Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Subject Characteristics
2.2. Biochemical Analysis
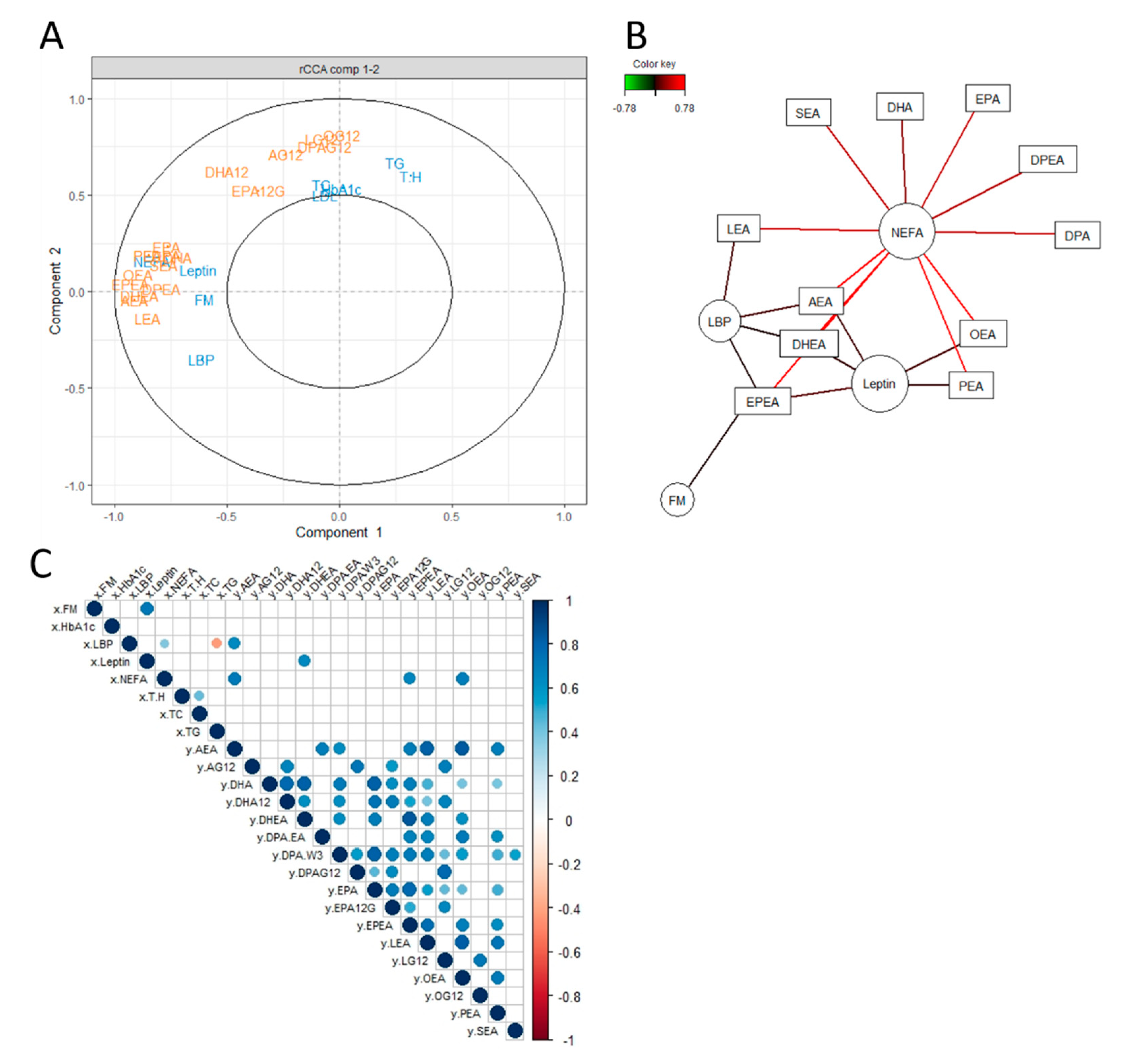
2.3. Explorative Multivariate Approaches
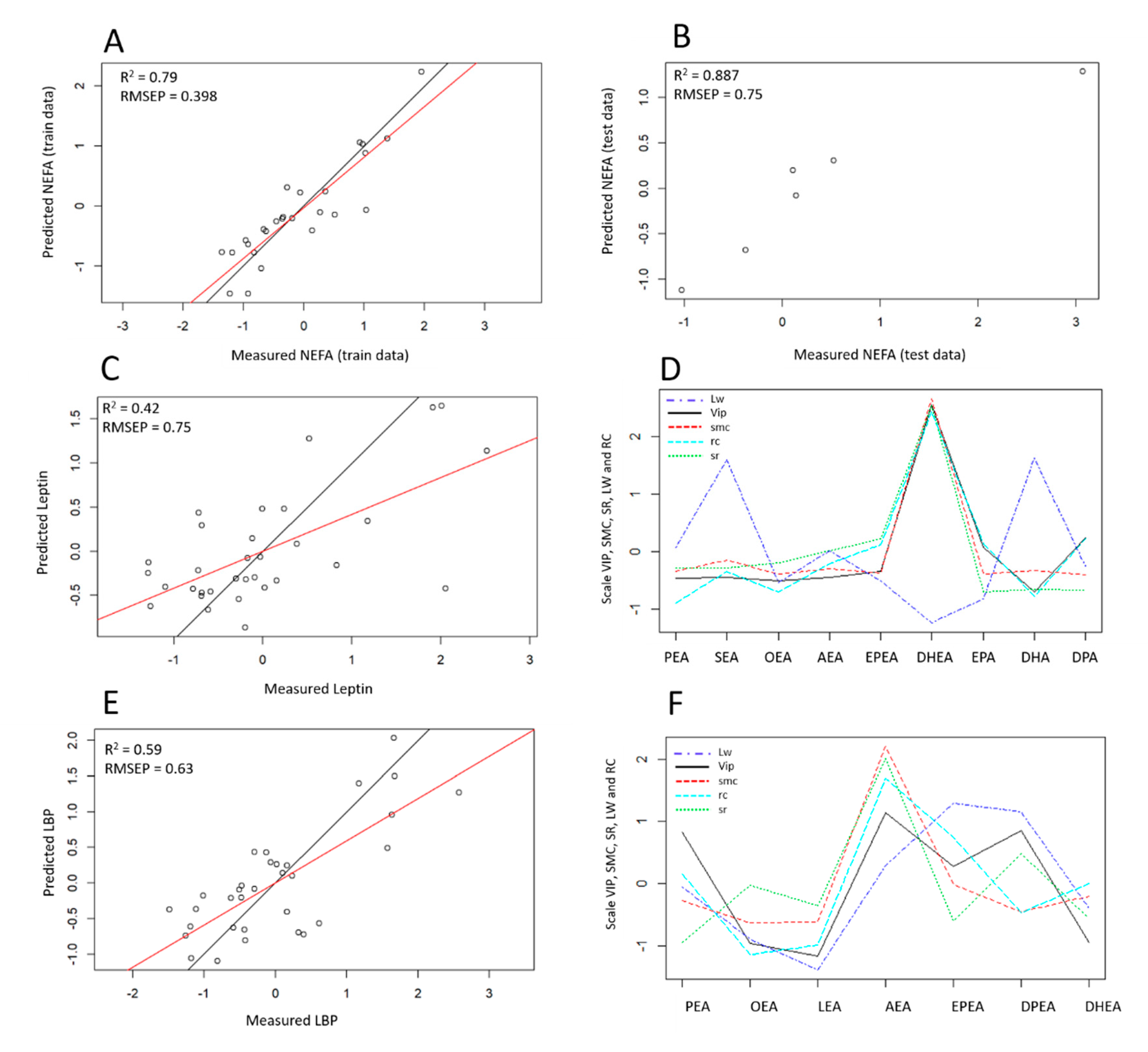
2.4. Variable-Response Modeling and Construction of Predictive Model
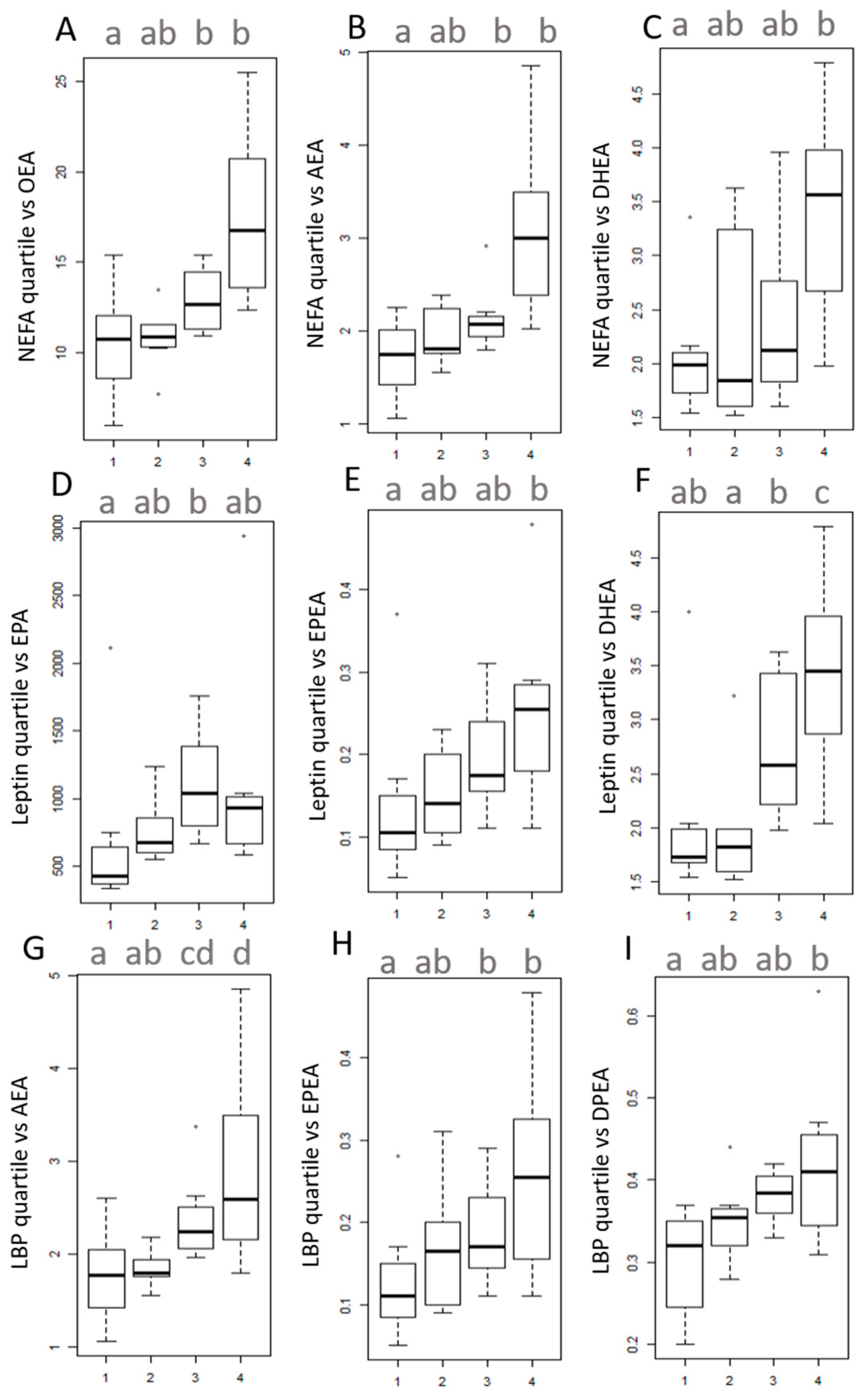
2.5. Univariate Analysis
3. Results
3.1. Unsupervised Exploratory Approaches to Unravel an Association between the eCB System and Certain Metabolic Parameters
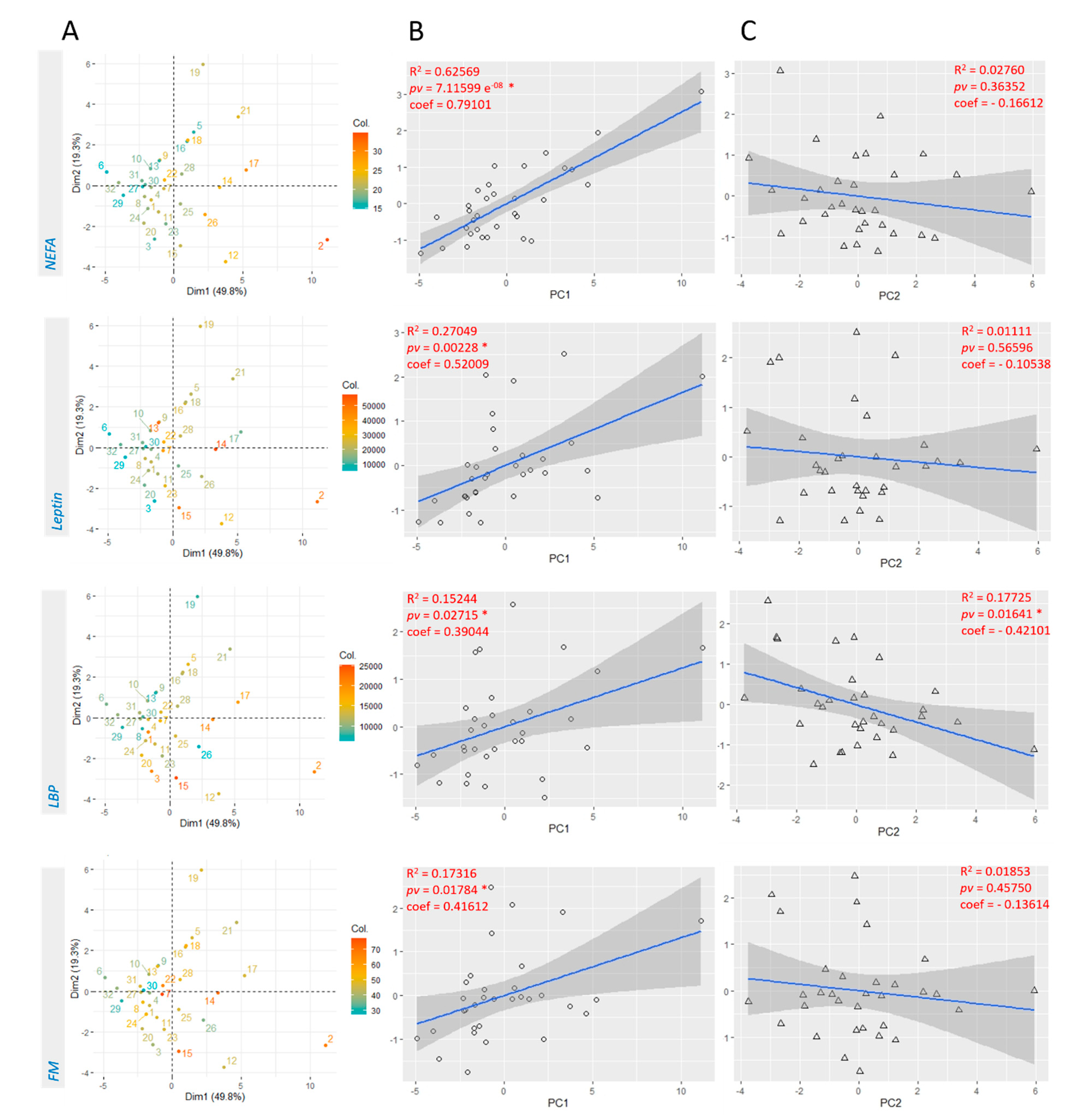
3.2. Linear Regression with Principal Components Analysis
3.3. Modeling Variable Response According to Selected eCBs and Construction of Predictive Models
3.3.1. NEFA Response Modeling
3.3.2. Leptin Response Modeling
3.3.3. LBP Response Modeling
3.4. Univariate Analysis on Quartiles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Marzo, V. The endocannabinoidome as a substrate for noneuphoric phytocannabinoid action and gut microbiome dysfunction in neuropsychiatric disorders. Dialogues Clin. Neurosci. 