate Cancer in Whole-Slide Images Through End-to-End Training With Image-Level Labels. IEEE Trans. Med. Imaging 2021, 40, 1817–1826. [Google Scholar] [CrossRef]
- Raciti, P.; Sue, J.; Ceballos, R.; Godrich, R.; Kunz, J.D.; Kapur, S.; Reuter, V.; Grady, L.; Kanan, C.; Klimstra, D.S.; et al. Novel Artificial Intelligence System Increases the Detection of Prostate Cancer in Whole Slide Images of Core Needle Biopsies. Mod. Pathol. 2020, 33, 2058–2066. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Tan, F.; Liu, Y.; Chen, P.-H.C.; Steiner, D.F.; Manoj, N.; Olson, N.; Smith, J.L.; Mohtashamian, A.; et al. Development and Validation of a Deep Learning Algorithm for Gleason Grading of Prostate Cancer From Biopsy Specimens. JAMA Oncol. 2020, 6, 1372–1380. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Albrecht Shach, A.; Shalev, V.; Vecsler, M.; et al. An Artificial Intelligence Algorithm for Prostate Cancer Diagnosis in Whole Slide Images of Core Needle Biopsies: A Blinded Clinical Validation and Deployment Study. Lancet Digit. Health 2020, 2, e407–e416. [Google Scholar] [CrossRef] [PubMed]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial Intelligence for Diagnosis and Grading of Prostate Cancer in Biopsies: A Population-Based, Diagnostic Study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Johnson, C.; Gaed, M.; Gómez, J.A.; Moussa, M.; Chin, J.L.; Pautler, S.; Bauman, G.S.; Ward, A.D. Histologic Tissue Components Provide Major Cues for Machine Learning-Based Prostate Cancer Detection and Grading on Prostatectomy Specimens. Sci. Rep. 2020, 10, 9911. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Sisk, A.; Ye, H.; Wallace, W.D.; Speier, W.; Arnold, C.W. A Multi-Resolution Model for Histopathology Image Classification and Localization with Multiple Instance Learning. Comput. Biol. Med. 2021, 131, 104253. [Google Scholar] [CrossRef]
- Silva-Rodríguez, J.; Schmidt, A.; Sales, M.A.; Molina, R.; Naranjo, V. Proportion Constrained Weakly Supervised Histopathology Image Classification. Comput. Biol. Med. 2022, 147, 105714. [Google Scholar] [CrossRef]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; Hulsbergen-van de Kaa, C.; Litjens, G. Automated Deep-Learning System for Gleason Grading of Prostate Cancer Using Biopsies: A Diagnostic Study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef]
- Sandeman, K.; Blom, S.; Koponen, V.; Manninen, A.; Juhila, J.; Rannikko, A.; Ropponen, T.; Mirtti, T. AI Model for Prostate Biopsies Predicts Cancer Survival. Diagnostics 2022, 12, 1031. [Google Scholar] [CrossRef]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-van de Kaa, C.; Bult, P.; van Ginneken, B.; van der Laak, J. Deep Learning as a Tool for Increased Accuracy and Efficiency of Histopathological Diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Kwak, J.T.; Hewitt, S.M. Nuclear Architecture Analysis of Prostate Cancer via Convolutional Neural Networks. IEEE Access 2017, 5, 18526–18533. [Google Scholar] [CrossRef]
- Kwak, J.T.; Hewitt, S.M. Lumen-Based Detection of Prostate Cancer via Convolutional Neural Networks. In Proceedings of the Medical Imaging 2017: Digital Pathology, Orlando, FL, USA, 1 March 2017; Volume 10140, pp. 47–52. [Google Scholar]
- Campanella, G.; Silva, V.W.K.; Fuchs, T.J. Terabyte-Scale Deep Multiple Instance Learning for Classification and Localization in Pathology. arXiv 2018, arXiv:1805.06983. [Google Scholar]
