Gut Microbiota and Neuroplasticity
Abstract
:1. Introduction
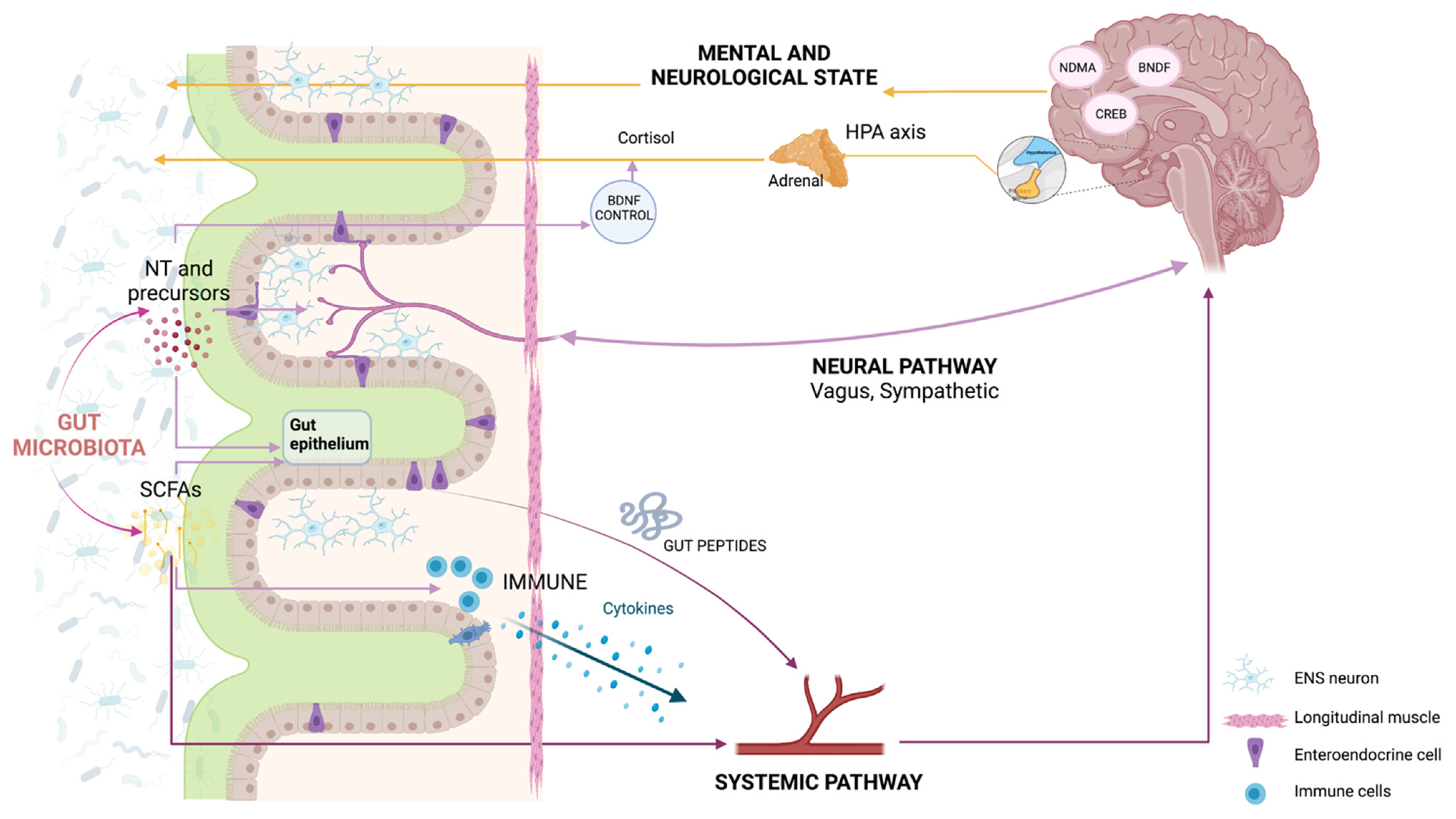
2. How Do the Microbiota and the Body Talk to Each Other?
3. Do the Gut Microbiota and Brain Talk to Each Other?
4. How Are Gut and Brain Diseases Related to Each Other?
5. How Does the Brain React to the Environment?
6. Can We Meet Our Own Microbiota?
7. Can We Treat Our Own Microbiota to Promote Neuroplasticity?
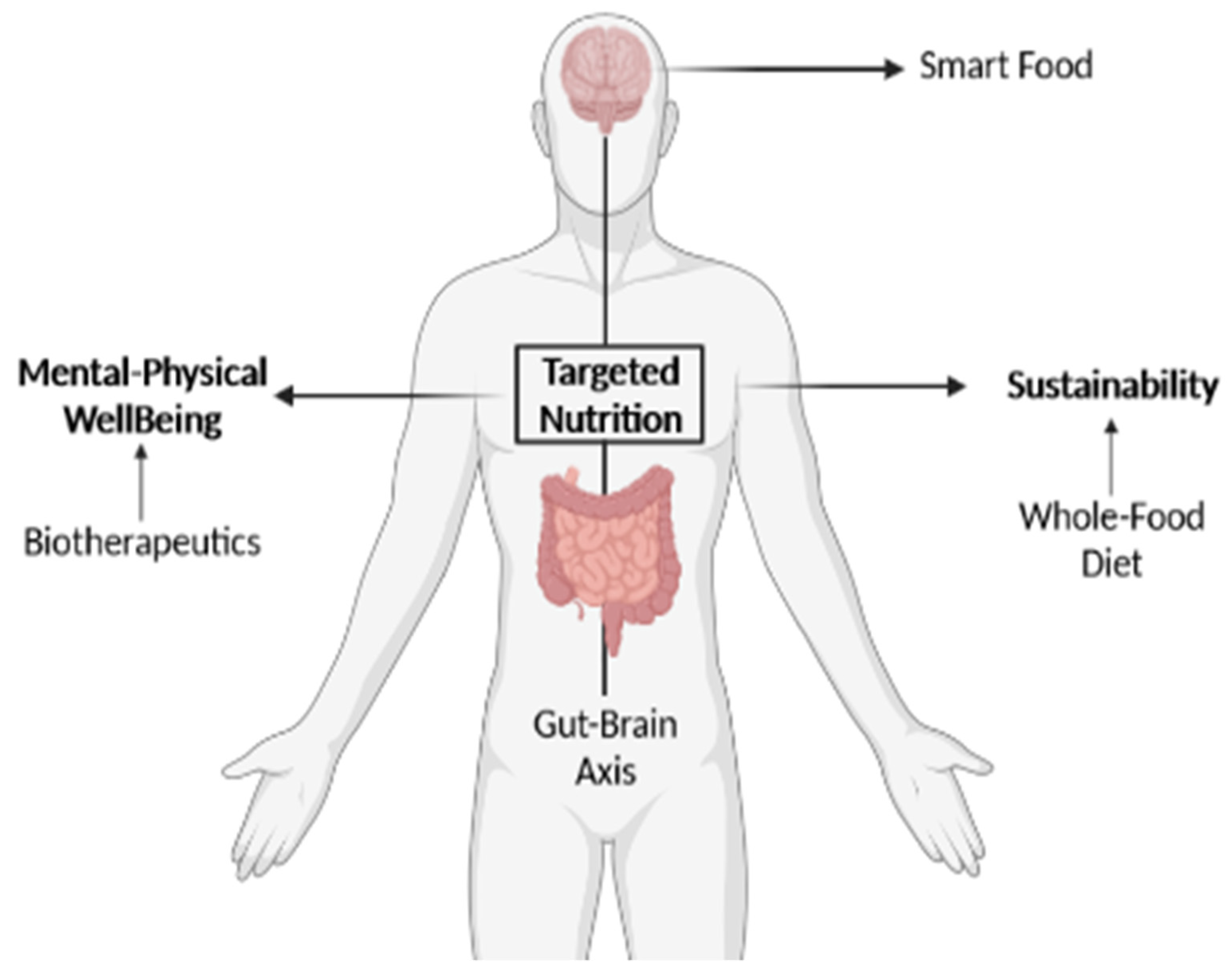
8. Can Targeted Nutrition Harness the Gut Microbiota and Promote Neuroplasticity?
9. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut microbes and the brain: Paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.C. Psychological co-morbidity in functional gastrointestinal disorders: Epidemiology, mechanisms and management. J. Neurogastroenterol. Motil. 2012, 18, 13–18. [Google Scholar] [CrossRef]
- Hugon, P.; Lagier, J.C.; Colson, P.; Bittar, F.; Raoult, D. Repertoire of human gut microbes. Microb. Pathog. 2017, 106, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, J.H.; Round, J.L. Snapshot: Microbiota effects on host physiology. Cell 2021, 184, 2796.e2791. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the teddy study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naveed, M.; Zhou, Q.-G.; Xu, C.; Taleb, A.; Meng, F.; Ahmed, B.; Zhang, Y.; Fukunaga, K.; Han, F. Gut-brain axis: A matter of concern in neuropsychiatric disorders…! Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 104, 110051. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Gambino, R.; Cassader, M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 2011, 62, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.; White, H.; Støy, S.; Bajaj, J.; Shawcross, D. Clinical science workshop: Targeting the gut-liver-brain axis. Metab. Brain Dis. 2015, 31, 1327–1337. [Google Scholar] [CrossRef] [Green Version]
- Fabbiano, S.; Suárez-Zamorano, N.; Chevalier, C.; Lazarević, V.; Kieser, S.; Rigo, D.; Leo, S.; Veyrat-Durebex, C.; Gaïa, N.; Maresca, M.; et al. Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab. 2018, 28, 907–921.e907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, P.; Blacher, E.; Elinav, E.; Pettersson, S. Our gut microbiome: The evolving inner self. Cell 2017, 171, 1481–1493. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Belkind-Gerson, J.; Graeme-Cook, F.; Winter, H. Enteric nervous system disease and recovery, plasticity, and regeneration. J. Pediatric Gastroenterol. Nutr. 2006, 42, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.J.; Balart, L.A.; Johnson, D.A. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin. Transl. Gastroenterol. 2015, 6, e91. [Google Scholar] [CrossRef] [PubMed]
- Anitha, M.; Vijay-Kumar, M.; Sitaraman, S.V.; Gewirtz, A.T.; Srinivasan, S. Gut microbial products regulate murine gastrointestinal motility via toll-like receptor 4 signaling. Gastroenterology 2012, 143, 1006–1016.e1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, P.A.; Matheis, F.; Schneeberger, M.; Kerner, Z.; Jové, V.; Mucida, D. Microbiota-modulated cart(+) enteric neurons autonomously regulate blood glucose. Science 2020, 370, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Rolig, A.S.; Mittge, E.K.; Ganz, J.; Troll, J.V.; Melancon, E.; Wiles, T.J.; Alligood, K.; Stephens, W.Z.; Eisen, J.S.; Guillemin, K. The enteric nervous system promotes intestinal health by constraining microbiota composition. PLoS Biol. 2017, 15, e2000689. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.D.; Wang, X.Y.; Parsons, S.P.; Khan, W.I.; Huizinga, J.D. Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G896–G907. [Google Scholar] [CrossRef]
