Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal
Abstract
:1. Introduction
2. Materials and Methods
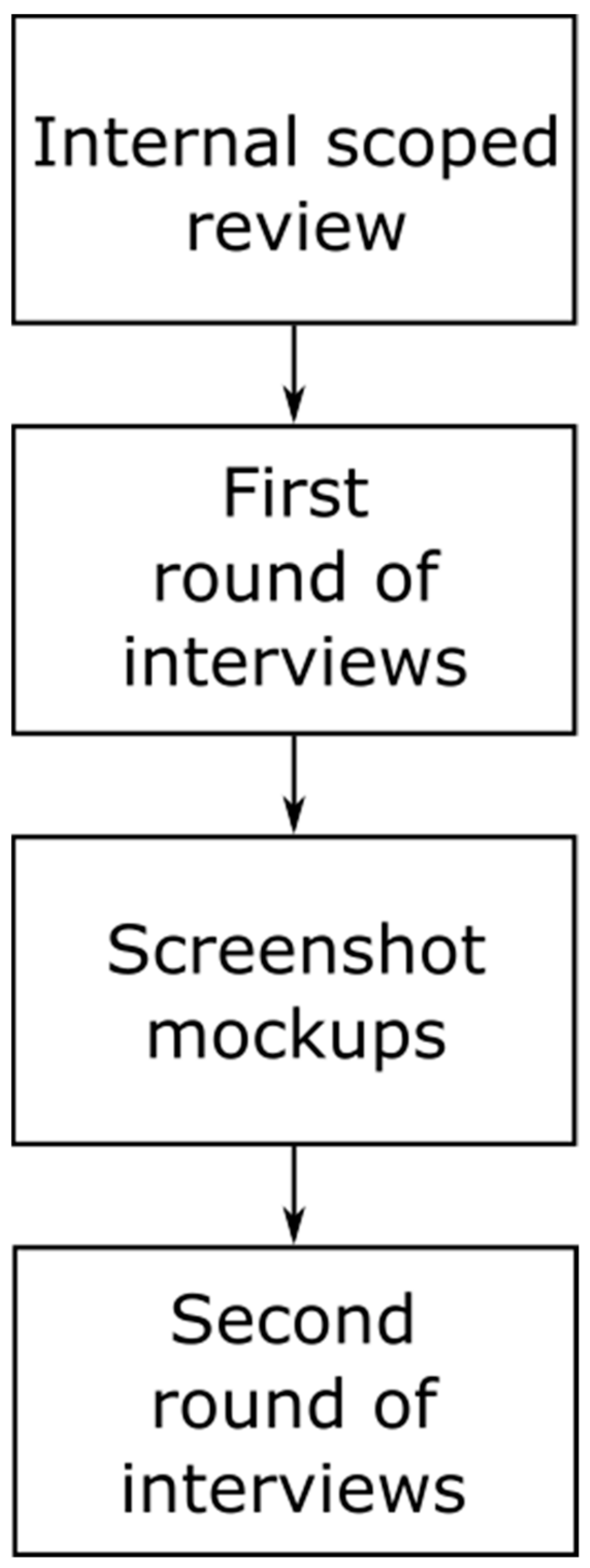
2.1. Details about Scoping Review
2.2. First and Second Round of Interviews
2.2.1. Structure and Purpose of the First Round of Interviews
2.2.2. Structure and Purpose of the Second Round of Interviews
- Localization and time of the sampling;
- Type of sampling (e.g., fine-needle aspiration biopsy);
- Distinction between fresh-frozen and formalin-fixed paraffin-embedded samples;
- Scope of sequencing (e.g., gene panel or whole-exome sequencing);
- Name and version of both the used panel and kit;
- Hyperlink to the corresponding product-specific website of the manufacturer.