2020, 22, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Plovier, H.; Van Hul, M.; Geurts, L.; Delzenne, N.M.; Druart, C.; Everard, A. Endocannabinoids--at the crossroads between the gut microbiota and host metabolism. Nat. Rev. Endocrinol. 2016, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, A.; Di Marzo, V.; Silvestri, C. The Expanded Endocannabinoid System/Endocannabinoidome as a Potential Target for Treating Diabetes Mellitus. Curr. Diabetes Rep. 2019, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat. Rev. Neurol. 2020, 16, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 2018, 17, 623–639. [Google Scholar] [CrossRef]
- Simon, V.; Cota, D. Mechanisms in Endocrinology: Endocannabinoids and metabolism: Past, present and future. Eur. J. Endocrinol. 2017, 176, R309–R324. [Google Scholar] [CrossRef]
- Silvestri, C.; Di Marzo, V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metab. 2013, 17, 475–490. [Google Scholar] [CrossRef]
- Di Marzo, V.; Silvestri, C. Lifestyle and Metabolic Syndrome: Contribution of the Endocannabinoidome. Nutrients 2019, 11, 1956. [Google Scholar] [CrossRef]
- Forte, N.; Fernández-Rilo, A.C.; Palomba, L.; Di Marzo, V.; Cristino, L. Obesity Affects the Microbiota-Gut-Brain Axis and the Regulation Thereof by Endocannabinoids and Related Mediators. Int. J. Mol. Sci. 2020, 21, 1554. [Google Scholar] [CrossRef]
- Osei-Hyiaman, D.; Liu, J.; Zhou, L.; Godlewski, G.; Harvey-White, J.; Jeong, W.I.; Bátkai, S.; Marsicano, G.; Lutz, B.; Buettner, C.; et al. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J. Clin. Investig. 2008, 118, 3160–3169. [Google Scholar] [CrossRef]
- Castonguay-Paradis, S.; Lacroix, S.; Rochefort, G.; Parent, L.; Perron, J.; Martin, C.; Lamarche, B.; Raymond, F.; Flamand, N.; Di Marzo, V.; et al. Dietary fatty acid intake and gut microbiota determine circulating endocannabinoidome signaling beyond the effect of body fat. Sci. Rep. 2020, 10, 15975. [Google Scholar] [CrossRef] [PubMed]
- Gatta-Cherifi, B.; Cota, D. New insights on the role of the endocannabinoid system in the regulation of energy balance. Int. J. Obes. 2016, 40, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Karwad, M.A.; Macpherson, T.; Wang, B.; Theophilidou, E.; Sarmad, S.; Barrett, D.A.; Larvin, M.; Wright, K.L.; Lund, J.N.; O’Sullivan, S.E. Oleoylethanolamine