- Ge, X.; Ding, C.; Zhao, W.; Xu, L.; Tian, H.; Gong, J.; Zhu, M.; Li, J.; Li, N. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J. Transl. Med. 2017, 15, 13. [Google Scholar] [CrossRef] [Green Version]
- Kabouridis, P.S.; Lasrado, R.; McCallum, S.; Chng, S.H.; Snippert, H.J.; Clevers, H.; Pettersson, S.; Pachnis, V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 2015, 85, 289–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, Y.; Castaño, Á.; Boeing, S.; Bon-Frauches, A.C.; Fung, C.; Fallesen, T.; de Agüero, M.G.; Yilmaz, B.; Lopes, R.; Huseynova, A.; et al. Neuronal programming by microbiota regulates intestinal physiology. Nature 2020, 578, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.M.; Bon-Frauches, A.C.; Keszthelyi, D.; Melotte, V.; Boesmans, W. The enteric nervous system in gastrointestinal disease etiology. Cell. Mol. Life Sci. CMLS 2021, 78, 4713–4733. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A. Gut feelings: The emerging biology of gut–brain communication. Nat. Rev. Neurosci. 2011, 12, 453–466. [Google Scholar] [CrossRef]
- Wiley, N.C.; Cryan, J.F.; Dinan, T.G.; Ross, R.P.; Stanton, C. Production of psychoactive metabolites by gut bacteria. Mod. Trends Psychiatry 2021, 32, 74–99. [Google Scholar] [PubMed]
- Chu, C.; Murdock, M.H.; Jing, D.; Won, T.H.; Chung, H.; Kressel, A.M.; Tsaava, T.; Addorisio, M.E.; Putzel, G.G.; Zhou, L.; et al. The microbiota regulate neuronal function and fear extinction learning. Nature 2019, 574, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Ogbonnaya, E.S.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; O’Leary, O.F. Adult hippocampal neurogenesis is regulated by the microbiome. Biol. Psychiatry 2015, 78, e7–e9. [Google Scholar] [CrossRef] [PubMed]
- Gars, A.; Ronczkowski, N.M.; Chassaing, B.; Castillo-Ruiz, A.; Forger, N.G. First encounters: Effects of the microbiota on neonatal brain development. Front. Cell. Neurosci. 2021, 15, 682505. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The microbiome and host behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Rincon, C.; Paré, J.-F.; Martyniuk, C.J.; Jannetty, S.K.; Harrison, C.; Fischer, A.; Dinis, A.; Keshari, V.; Novak, R.; Levin, M. An in vivo brain–bacteria interface: The developing brain as a key regulator of innate immunity. NPJ Regen. Med. 2020, 5, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, P.; Keijzer, F.; Arendt, D.; Levin, M. Reframing cognition: Getting down to biological basics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190750. [Google Scholar] [CrossRef]
- Sarkar, A.; Lehto, S.M.; Harty, S.; Dinan, T.G.; Cryan, J.F.; Burnet, P.W. Psychobiotics and the manipulation of bacteria-gut-brain signals. Trends Neurosci. 2016, 39, 763–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Lomasney, K.W.; Houston, A.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; Hyland, N.P. Selective influence of host microbiota on camp-mediated ion transport in mouse colon. Neurogastroenterol. Motil. 2014, 26, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Kunze, W.A.; Mao, Y.K.; Wang, B.; Huizinga, J.D.; Ma, X.; Forsythe, P.; Bienenstock, J. Lactobacillus reuteri enhances excitability of colonic ah neurons by inhibiting calcium-dependent potassium channel opening. J. Cell Mol. Med. 2009, 13, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jacob, E. Learning from bacteria about natural information processing. Ann. N. Y. Acad. Sci. 2009, 1178, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jacob, E.; Levine, H. Self-engineering capabilities of bacteria. J. R. Soc. Interface 2006, 3, 197–214. [Google Scholar] [CrossRef] [Green Version]