2.3. Low-Fidelity Mockup Demonstrator
2.4. Consultation with Main Developers of MSKCC
2.5. Ethical Approval
3. Results
3.1. Overview of Scoped Review
3.2. Details about Interviewees
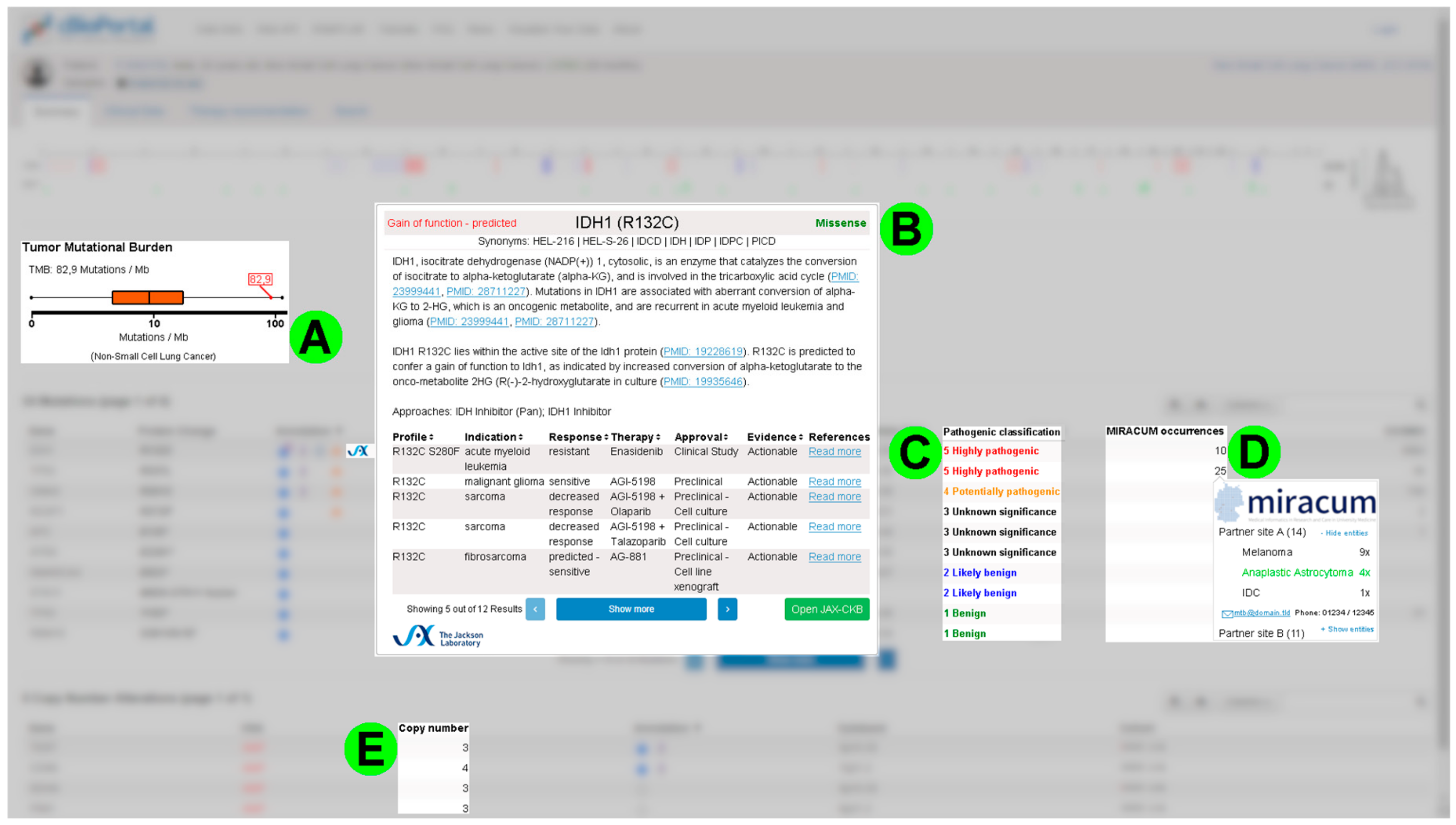
3.3. Requirements from the First Round of Interviews and Screenshot Mockups
- Highlight mutations with existing treatment options;
- Display information about general availability of a specific drug in Germany;
- Point out mutations causing treatment resistance;
- Mark germline mutations;
- Display variant allele frequencies alongside corresponding coverage;
- Integrate the database “Clinical Interpretations of Variants in Cancer” (CIViC) [19];
- Visualize mRNA expression data.