- Raciti, P.; Sue, J.; Retamero, J.A.; Ceballos, R.; Godrich, R.; Kunz, J.D.; Casson, A.; Thiagarajan, D.; Ebrahimzadeh, Z.; Viret, J.; et al. Clinical Validation of Artificial Intelligence–Augmented Pathology Diagnosis Demonstrates Significant Gains in Diagnostic Accuracy in Prostate Cancer Detection. Arch. Pathol. Lab. Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.M.; Pereira, E.M.; Salles, P.G.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Ferreira, C.G.; et al. Independent Real-world Application of a Clinical-grade Automated Prostate Cancer Detection System. J. Pathol. 2021, 254, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Perincheri, S.; Levi, A.W.; Celli, R.; Gershkovich, P.; Rimm, D.; Morrow, J.S.; Rothrock, B.; Raciti, P.; Klimstra, D.; Sinard, J. An Independent Assessment of an Artificial Intelligence System for Prostate Cancer Detection Shows Strong Diagnostic Accuracy. Mod. Pathol. 2021, 34, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kalaw, E.M.; Jie, W.; Al-Shabi, M.; Wong, C.F.; Giron, D.M.; Chong, K.-T.; Tan, M.; Zeng, Z.; Lee, H.K. Cribriform Pattern Detection in Prostate Histopathological Images Using Deep Learning Models. arXiv 2019, arXiv:1910.04030. [Google Scholar]
- Ambrosini, P.; Hollemans, E.; Kweldam, C.F.; van Leenders, G.J.L.H.; Stallinga, S.; Vos, F. Automated Detection of Cribriform Growth Patterns in Prostate Histology Images. Sci. Rep. 2020, 10, 14904. [Google Scholar] [CrossRef]
- Silva-Rodríguez, J.; Colomer, A.; Sales, M.A.; Molina, R.; Naranjo, V. Going Deeper through the Gleason Scoring Scale: An Automatic End-to-End System for Histology Prostate Grading and Cribriform Pattern Detection. Comput. Methods Programs Biomed. 2020, 195, 105637. [Google Scholar] [CrossRef] [PubMed]
- Tsuneki, M.; Abe, M.; Kanavati, F. A Deep Learning Model for Prostate Adenocarcinoma Classification in Needle Biopsy Whole-Slide Images Using Transfer Learning. Diagnostics 2022, 12, 768. [Google Scholar] [CrossRef]
- Tsuneki, M.; Abe, M.; Kanavati, F. Transfer Learning for Adenocarcinoma Classifications in the Transurethral Resection of Prostate Whole-Slide Images. Cancers 2022, 14, 4744. [Google Scholar] [CrossRef]
- García, G.; Colomer, A.; Naranjo, V. First-Stage Prostate Cancer Identification on Histopathological Images: Hand-Driven versus Automatic Learning. Entropy 2019, 21, 356. [Google Scholar] [CrossRef]
- Jones, A.D.; Graff, J.P.; Darrow, M.; Borowsky, A.; Olson, K.A.; Gandour-Edwards, R.; Mitra, A.D.; Wei, D.; Gao, G.; Durbin-Johnson, B.; et al. Impact of Pre-Analytic Variables on Deep Learning Accuracy in Histopathology. Histopathology 2019, 75, 39–53. [Google Scholar] [CrossRef]
- Bukhari, S.U.; Mehtab, U.; Hussain, S.; Syed, A.; Armaghan, S.; Shah, S. The Assessment of Deep Learning Computer Vision Algorithms for the Diagnosis of Prostatic Adenocarcinoma. Ann. Clin. Anal. Med. 2021, 12 (Suppl. 1), S31–S34. [Google Scholar] [CrossRef]