- Busch, W.; Benfey, P.N. Information processing without brains—the power of intercellular regulators in plants. Development 2010, 137, 1215–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Suel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Ben Jacob, E.; Becker, I.; Shapira, Y.; Levine, H. Bacterial linguistic communication and social intelligence. Trends Microbiol. 2004, 12, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Beagle, S.D.; Lockless, S.W. Microbiology: Electrical signalling goes bacterial. Nature 2015, 527, 44–45. [Google Scholar] [CrossRef] [PubMed]
- Lyon, P. The cognitive cell: Bacterial behavior reconsidered. Front. Microbiol. 2015, 6, 264. [Google Scholar] [CrossRef] [PubMed]
- Westerhoff, H.V.; Brooks, A.N.; Simeonidis, E.; Garcia-Contreras, R.; He, F.; Boogerd, F.C.; Jackson, V.J.; Goncharuk, V.; Kolodkin, A. Macromolecular networks and intelligence in microorganisms. Front. Microbiol. 2014, 5, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, C.R.; MacDonald, H.; Mann, R.P.; Marshall, J.A.; Latty, T.; Garnier, S. Decision-making without a brain: How an amoeboid organism solves the two-armed bandit. J. R. Soc. Interface 2016, 13, 20160030. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, T.; Tero, A.; Nakagaki, T.; Kuramoto, Y. Amoebae anticipate periodic events. Phys. Rev. Lett. 2008, 100, 018101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbonnet, L.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Microbiota is essential for social development in the mouse. Mol. Psychiatry 2014, 19, 146–148. [Google Scholar] [CrossRef]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.Y. The treatment of chronic hepatic encephalopathy. Hepatogastroenterology 1991, 38, 377–387. [Google Scholar] [PubMed]
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and its discontents. mBio 2017, 8, e01492-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, R.N.; Bearss, K.E.; Lettinga, J.; Erickson, C.; Rodowski, M.; Aman, M.G.; McCracken, J.T.; McDougle, C.J.; Tierney, E.; Vitiello, B.; et al. Gastrointestinal symptoms in a sample of children with pervasive developmental disorders. J. Autism Dev. Disord. 2009, 39, 405–413. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Wu, F.; Zheng, H.; Peng, Q.; Zhou, H. Altered composition and function of intestinal microbiota in autism spectrum disorders: A systematic review. Transl. Psychiatry 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; Comegna, M.; Buommino, E.; et al. Gut microbiota features in young children with autism spectrum disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium akkermansia muciniphila and bifidobacterium spp. In feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Parracho, H.M.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J. Med. Microbiol. 2005, 54, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Bolte, E.R. Autism and clostridium tetani. Med. Hypotheses 1998, 51, 133–144. [Google Scholar] [CrossRef]