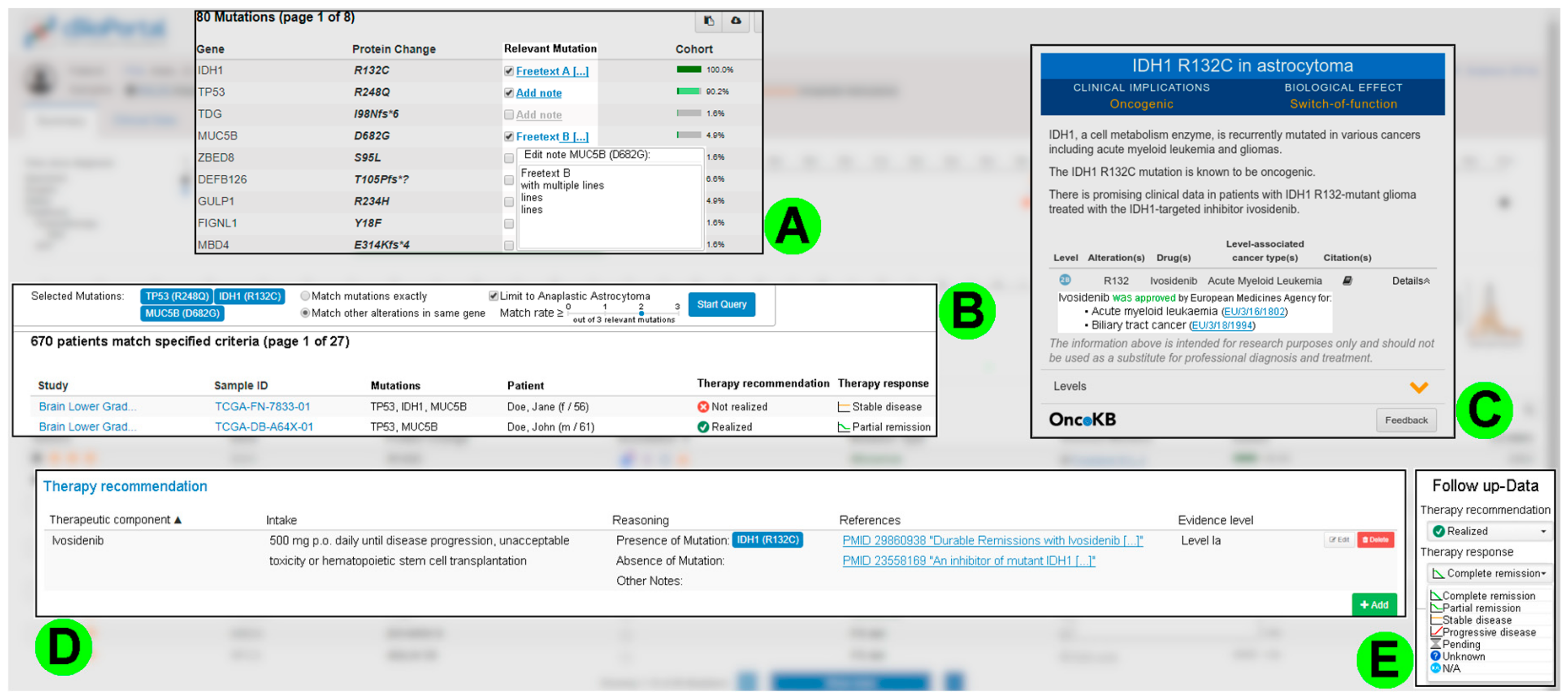
3.4. Consolidated Requirements from the Second Round of Interviews
3.4.1. Improving Patient Case Analysis
3.4.2. Supporting the Development and Recording of a Therapy Recommendation
3.4.3. Requirements for IT Infrastructure
4. Discussion
4.1. Results and Future Work
4.2. Related Work
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application programming interface |
| cBioPortal | cBio Cancer Genomics Portal |
| CIViC | Clinical Interpretation of Variants in Cancer (knowledgebase) |
| CNV | Copy Number Variation |
| EHR | Electronic Health Record |
| EMA | European Medicines Agency |
| FHIR | Fast Healthcare Interoperability Resources |
| GIMP | GNU Image Manipulation Program |
| gnomAD | Genome Aggregation Database |
| HIS | Hospital Information System |
| IT | Information technology |
| JAX-CKB | Jackson Laboratory Clinical Knowledgebase |
| JSON | JavaScript Object Notation |
| MIRACUM | Medical Informatics in Research and Care in University Medicine |
| MSKCC | Memorial Sloan Kettering Cancer Center |
| MTB | Molecular Tumor Board |
| NGS | Next-generation sequencing |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analysis |
| RFC | Request for Comments |
| TMB | Tumor Mutational Burden |
References
- Parker, B.A.; Schwaederlé, M.; Scur, M.D.; Boles, S.G.; Helsten, T.; Subramanian, R.; Schwab, R.B.; Kurzrock, R. Breast Cancer Experience of the Molecular Tumor Board at the University of California, San Diego Moores Cancer Center. J. Oncol. Pract. 