- Krajňanský, V.; Gallo, M.; Nenutil, R.; Němeček, M.; Holub, P.; Brázdil, T. Shedding Light on the Black Box of a Neural Network Used to Detect Prostate Cancer in Whole Slide Images by Occlusion-Based Explainability. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, C.-M.; Huang, Y.-S.; Fang, P.-W.; Liang, C.-W.; Chang, R.-F. A Computer-Aided Diagnosis System for Differentiation and Delineation of Malignant Regions on Whole-Slide Prostate Histopathology Image Using Spatial Statistics and Multidimensional DenseNet. Med. Phys. 2020, 47, 1021–1033. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Karimi, D.; Nir, G.; Fazli, L.; Black, P.C.; Goldenberg, L.; Salcudean, S.E. Deep Learning-Based Gleason Grading of Prostate Cancer From Histopathology Images—Role of Multiscale Decision Aggregation and Data Augmentation. IEEE J. Biomed. Health Inform. 2020, 24, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Kott, O.; Linsley, D.; Amin, A.; Karagounis, A.; Jeffers, C.; Golijanin, D.; Serre, T.; Gershman, B. Development of a Deep Learning Algorithm for the Histopathologic Diagnosis and Gleason Grading of Prostate Cancer Biopsies: A Pilot Study. Eur. Urol. Focus 2021, 7, 347–351. [Google Scholar] [CrossRef]
- Kallen, H.; Molin, J.; Heyden, A.; Lundstrom, C.; Astrom, K. Towards Grading Gleason Score Using Generically Trained Deep Convolutional Neural Networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1163–1167. [Google Scholar]
- Poojitha, U.P.; Lal Sharma, S. Hybrid Unified Deep Learning Network for Highly Precise Gleason Grading of Prostate Cancer. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 2019, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Otálora, S.; Marini, N.; Müller, H.; Atzori, M. Combining Weakly and Strongly Supervised Learning Improves Strong Supervision in Gleason Pattern Classification. BMC Med. Imaging 2021, 21, 77. [Google Scholar] [CrossRef]
- Marini, N.; Otálora, S.; Müller, H.; Atzori, M. Semi-Supervised Training of Deep Convolutional Neural Networks with Heterogeneous Data and Few Local Annotations: An Experiment on Prostate Histopathology Image Classification. Med. Image Anal. 2021, 73, 102165. [Google Scholar] [CrossRef]
- Lucas, M.; Jansen, I.; Savci-Heijink, C.D.; Meijer, S.L.; de Boer, O.J.; van Leeuwen, T.G.; de Bruin, D.M.; Marquering, H.A. Deep Learning for Automatic Gleason Pattern Classification for Grade Group Determination of Prostate Biopsies. Virchows Arch. Int. J. Pathol. 2019, 475, 77–83. [Google Scholar] [CrossRef]
- Li, J.; Sarma, K.V.; Chung Ho, K.; Gertych, A.; Knudsen, B.S.; Arnold, C.W. A Multi-Scale U-Net for Semantic Segmentation of Histological Images from Radical Prostatectomies. AMIA Annu. Symp. Proc. AMIA Symp. 2017, 2017, 1140–1148. [Google Scholar]
- Lokhande, A.; Bonthu, S.; Singhal, N. Carcino-Net: A Deep Learning Framework for Automated Gleason Grading of Prostate Biopsies. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1380–1383. [Google Scholar]
- Hassan, T.; Shafay, M.; Hassan, B.; Akram, M.U.; ElBaz, A.; Werghi, N. Knowledge Distillation Driven Instance Segmentation for Grading Prostate Cancer. Comput. Biol. Med. 2022, 150, 106124. [Google Scholar] [CrossRef]