- Sandler, R.H.; Finegold, S.M.; Bolte, E.R.; Buchanan, C.P.; Maxwell, A.P.; Väisänen, M.L.; Nelson, M.N.; Wexler, H.M. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J. Child Neurol. 2000, 15, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.-H.; Woo, Y.S.; Lee, S.-Y.; Bahk, W.-M. The brain-gut-microbiome axis in psychiatry. Int. J. Mol. Sci. 2020, 21, 7122. [Google Scholar] [CrossRef] [PubMed]
- Bastiaanssen, T.F.S.; Cussotto, S.; Claesson, M.J.; Clarke, G.; Dinan, T.G.; Cryan, J.F. Gutted! Unraveling the role of the microbiome in major depressive disorder. Harv. Rev. Psychiatry 2020, 28, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Masand, P.S.; Kaplan, D.; Bhandary, A.; Hendricks, S. The relationship between schizophrenia and irritable bowel syndrome (ibs). Schizophr. Res. 1997, 23, 265–268. [Google Scholar] [CrossRef]
- Catassi, C.; Bai, J.C.; Bonaz, B.; Bouma, G.; Calabrò, A.; Carroccio, A.; Castillejo, G.; Ciacci, C.; Cristofori, F.; Dolinsek, J.; et al. Non-celiac gluten sensitivity: The new frontier of gluten related disorders. Nutrients 2013, 5, 3839–3853. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E.; Maukonen, J.; Hyytiäinen, T.; Kieseppä, T.; Orešič, M.; Sabunciyan, S.; Mantere, O.; Saarela, M.; Yolken, R.; Suvisaari, J. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr. Res. 2018, 192, 398–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, C.M.; Gilbert-Jaramillo, J.; Westman, E.C. The ketogenic diet and remission of psychotic symptoms in schizophrenia: Two case studies. Schizophr. Res. 2019, 208, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Tregellas, J.R.; Smucny, J.; Legget, K.T.; Stevens, K.E. Effects of a ketogenic diet on auditory gating in dba/2 mice: A proof-of-concept study. Schizophr. Res. 2015, 169, 351–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keshavarzian, A.; Green, S.J.; Engen, P.A.; Voigt, R.M.; Naqib, A.; Forsyth, C.B.; Mutlu, E.; Shannon, K.M. Colonic bacterial composition in parkinson’s disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Bedarf, J.R.; Hildebrand, F.; Coelho, L.P.; Sunagawa, S.; Bahram, M.; Goeser, F.; Bork, P.; Wüllner, U. Functional implications of microbial and viral gut metagenome changes in early stage l-dopa-naïve parkinson’s disease patients. Genome Med. 2017, 9, 39. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Debelius, J.W.; Morton, J.T.; Wissemann, W.T.; Lewis, M.R.; Wallen, Z.D.; Peddada, S.D.; Factor, S.A.; Molho, E.; Zabetian, C.P.; et al. Parkinson’s disease and parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef]
- Kong, G.; Cao, K.-A.L.; Judd, L.M.; Li, S.; Renoir, T.; Hannan, A.J. Microbiome profiling reveals gut dysbiosis in a transgenic mouse model of huntington’s disease. Neurobiol. Dis. 2020, 135, 104268. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, X.; Yang, S.; Meng, F.; Wang, X.; Wei, H.; Chen, T. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front. Microbiol. 2016, 7, 1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.G.; Wu, S.; Yi, J.; Xia, Y.; Jin, D.; Zhou, J.; Sun, J. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin. Ther. 2017, 39, 322–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.-C.; Cao, Z.-S.; Chang, K.-M.; Juang, J.-L. Intestinal microbial dysbiosis aggravates the progression of alzheimer’s disease in drosophila. Nat. Commun. 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Von Bernhardi, R.; Bernhardi, L.E.; Eugenín, J. What is neural plasticity? Adv. Exp. Med. Biol. 2017, 1015, 1–15. [Google Scholar] [PubMed]