2015, 11, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Bryce, A.H.; Egan, J.B.; Borad, M.J.; Stewart, A.K.; Nowakowski, G.S.; Chanan-Khan, A.; Patnaik, M.M.; Ansell, S.M.; Banck, M.S.; Robinson, S.I.; et al. Experience with precision genomics and tumor board, indicates frequent target identification, but barriers to delivery. Oncotarget 2017, 8, 27145–27154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoefflin, R.; Geißler, A.-L.; Fritsch, R.; Claus, R.; Wehrle, J.; Metzger, P.; Reiser, M.; Mehmed, L.; Fauth, L.; Heiland, D.H.; et al. Personalized Clinical Decision Making Through Implementation of a Molecular Tumor Board: A German Single-Center Experience. JCO Precis. Oncol. 2018, 1–16. [Google Scholar] [CrossRef]
- Perera-Bel, J.; Hutter, B.; Heining, C.; Bleckmann, A.; Fröhlich, M.; Fröhling, S.; Glimm, H.; Brors, B.; Beißbarth, T. From somatic variants towards precision oncology: Evidence-driven reporting of treatment options in molecular tumor boards. Genome Med. 2018, 10, 18:1–18:15. [Google Scholar] [CrossRef] [PubMed]
- Hinderer, M.; Boerries, M.; Haller, F.; Wagner, S.; Sollfrank, S.; Acker, T.; Prokosch, H.-U.; Christoph, J. Supporting Molecular Tumor Boards in Molecular-Guided Decision-Making - The Current Status of Five German University Hospitals. Stud. Health Technol. Inform. 2017, 236, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Prokosch, H.-U.; Acker, T.; Bernarding, J.; Binder, H.; Boeker, M.; Boerries, M.; Daumke, P.; Ganslandt, T.; Hesser, J.; Höning, G.; et al. MIRACUM: Medical Informatics in Research and Care in University Medicine. Methods Inf. Med. 2018, 57, e82–e91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzger, P.; Scheible, R.; Hess, M.; Boeker, M.; Andrieux, G.; Börries, P. MIRACUM-Pipe (AG-Boerries/MIRACUM-Pipe Repository). Available online: https://github.com/AG-Boerries/MIRACUM-Pipe (accessed on 11 August 2019).