- Li, J.; Speier, W.; Ho, K.C.; Sarma, K.V.; Gertych, A.; Knudsen, B.S.; Arnold, C.W. An EM-Based Semi-Supervised Deep Learning Approach for Semantic Segmentation of Histopathological Images from Radical Prostatectomies. Comput. Med. Imaging Graph. 2018, 69, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.S.; Jin, M.-S.; Park, J.H.; Lee, S.; Cho, J.; Oh, S.; Kwak, T.-Y.; Woo, J.I.; Mun, Y.; Kim, S.W.; et al. Automated Gleason Scoring and Tumor Quantification in Prostate Core Needle Biopsy Images Using Deep Neural Networks and Its Comparison with Pathologist-Based Assessment. Cancers 2019, 11, 1860. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer. npj Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Jimenez-del-Toro, O.; Atzori, M.; Andersson, M.; Eurén, K.; Hedlund, M.; Rönnquist, P.; Müller, H. Convolutional Neural Networks for an Automatic Classification of Prostate Tissue Slides with High-Grade Gleason Score. In Proceedings of the Medical Imaging 2017: Digital Pathology, Orlando, FL, USA, 5 April 2017. [Google Scholar] [CrossRef]
- Jung, M.; Jin, M.-S.; Kim, C.; Lee, C.; Nikas, I.P.; Park, J.H.; Ryu, H.S. Artificial Intelligence System Shows Performance at the Level of Uropathologists for the Detection and Grading of Prostate Cancer in Core Needle Biopsy: An Independent External Validation Study. Mod. Pathol. 2022, 35, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Kartasalo, K.; Chen, P.-H.C.; Ström, P.; Pinckaers, H.; Nagpal, K.; Cai, Y.; Steiner, D.F.; van Boven, H.; Vink, R.; et al. Artificial Intelligence for Diagnosis and Gleason Grading of Prostate Cancer: The PANDA Challenge. Nat. Med. 2022, 28, 154–163. [Google Scholar] [CrossRef]
- Marrón-Esquivel, J.M.; Duran-Lopez, L.; Linares-Barranco, A.; Dominguez-Morales, J.P. A Comparative Study of the Inter-Observer Variability on Gleason Grading against Deep Learning-Based Approaches for Prostate Cancer. Comput. Biol. Med. 2023, 159, 106856. [Google Scholar] [CrossRef] [PubMed]
- Hammouda, K.; Khalifa, F.; El-Melegy, M.; Ghazal, M.; Darwish, H.E.; Abou El-Ghar, M.; El-Baz, A. A Deep Learning Pipeline for Grade Groups Classification Using Digitized Prostate Biopsy Specimens. Sensors 2021, 21, 6708. [Google Scholar] [CrossRef]
- Marginean, F.; Arvidsson, I.; Simoulis, A.; Christian Overgaard, N.; Åström, K.; Heyden, A.; Bjartell, A.; Krzyzanowska, A. An Artificial Intelligence-Based Support Tool for Automation and Standardisation of Gleason Grading in Prostate Biopsies. Eur. Urol. Focus 2021, 7, 995–1001. [Google Scholar] [CrossRef]
- Huang, W.; Randhawa, R.; Jain, P.; Hubbard, S.; Eickhoff, J.; Kummar, S.; Wilding, G.; Basu, H.; Roy, R. A Novel Artificial Intelligence-Powered Method for Prediction of Early Recurrence of Prostate Cancer After Prostatectomy and Cancer Drivers. JCO Clin. Cancer Inform. 2022, 6, e2100131. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Arora, A.; Kumar, A.; Gupta, S.; Sethi, A.; Gann, P.H. Convolutional Neural Networks for Prostate Cancer Recurrence Prediction. In Proceedings of the Medical Imaging 2017: Digital Pathology, Orlando, FL, USA, 1 March 2017; Volume 10140, pp. 106–117. [Google Scholar]
- Pinckaers, H.; van Ipenburg, J.; Melamed, J.; De Marzo, A.; Platz, E.A.; van Ginneken, B.; van der Laak, J.; Litjens, G. Predicting Biochemical Recurrence of Prostate Cancer with Artificial Intelligence. Commun. Med. 2022, 2, 64. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Karagoz, K.; Gatza, M.L.; Singer, E.A.; Sadimin, E.; Foran, D.J.; Qi, X. Recurrence Analysis on Prostate Cancer Patients with Gleason Score 7 Using Integrated Histopathology Whole-Slide Images and Genomic Data through Deep Neural Networks. J. Med. Imaging 2018, 5, 047501. [Google Scholar] [CrossRef] [PubMed]