- Citri, A.; Malenka, R.C. Synaptic plasticity: Multiple forms, functions, and mechanisms. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2008, 33, 18–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampaio-Baptista, C.; Johansen-Berg, H. White matter plasticity in the adult brain. Neuron 2017, 96, 1239–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, W.C.; Bear, M.F. Metaplasticity: The plasticity of synaptic plasticity. Trends Neurosci. 1996, 19, 126–130. [Google Scholar] [CrossRef]
- Tsodyks, M.V.; Markram, H. The neural code between neocortical pyramidal neurons depends on neurotransmitter release probability. Proc. Natl. Acad. Sci. USA 1997, 94, 719–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkwood, A.; Dudek, S.M.; Gold, J.T.; Aizenman, C.D.; Bear, M.F. Common forms of synaptic plasticity in the hippocampus and neocortex in vitro. Science 1993, 260, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Hoban, A.E.; Stilling, R.M.; Moloney, G.; Shanahan, F.; Dinan, T.G.; Clarke, G.; Cryan, J.F. The microbiome regulates amygdala-dependent fear recall. Mol. Psychiatry 2018, 23, 1134–1144. [Google Scholar] [CrossRef]
- Stilling, R.M.; Moloney, G.M.; Ryan, F.J.; Hoban, A.E.; Bastiaanssen, T.F.S.; Shanahan, F.; Clarke, G.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Social interaction-induced activation of rna splicing in the amygdala of microbiome-deficient mice. eLife 2018, 7, e33070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luczynski, P.; Whelan, S.O.; O’Sullivan, C.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. Eur. J. Neurosci. 2016, 44, 2654–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darch, H.T.; Collins, M.K.; O’Riordan, K.J.; Cryan, J.F. Microbial memories: Sex-dependent impact of the gut microbiome on hippocampal plasticity. Eur. J. Neurosci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gubert, C.; Hannan, A.J. Plastic brains and gastrointestinal strains: The microbiota–gut–brain axis as a modulator of cellular plasticity and cognitive function (commentary on darch et al., 2021). Eur. J. Neurosci. 2021. [Google Scholar] [CrossRef]
- Hara, Y. Brain plasticity and rehabilitation in stroke patients. J. Nippon. Med. Sch. 2015, 82, 4–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quigley, E.M.M. Microbiota-brain-gut axis and neurodegenerative diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Wiley, N.C.; Dinan, T.G.; Ross, R.P.; Stanton, C.; Clarke, G.; Cryan, J.F. The microbiota-gut-brain axis as a key regulator of neural function and the stress response: Implications for human and animal health 1,2. J. Anim. Sci. 2017, 95, 3225–3246. [Google Scholar] [PubMed]
- Dinan, T.G.; Cryan, J.F. Gut instincts: Microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long-Smith, C.; O’Riordan, K.J.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota-gut-brain axis: New therapeutic opportunities. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 477–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Srikantha, P.; Mohajeri, M.H. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [Green Version]
- Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Zhou, W.; Buck, G.A.; Snyder, M.P.; Strauss, J.F.; Weinstock, G.M.; et al. The integrative human microbiome project. Nature 2019, 569, 641–648. [Google Scholar]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ’unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biteen, J.S.; Blainey, P.C.; Cardon, Z.G.; Chun, M.; Church, G.M.; Dorrestein, P.C.; Fraser, S.E.; Gilbert, J.A.; Jansson, J.K.; Knight, R.; et al. Tools for the microbiome: Nano and beyond. ACS Nano 2016, 10, 6–37. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef] [Green Version]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilo, S.; Rossman, H.; Segal, E. Axes of a revolution: Challenges and promises of big data in healthcare. Nat. Med. 2020, 26, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinan, T.G.; Stanton, C.; Cryan, J.F. Psychobiotics: A novel class of psychotropic. Biol. Psychiatry 2013, 74, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, J.; Walton, G.; Van Hemert, S.; Williams, C.; Lamport, D. The effect of probiotics on cognitive function across the human lifespan: A systematic review. Neurosci. Biobehav. Rev. 2021, 128, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Silk, D.B.; Davis, A.; Vulevic, J.; Tzortzis, G.; Gibson, G.R. Clinical trial: The effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment. Pharmacol. Ther. 2009, 29, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Cowen, P.J.; Harmer, C.J.; Tzortzis, G.; Errington, S.; Burnet, P.W. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology 2015, 232, 1793–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gareau, M.G.; Wine, E.; Rodrigues, D.M.; Cho, J.H.; Whary, M.T.; Philpott, D.J.; Macqueen, G.; Sherman, P.M. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 2011, 60, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cao, S.; Zhang, X. Modulation of gut microbiota–brain axis by probiotics, prebiotics, and diet. J. Agric. Food Chem. 2015, 63, 7885–7895. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.L.; Lin, T.L.; Chang, C.J.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3. [Google Scholar] [CrossRef]
- Allegretti, J.R.; Mullish, B.H.; Kelly, C.; Fischer, M. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet 2019, 394, 420–431. [Google Scholar] [CrossRef]
- Dardiotis, E.; Tsouris, Z.; Mentis, A.A.; Siokas, V.; Michalopoulou, A.; Sokratous, M.; Dastamani, M.; Bogdanos, D.P.; Deretzi, G.; Kountouras, J.H. Pylori and parkinson’s disease: Meta-analyses including clinical severity. Clin. Neurol Neurosurg. 2018, 175, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics-a step beyond pre- and probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Toussaint, N.C.; Chen, S.P.; Ratner, A.J.; Whittier, S.; Wang, T.C.; Wang, H.H.; Abrams, J.A. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: A crossover trial. Gastroenterology 2015, 149, 883–885.e889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz-Guerrero, G.; Amador-Muñoz, D.; Calderón-Ospina, C.A.; López-Fuentes, D.; Nava Mesa, M.O. Proton pump inhibitors and dementia: Physiopathological mechanisms and clinical consequences. Neural Plast. 2018, 2018, 5257285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Oudenhove, L.; McKie, S.; Lassman, D.; Uddin, B.; Paine, P.; Coen, S.; Gregory, L.; Tack, J.; Aziz, Q. Fatty acid-induced gut-brain signaling attenuates neural and behavioral effects of sad emotion in humans. J. Clin. Investig. 2011, 121, 3094–3099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillisch, K.; Labus, J.; Kilpatrick, L.; Jiang, Z.; Stains, J.; Ebrat, B.; Guyonnet, D.; Legrain-Raspaud, S.; Trotin, B.; Naliboff, B.; et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013, 144, 1394–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Baarlen, P.; Troost, F.; van der Meer, C.; Hooiveld, G.; Boekschoten, M.; Brummer, R.J.M.; Kleerebezem, M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 4562–4569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-H.; An, H.J.; Garrido, D.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Proteomic analysis of bifidobacterium longum subsp. Infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS ONE 2013, 8, e57535. [Google Scholar] [CrossRef] [PubMed]