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Shneiderman, B. Inventing Discovery Tools: Combining Information Visualization with Data Mining. In Discovery science, Proceedings of the 4th International Conference, DS 2001, Washington, DC, USA, 25–28 November 2001; Jantke, K.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 17–28. ISBN 978-3-540-42956-2. [Google Scholar]
- Omta, W.A.; de Nobel, J.; Klumperman, J.; Egan, D.A.; Spruit, M.R.; Brinkhuis, M.J.S. Improving Comprehension Efficiency of High Content Screening Data Through Interactive Visualizations. Assay Drug Dev. Technol. 2017, 15, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halfmann, M.; Stenzhorn, H.; Gerjets, P.; Kohlbacher, O.; Oestermeier, U. User-Driven Development of a Novel Molecular Tumor Board Support Tool. In Data Integration in the Life Sciences; Auer, S., Vidal, M.-E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; ISBN 978-3-030-06015-2. [Google Scholar]
- Fegeler, C.; Zsebedits, D.; Bochum, S.; Finkeisen, D.; Martens, U.M. Implementierung eines IT-gestützten molekularen Tumorboards in der Regelversorgung. FORUM 2018, 5, 322–328. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009, 6, e1000097:1–e1000097:6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009, 6, e1000100:1–e1000100:28. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- SOPHiA GENETICS. Available online: https://www.sophiagenetics.com/ (accessed on 29 May 2019).
- Dubovenko, A.; Nikolsky, Y.; Rakhmatulin, E.; Nikolskaya, T. Functional Analysis of OMICs Data and Small Molecule Compounds in an Integrated "Knowledge-Based" Platform. Methods Mol. Biol. 2017, 1613, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Spies, N.C.; Krysiak, K.; McMichael, J.F.; Coffman, A.C.; Danos, A.M.; Ainscough, B.J.; Ramirez, C.A.; Rieke, D.T.; Kujan, L.; et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 2017, 49, 170–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Jackson Laboratory. JAX Clinical Knowledgebase - Disclaimer. Available online: https://ckb.jax.org/about/disclaimer (accessed on 18 August 2019).
- Patterson, S.E.; Liu, R.; Statz, C.M.; Durkin, D.; Lakshminarayana, A.; Mockus, S.M. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum. Genomics 2016, 10, 4:1–4:13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. National Library of Medicine. ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 10 August 2019).
- HL7. Welcome to FHIR. Available online: https://www.hl7.org/fhir/ (accessed on 30 August 2019).
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- GitHub User Leexgh. Pull Request: Add Gnomad Column #2064. Available online: https://github.com/cBioPortal/cbioportal-frontend/pull/2064 (accessed on 15 August 2019).
- GitHub User Leexgh. Pull Request: Add dbsnp Column to Mutation Table #2502. Available online: https://github.com/cBioPortal/cbioportal-frontend/pull/2502 (accessed on 15 August 2019).
- GitHub User Leexgh. Pull Request: Add Clinvar to Patient Page #2596. Available online: https://github.com/cBioPortal/cbioportal-frontend/pull/2596 (accessed on 15 August 2019).
- GitHub User Jjgao. Issue: Adding Clinical Trials Matching in Patient View #6444. Available online: https://github.com/cBioPortal/cbioportal/issues/6444 (accessed on 15 August 2019).
- GitHub User Pvannierop. Pull Request: Integration of treatment response data in OncoPrint tab #2053. Available online: https://github.com/cBioPortal/cbioportal-frontend/pull/2053 (accessed on 15 August 2019).
- GitHub User Pvannierop. Pull Request: Integration of Treatment Response Data in PlotsTab (Incl. New Waterfall Plot) #2055. Available online: https://github.com/cBioPortal/cbioportal-frontend/pull/2055 (accessed on 15 August 2019).
- GitHub User Kjgao. Issue: PDF of Patient View Page #6446. Available online: https://github.com/cBioPortal/cbioportal/issues/6446 (accessed on 15 August 2019).
- Lukasse, P.; van Hagen, S. RFC45: Gene Panel Information in Patient View. Available online: https://docs.google.com/document/d/1X7dA_wJFtv5xJO1oHCSt8DUdTmk07RexvHUpjCJsSM4/edit (accessed on 15 August 2019).