- Wessels, F.; Schmitt, M.; Krieghoff-Henning, E.; Jutzi, T.; Worst, T.S.; Waldbillig, F.; Neuberger, M.; Maron, R.C.; Steeg, M.; Gaiser, T.; et al. Deep Learning Approach to Predict Lymph Node Metastasis Directly from Primary Tumour Histology in Prostate Cancer. BJU Int. 2021, 128, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Feng, J.; van der Wal, D.; Huang, S.-C.; Simko, J.P.; DeVries, S.; Chen, E.; Schaeffer, E.M.; Morgan, T.M.; Sun, Y.; et al. Prostate Cancer Therapy Personalization via Multi-Modal Deep Learning on Randomized Phase III Clinical Trials. Npj Digit. Med. 2022, 5, 71. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, Y.; Weitz, P.; Lindberg, J.; Hartman, J.; Wang, W.; Egevad, L.; Grönberg, H.; Eklund, M.; Rantalainen, M. Using Deep Learning to Detect Patients at Risk for Prostate Cancer despite Benign Biopsies. iScience 2022, 25, 104663. [Google Scholar] [CrossRef]
- Leo, P.; Chandramouli, S.; Farré, X.; Elliott, R.; Janowczyk, A.; Bera, K.; Fu, P.; Janaki, N.; El-Fahmawi, A.; Shahait, M.; et al. Computationally Derived Cribriform Area Index from Prostate Cancer Hematoxylin and Eosin Images Is Associated with Biochemical Recurrence Following Radical Prostatectomy and Is Most Prognostic in Gleason Grade Group 2. Eur. Urol. Focus 2021, 7, 722–732. [Google Scholar] [CrossRef]
- Ren, J.; Singer, E.A.; Sadimin, E.; Foran, D.J.; Qi, X. Statistical Analysis of Survival Models Using Feature Quantification on Prostate Cancer Histopathological Images. J. Pathol. Inform. 2019, 10, 30. [Google Scholar] [CrossRef]
- Schmauch, B.; Romagnoni, A.; Pronier, E.; Saillard, C.; Maillé, P.; Calderaro, J.; Kamoun, A.; Sefta, M.; Toldo, S.; Zaslavskiy, M.; et al. A Deep Learning Model to Predict RNA-Seq Expression of Tumours from Whole Slide Images. Nat. Commun. 2020, 11, 3877. [Google Scholar] [CrossRef]
- Weitz, P.; Wang, Y.; Kartasalo, K.; Egevad, L.; Lindberg, J.; Grönberg, H.; Eklund, M.; Rantalainen, M. Transcriptome-Wide Prediction of Prostate Cancer Gene Expression from Histopathology Images Using Co-Expression-Based Convolutional Neural Networks. Bioinformatics 2022, 38, 3462–3469. [Google Scholar] [CrossRef]
- Chelebian, E.; Avenel, C.; Kartasalo, K.; Marklund, M.; Tanoglidi, A.; Mirtti, T.; Colling, R.; Erickson, A.; Lamb, A.D.; Lundeberg, J.; et al. Morphological Features Extracted by AI Associated with Spatial Transcriptomics in Prostate Cancer. Cancers 2021, 13, 4837. [Google Scholar] [CrossRef] [PubMed]
- Schaumberg, A.J.; Rubin, M.A.; Fuchs, T.J. H&E-Stained Whole Slide Image Deep Learning Predicts SPOP Mutation State in Prostate Cancer. bioRxiv 2018. [Google Scholar] [CrossRef]
- Dadhania, V.; Gonzalez, D.; Yousif, M.; Cheng, J.; Morgan, T.M.; Spratt, D.E.; Reichert, Z.R.; Mannan, R.; Wang, X.; Chinnaiyan, A.; et al. Leveraging Artificial Intelligence to Predict ERG Gene Fusion Status in Prostate Cancer. BMC Cancer 2022, 22, 494. [Google Scholar] [CrossRef] [PubMed]
- Vahadane, A.; Peng, T.; Sethi, A.; Albarqouni, S.; Wang, L.; Baust, M.; Steiger, K.; Schlitter, A.M.; Esposito, I.; Navab, N. Structure-Preserving Color Normalization and Sparse Stain Separation for Histological Images. IEEE Trans. Med. Imaging 2016, 35, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A Method for Normalizing Histology Slides for Quantitative Analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the Effects of Data Augmentation and Stain Color Normalization in Convolutional Neural Networks for Computational Pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed]