- Konjevod, M.; Nikolac Perkovic, M.; Sáiz, J.; Svob Strac, D.; Barbas, C.; Rojo, D. Metabolomics analysis of microbiota-gut-brain axis in neurodegenerative and psychiatric diseases. J. Pharm. Biomed. Anal. 2021, 194, 113681. [Google Scholar] [CrossRef] [PubMed]
- Marler, S.; Ferguson, B.J.; Lee, E.B.; Peters, B.; Williams, K.C.; McDonnell, E.; Macklin, E.A.; Levitt, P.; Gillespie, C.H.; Anderson, G.M.; et al. Brief report: Whole blood serotonin levels and gastrointestinal symptoms in autism spectrum disorder. J. Autism Dev. Disord. 2016, 46, 1124–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rio, D.; Zimetti, F.; Caffarra, P.; Tassotti, M.; Bernini, F.; Brighenti, F.; Zini, A.; Zanotti, I. The gut microbial metabolite trimethylamine-n-oxide is present in human cerebrospinal fluid. Nutrients 2017, 9, 1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spichak, S.; Bastiaanssen, T.F.S.; Berding, K.; Vlckova, K.; Clarke, G.; Dinan, T.G.; Cryan, J.F. Mining microbes for mental health: Determining the role of microbial metabolic pathways in human brain health and disease. Neurosci. Biobehav. Rev. 2021, 125, 698–761. [Google Scholar] [CrossRef] [PubMed]
- Caputi, V.; Giron, M.C. Microbiome-gut-brain axis and toll-like receptors in parkinson’s disease. Int. J. Mol. Sci. 2018, 19, 1689. [Google Scholar] [CrossRef] [Green Version]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Gubert, C.; Kong, G.; Renoir, T.; Hannan, A.J. Exercise, diet and stress as modulators of gut microbiota: Implications for neurodegenerative diseases. Neurobiol. Dis. 2020, 134, 104621. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zafar, S.; Salih Ibrahim, R.M.; Chi, H.L.; Xiao, T.; Xia, W.J.; Li, H.B.; Kang, Y.M. Exercise and food supplement of vitamin c ameliorate hypertension through improvement of gut microflora in the spontaneously hypertensive rats. Life Sci. 2021, 269, 119097. [Google Scholar] [CrossRef]
- Dalton, A.; Mermier, C.; Zuhl, M. Exercise influence on the microbiome-gut-brain axis. Gut Microbes 2019, 10, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise alters gut microbiota composition and function in lean and obese humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.S.; Jeraldo, P.R.; Kurti, A.; Miller, M.E.; Cook, M.D.; Whitlock, K.; Goldenfeld, N.; Woods, J.A.; White, B.A.; Chia, N.; et al. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol. Neurodegener. 2014, 9, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhu, K.V.; Sherwin, E.; Schellekens, H.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Feeding the microbiota-gut-brain axis: Diet, microbiome, and neuropsychiatry. Transl. Res. 2017, 179, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhu, X.; Zeng, M.; Qi, L.; Tang, X.; Wang, D.; Zhang, M.; Xie, Y.; Li, H.; Yang, X.; et al. A diet high in sugar and fat influences neurotransmitter metabolism and then affects brain function by altering the gut microbiota. Transl. Psychiatry 2021, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Marx, W.; Moseley, G.; Berk, M.; Jacka, F. Nutritional psychiatry: The present state of the evidence. Proc. Nutr. Soc. 2017, 76, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinan, T.G.; Butler, M.I.; Cryan, J.F. Psychobiotics: Evolution of novel antidepressants. Mod. Trends Psychiatry 2021, 32, 134–143. [Google Scholar] [PubMed]