- Lindsay, J.; Fitz, C.D.V.; Zwiesler, Z.; Kumari, P.; van der Veen, B.; Monrose, T.; Mazor, T.; Barry, S.; Albayrak, A.; Tung, M.; et al. MatchMiner: An Open Source Computational Platform for Real-Time Matching of Cancer Patients to Precision Medicine Clinical Trials Using Genomic and Clinical Criteria. Available online: https://www.biorxiv.org/content/10.1101/199489v3 (accessed on 20 January 2020).
- GitHub User Victoria34. Pull Request: Matchminer Proxy Controller #5679. Available online: https://github.com/cBioPortal/cbioportal/pull/5679 (accessed on 21 January 2019).
- MEONA GmbH. Meona. Available online: https://www.meona.de/ (accessed on 20 January 2020).
- Unberath, P. RFC50: Add Support for Additional Arbitrary Variant Annotation. Available online: https://docs.google.com/document/d/1Pybk4_-lrirKJZ_cH64riZBRdWXdkJnCQqzx1O2fjRo/edit#heading=h.oyj1ec8k7lgx (accessed on 18 August 2019).
- German Federal Ministry of Health. Updated Standardized Oncological Basic Data Set of the Consortium of German Tumor Centers e.V. (ADT) and the Society of Epidemiological Cancer Registries in Germany e.V. (GEKID) (“Aktualisierter Einheitlicher Onkologischer Basisdatensatz der Arbeitsgemeinschaft Deutscher Tumorzentren e.V. (ADT) und der Gesellschaft der Epidemiologischen Krebsregister in Deutschland e.V. (GEKID)”). Available online: https://www.bundesanzeiger.de/ (accessed on 9 March 2019).
- OncoTree. Available online: https://github.com/cBioPortal/oncotree (accessed on 19 March 2019).
- German Federal Ministry of Justice and Consumer Protection. German Federal Data Protection Act (“Bundesdatenschutzgesetz / BDSG”). Available online: https://www.gesetze-im-internet.de/englisch_bdsg/englisch_bdsg.pdf (accessed on 4 April 2019).
- REGULATION (EU) 2016/679 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL - of 27 April 2016 - On the Protection of Natural Persons with Regard to the Processing of Personal Data and On the Free Movement of Such Data, and Repealing Directive 95/46/EC (General Data Protection Regulation); GDPR, 2016; European Parliament: Brussels, Belgium, 2016.
- German Federal Ministry of Justice and Consumer Protection. German Medical Devices Act (“Gesetz über Medizinprodukte - MPG”). Available online: https://www.gesetze-im-internet.de/mpg/ (accessed on 18 August 2019).
- YouTube-User PersOnS. Interface Prototype Demo—YouTube. Available online: https://www.youtube.com/watch?v=VXD3Rap11zg (accessed on 19 August 2019).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buechner, P.; Hinderer, M.; Unberath, P.; Metzger, P.; Boeker, M.; Acker, T.; Haller, F.; Mack, E.; Nowak, D.; Paret, C.; et al. Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal. Diagnostics 2020, 10, 93. https://doi.org/10.3390/diagnostics10020093
Buechner P, Hinderer M, Unberath P, Metzger P, Boeker M, Acker T, Haller F, Mack E, Nowak D, Paret C, et al. Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal. Diagnostics. 2020; 10(2):93. https://doi.org/10.3390/diagnostics10020093
Chicago/Turabian StyleBuechner, Philipp, Marc Hinderer, Philipp Unberath, Patrick Metzger, Martin Boeker, Till Acker, Florian Haller, Elisabeth Mack, Daniel Nowak, Claudia Paret, and et al. 2020. "Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal" Diagnostics 10, no. 2: 93. https://doi.org/10.3390/diagnostics10020093
APA StyleBuechner, P., Hinderer, M., Unberath, P., Metzger, P., Boeker, M., Acker, T., Haller, F., Mack, E., Nowak, D., Paret, C., Schanze, D., von Bubnoff, N., Wagner, S., Busch, H., Boerries, M., & Christoph, J. (2020). Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal. Diagnostics, 10(2), 93. https://doi.org/10.3390/diagnostics10020093