- Tellez, D.; Litjens, G.; van der Laak, J.; Ciompi, F. Neural Image Compression for Gigapixel Histopathology Image Analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 567–578. [Google Scholar] [CrossRef]
- McKay, F.; Williams, B.J.; Prestwich, G.; Bansal, D.; Hallowell, N.; Treanor, D. The Ethical Challenges of Artificial Intelligence-Driven Digital Pathology. J. Pathol. Clin. Res. 2022, 8, 209–216. [Google Scholar] [CrossRef]
- Thompson, N.; Greenewald, K.; Lee, K.; Manso, G.F. The Computational Limits of Deep Learning. In Proceedings of the Ninth Computing within Limits 2023, Virtual, 14 June 2023. [Google Scholar]
| Gleason ISUP Group | Gleason Score |
|---|---|
| 1 | Score = 6 |
| 2 | Score 7 (3 + 4) |
| 3 | Score 7 (4 + 3) |
| 4 | Score 8 (4 + 4, 5 + 3, 3 + 5) |
| 5 | Score 9 or 10 |
| Quadas-2 Category | Adaptation | Criteria |
|---|---|---|
| Patient selection | Data source | Existence of exclusion criteria |
| Data publicly available | ||
| Reference to demographics and stratification | ||
| Sufficient amount of training data * | ||
| Data process | Normalization process properly described | |
| Strategy for data imbalance | ||
| Index Test | Model | Description of model task |
| Description of model output | ||
| Description of architecture | ||
| Definition of hyperparameter values | ||
| Code publicly available | ||
| Reference standard | Ground truth | Ground truth defined in accordance with good clinical practice |
| Ground truth defined by multiple experts | ||
| Flow and timing | Analysis | Description of splitting methods |
| Definition of a metric appropriate to the analysis | ||
| Relevance of algorithms with state of the art | ||
| Use of external testing cohort | ||
| Explainability of the model |
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Schömig-Markiefka, 2021 [16] | InceptionResNetV2 | Already trained | Subset of TCGA slides | 686 WSIs | Impact of artifacts on tissue classification | Tissue classification performance decreases with the appearance of artifacts |
| Haghighat, 2022 [17] | ResNet18 PathProfiler | 198 WSIs | 3819 WSIs | None | Prediction of tile-level usability | AUC: 0.94 (IV) |
| Prediction of slide-level usability | AUC: 0.987 (IV) PC: 0.889 (IV) | |||||
| Brendel, 2022 [18] | ResNet34-IBN | 4984 WSIs TCGA (6 cancer types) | 866 WSIs TCGA (6 cancer types) | 78 WSIs | Cancer and tissue type prediction | F1: 1 (IV) F1: 0.83 (EV) |
| Tumor purity above 70% | AUC: 0.8 (IV) | |||||
| Anghel, 2019 [19] | VGG-like network | 96 WSIs | 14 WSIs | None | Improvement of cancer detection when using poor-quality WSIs | F1 (base): 0.79 F1 (best): 0.87 |
| Otàlora, 2019 [20] | MobileNet [21] + 2 layers | 3540 TMAs | 1321 TMAs | 3961 TMAs | Impact of normalization on GP classification | AUC (base): 0.56 (IV) & 0.48 (EV) AUC (best): 0.84 (IV) & 0.69 (EV) |
| Rana, 2020 [22] | GANs | 148 WSIs | 13 WSIs | None | Comparison of dye and computationally stained images | : 0.96 Structural similarity index: 0.9 |
| Comparison of unstained and computationally destained images | : 0.96 Structural similarity index: 0.9 | |||||