- Allen, S.J.; Wareham, K.; Wang, D.; Bradley, C.; Hutchings, H.; Harris, W.; Dhar, A.; Brown, H.; Foden, A.; Gravenor, M.B.; et al. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and clostridium difficile diarrhoea in older inpatients (placide): A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2013, 382, 1249–1257. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.; Grønborg, T.K.; Sørensen, M.J.; Schendel, D.; Parner, E.T.; Pedersen, L.H.; Vestergaard, M. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA 2013, 309, 1696–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquette, A.G.; Hood, L.; Price, N.D.; Sadovsky, Y. Deep phenotyping during pregnancy for predictive and preventive medicine. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Vasilevsky, N.A.; Engelstad, M.; Foster, E.; McMurry, J.; Aymé, S.; Baynam, G.; Bello, S.M.; Boerkoel, C.F.; Boycott, K.M.; et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017, 45, D865–D876. [Google Scholar] [CrossRef] [PubMed]
- Surana, N.K.; Kasper, D.L. Moving beyond microbiome-wide associations to causal microbe identification. Nature 2017, 552, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Zihler Berner, A.; Fuentes, S.; Dostal, A.; Payne, A.N.; Vazquez Gutierrez, P.; Chassard, C.; Grattepanche, F.; de Vos, W.M.; Lacroix, C. Novel polyfermentor intestinal model (polyferms) for controlled ecological studies: Validation and effect of ph. PLoS ONE 2013, 8, e77772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera-Quintanar, L.; Ortuño-Sahagún, D.; Franco-Arroyo, N.N.; Viveros-Paredes, J.M.; Zepeda-Morales, A.S.; Lopez-Roa, R.I. The human microbiota and obesity: A literature systematic review of in vivo models and technical approaches. Int. J. Mol. Sci. 2018, 19, 3827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenstein, M. The hunt for a healthy microbiome. Nature 2020, 577, S6–S8. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Prindle, A.; Humphries, J.; Gabalda-Sagarra, M.; Asally, M.; Lee, D.Y.; Ly, S.; Garcia-Ojalvo, J.; Suel, G.M. Metabolic co-dependence gives rise to collective oscillations within biofilms. Nature 2015, 523, 550–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, M. Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 2021, 184, 1971–1989. [Google Scholar] [CrossRef] [PubMed]
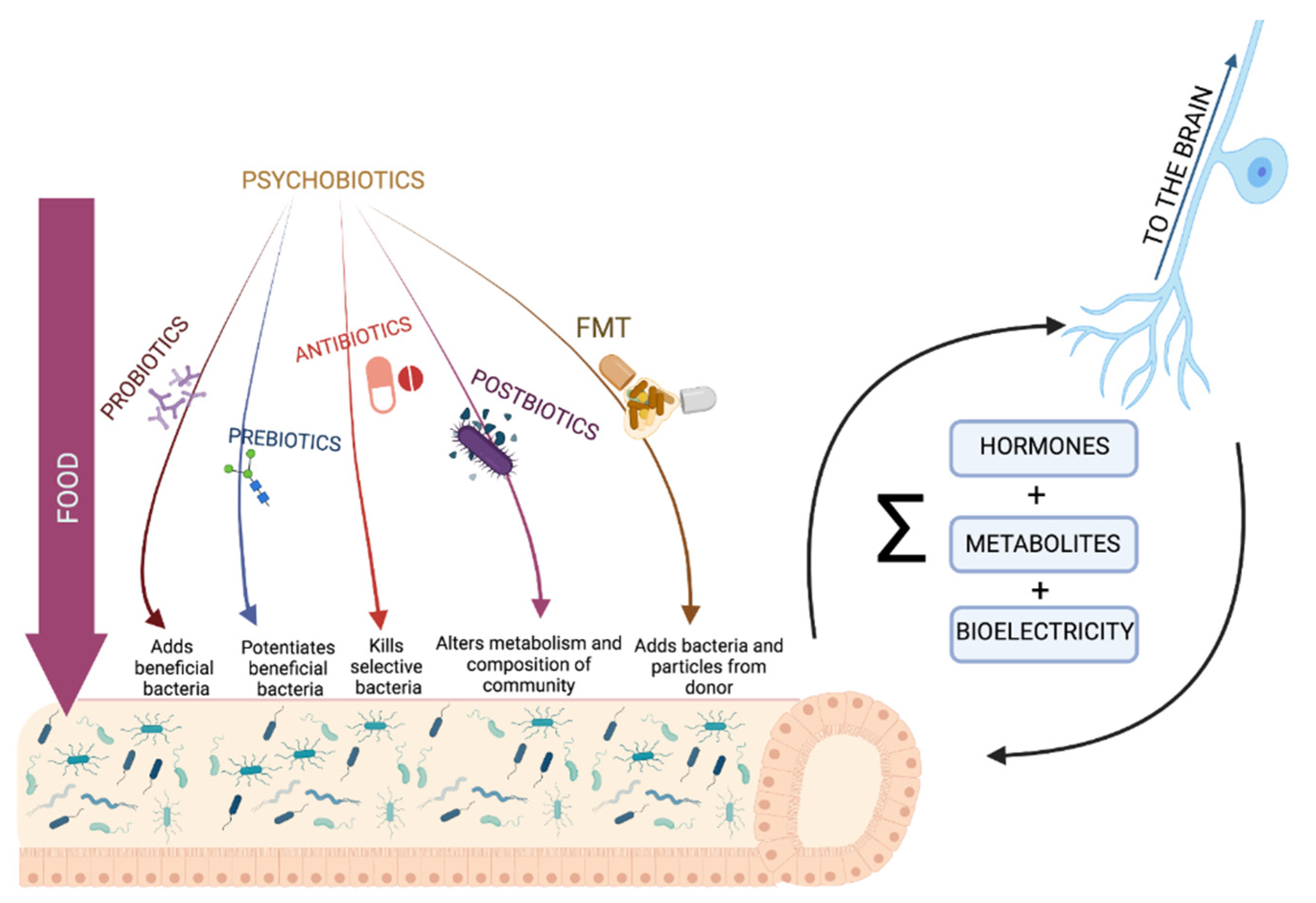
| Therapy | Results | References |
|---|---|---|
| Probiotics |
| [99,103,118,119] |
| Prebiotics |
| [103,119,120] |
| FMT |
| [103,121] |
| Antibiotics |
| [99,103,119,122] |
| Postbiotics |
| [103,123] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murciano-Brea, J.; Garcia-Montes, M.; Geuna, S.; Herrera-Rincon, C. Gut Microbiota and Neuroplasticity. Cells 2021, 10, 2084. https://doi.org/10.3390/cells10082084
Murciano-Brea J, Garcia-Montes M, Geuna S, Herrera-Rincon C. Gut Microbiota and Neuroplasticity. Cells. 2021; 10(8):2084. https://doi.org/10.3390/cells10082084
Chicago/Turabian StyleMurciano-Brea, Julia, Martin Garcia-Montes, Stefano Geuna, and Celia Herrera-Rincon. 2021. "Gut Microbiota and Neuroplasticity" Cells 10, no. 8: 2084. https://doi.org/10.3390/cells10082084