| Sethi, 2016 [23] | Custom CNN | 20 WSIs | 10 WSIs | None | Epithelial-stromal segmentation | AUC: 0.96 (Vahadane) AUC: 0.95 (Khan) |
| Swiderska-Chadaj, 2020 [24] | U-Net GANs | 324 WSIs | 258 WSIs | 85 WSIs | Impact of normalization on cancer detection | AUC: 0.92 AUC: 0.98 (GAN) |
| 50 WSIs | AUC: 0.83 AUC: 0.97 (GAN) | |||||
| Salvi, 2021 [25] | Inceptionv3 pre-trained on ImageNet | 400 WSIs | 100 WSIs | None | WSI-level classification algorithm performance adding normalization and tile selection | Sens (base): 0.94 Spec (base): 0.68 Sens (best): 1 Spec (best): 0.98 |
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Ren, 2017 [26] | U-Net | 22 WSIs | 5-fold cross val | None | Gland Segmentation in H&E slides | F1: 0.84 |
| Li, 2019 [29] | R-CNN | 40 patients | 5-fold cross val | None | Epithelial cell detection | AUC: 0.998 |
| Bulten, 2019 [28] | U-Net | 20 WSIs | 5 WSIs | None | Epithelium segmentation based on IHC | IoU: 0.854 F1: 0.915 |
| Bukowy, 2020 [30] | SegNet (VGG16) | 10 WSIs | 6 WSIs | None | Weakly- and strongly-annotated segmentation | : 0.85 (strong) : 0.93 (weak fine-tuned strong) |
| 140 WSIs | Epithelium segmentation with a combination of 3 models | : 0.86 (DL) | ||||
| Salvi, 2021 [27] | U-Net + post-processing | 100 patients | 50 patients | None | Gland segmentation | Dice: 0.901 U-Net + post-processing Dice: 0.892 U-Net only |
| First Author, Year Reference | DL Architecture | Training Cohort | IV Cohort | EV Cohort | Aim | Results |
|---|---|---|---|---|---|---|
| Litjens, 2016 [43] | Custom CNN | 150 patients | 75 patients | None | Cancer detection at the pixel level | AUC: 0.99 |
| Kwak, 2017 [44] | Custom CNN | 162 TMAs | 491 TMAs | None | Cancer detection at the sample level | AUC: 0.974 |
| Kwak, 2017 [45] | Custom CNN | 162 TMAs | 185 TMAs | None | Cancer detection at the sample level | AUC: 0.95 |
| Campanella, 2018 [46] | ResNet34 and VGG11-BN (MIL) | 12610 WSIs | 1824 WSIs | None | Cancer detection at the WSI level | AUC: 0.98 |
| Campanella, 2019 [32] | RNN (MIL) | 12 132 WSIs | 1784 WSIs | 12 727 WSIs | Cancer detection at the WSI level | AUC: 0.991 (IV) & 0.932 (EV) |
| ResNet34 (MIL) | AUC: 0.986 (IV) | |||||
| Garcià, 2019 [55] | VGG19 | 6195 glands (from 35 WSIs) | 5-fold cross val | None | Malignancy gland classification | AUC: 0.889 |
| Singh, 2019 [50] | ResNet22 | 749 WSIs | 3-fold cross val | None | Cribriform pattern detection at the tile level | ACC: 0.88 |
| Jones, 2019 [56] | ResNet50 & SqueezeNet | 1000 tiles from 10 WSIs | 200 tiles from 10 WSIs | 70 tiles from unknown number of WSIs | Malignancy detection at the tile level | : 0.96 (IV) & 0.78 (EV) |
| Duong, 2019 [31] | ResNet50 and multiscale embedding | 602 TMAs | 303 TMAs | None | TMA classification using only ×10 magnification | AUC: 0.961 |
| TMA classification with multi-scale embedding | AUC: 0.971 | |||||
| Raciti, 2020 [34] | PaigeProstate | Pre-trained [32] | Pre-trained [32] | 232 biopsies | Malignancy classification at the WSI level | Sens: 0.96 Spec: 0.98 |
| Improvement of pathologist’s classification | : 0.738 to 0.900 : 0.966 to 0.952 | |||||
| Han, 2020 [38] | AlexNet | 286 WSIs from 68 patients | Leave one out cross val | None | Cancer classification at the WSI level | AUC: 0.98 |
| Ambrosini, 2020 [51] | Custom CNN | 128 WSIs | 8-fold cross val | None | Cribriform pattern detection for biopsies | : 0.8 |
| Bukhari, 2021 [57] | ResNet50 | 640 WSIs | 162 WSIs | None | Cancer/hyperplasia detection at the tile level | F1: 1 ACC: 0.995 |
| Pinckaers, 2021 [33] | ResNet-34 (MIL) | 5209 biopsies | 535 biopsies | 205 biopsies | Cancer detection for biopsies | AUC: 0.99 (IV) & 0.799 (EV) |
| Streaming CNN | AUC: 0.992 (IV) & 0.902 (EV) | |||||
| Perincheri, 2021 [49] | Paige Prostate | Pre-trained [34] | Pre-trained [34] | 1876 biopsies from 116 patients | Paige classification evaluation | 110/118 patients were correctly classified |
| Da Silva, 2021 [48] | PaigeProstate | Pre-trained [34] | Pre-trained [34] | 600 biopsies from 100 patients | Malignancy classification for biopsies | Sens: 0.99 Spec: 0.93 |
| Malignancy classification for patients | Sens: 1 Spec: 0.78 | |||||
| Raciti, 2022 [47] | PaigeProstate | Pre-trained [34] | Pre-trained [34] | 610 biopsies | Cancer detection for patients | Sens: 0.974 Spec: 0.948 AUC: 0.99 |
| Krajnansky, 2022 [58] | VGG16-mode | 156 biopsies from 262 WSIs | Ten biopsies from 87 WSIs | None | Malignancy detection for biopsies | FROC: 0.944 |
| Malignancy detection for patients | AUC: 1 | |||||
| Tsuneki, 2022 [53] | EfficientNetB1 pre-trained on colon | 1182 needle biopsies | 1244 TURP biopsies 500 needle biopsies | 767 WSIs | Cancer detection for classic and TURP biopsies | AUC: 0.967 (IV) & 0.987 (EV) AUC (TURP): 0.845 |
| EfficientNetB1 pre-trained on ImageNet | AUC: 0.971 (IV) & 0.945 (EV) AUC (TURP): 0.803 | |||||
| Tsuneki, 2022 [54] | EfficientNetB1 pre-trained on ImageNet | 1060 TURP biopsies | 500 needle biopsies, 500 TURP | 768 WSIs | Cancer detection in classic and TURP biopsies | AUC: 0.885 (IV) TURP AUC: 0.779 (IV) & 0.639 (EV) biopsies |
| EfficientNetB1 pre-trained on colon | AUC: 0.947 (IV) TURP AUC: 0.913 (IV) & 0.947 (EV) biopsies | |||||
| Chen, 2022 [59] | DenseNet | 29 WSIs | 3 WSIs | None | Classification of tissue malignancy | AUC: 0.98 (proposed method) AUC: 0.90 (DenseNet-121) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabilloud, N.; Allaume, P.; Acosta, O.; De Crevoisier, R.; Bourgade, R.; Loussouarn, D.; Rioux-Leclercq, N.; Khene, Z.-e.; Mathieu, R.; Bensalah, K.; et al. Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review. Diagnostics 2023, 13, 2676. https://doi.org/10.3390/diagnostics13162676
Rabilloud N, Allaume P, Acosta O, De Crevoisier R, Bourgade R, Loussouarn D, Rioux-Leclercq N, Khene Z-e, Mathieu R, Bensalah K, et al. Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review. Diagnostics. 2023; 13(16):2676. https://doi.org/10.3390/diagnostics13162676
Chicago/Turabian StyleRabilloud, Noémie, Pierre Allaume, Oscar Acosta, Renaud De Crevoisier, Raphael Bourgade, Delphine Loussouarn, Nathalie Rioux-Leclercq, Zine-eddine Khene, Romain Mathieu, Karim Bensalah, and et al. 2023. "Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review" Diagnostics 13, no. 16: 2676. https://doi.org/10.3390/diagnostics13162676
APA StyleRabilloud, N., Allaume, P., Acosta, O., De Crevoisier, R., Bourgade, R., Loussouarn, D., Rioux-Leclercq, N., Khene, Z.-e., Mathieu, R., Bensalah, K., Pecot, T., & Kammerer-Jacquet, S.-F. (2023). Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review. Diagnostics, 13(16), 2676. https://doi.org/10.3390/diagnostics13162676