and palmitoylethanolamine modulate intestinal permeability in vitro via TRPV1 and PPARalpha. FASEB J. 2017, 31, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, F.; Romano, B.; Petrosino, S.; Pagano, E.; Capasso, R.; Coppola, D.; Battista, G.; Orlando, P.; Di Marzo, V.; Izzo, A.A. Palmitoylethanolamide, a naturally occurring lipid, is an orally effective intestinal anti-inflammatory agent. Br. J. Pharmacol. 2015, 172, 142–158. [Google Scholar] [CrossRef]
- Syed, S.K.; Bui, H.H.; Beavers, L.S.; Farb, T.B.; Ficorilli, J.; Chesterfield, A.K.; Kuo, M.S.; Bokvist, K.; Barrett, D.G.; Efanov, A.M. Regulation of GPR119 receptor activity with endocannabinoid-like lipids. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1469–E1478. [Google Scholar] [CrossRef]
- Overton, H.A.; Babbs, A.J.; Doel, S.M.; Fyfe, M.C.; Gardner, L.S.; Griffin, G.; Jackson, H.C.; Procter, M.J.; Rasamison, C.M.; Tang-Christensen, M.; et al. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006, 3, 167–175. [Google Scholar] [CrossRef]
- Godlewski, G.; Offertaler, L.; Wagner, J.A.; Kunos, G. Receptors for acylethanolamides-GPR55 and GPR119. Prostaglandins Other Lipid Mediat. 2009, 89, 105–111. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Di Marzo, V.; Petrosino, S. Endocannabinoids and endocannabinoid-related mediators: Targets, metabolism and role in neurological disorders. Prog. Lipid Res. 2016, 62, 107–128. [Google Scholar] [CrossRef]
- Meng, C.; Zeleznik, O.A.; Thallinger, G.G.; Kuster, B.; Gholami, A.M.; Culhane, A.C. Dimension reduction techniques for the integrative analysis of multi-omics data. Briefings Bioinform. 2016, 17, 628–641. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.; Tonjes, A.; Stumvoll, M.; Thiery, J.; Kratzsch, J. Assay-dependent variability of serum insulin levels during oral glucose tolerance test: Influence on reference intervals for insulin and on cut-off values for insulin sensitivity indices. Clin. Chem. Lab. Med. 2008, 46, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Borza, D.; Karmali, R.; Andre, S.; Valsamis, J.; Cytryn, E.; Hermans, M.P.; Beyer, I. Influence of assay-dependent variability of serum insulin levels on insulin sensitivity indices. Clin. Chem. Lab. Med. 2008, 46, 1655–1656. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Archambault, A.S.; Dumais, É.; Martin, C.; Blanchet, M.R.; Bissonnette, E.; Ohashi, N.; Yamamoto, K.; Itoh, T.; Laviolette, M.; et al. Endocannabinoid hydrolysis inhibition unmasks that unsaturated fatty acids induce a robust biosynthesis of 2-arachidonoyl-glycerol and its congeners in human myeloid leukocytes. FASEB J. 2020, 34, 4253–4265. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Plovier, H.; Rastelli, M.; Van Hul, M.; de Wouters d’Oplinter, A.; Geurts, L.; Druart, C.; Robine, S.; Delzenne, N.M.; Muccioli, G.G.; et al. Intestinal epithelial N-acylphosphatidylethanolamine phospholipase D links dietary fat to metabolic adaptations in obesity and steatosis. Nat. Commun. 2019, 10, 457. [Google Scholar] [CrossRef] [PubMed]
- Le Cao, K.-A.R.; Gonzalez, I.; Dejean, S.; Gautier, B.; Bartolo, F.; Monget, P.; Coquery, J.; Yao, F.; Liquet, B. mixOmics: Omics Data Integration Project. R Package Version 6.1.1. Available online: https://CRAN.R-project.org/package=mixOmics (accessed on 17 November 2020).
- Gonzalez, I.; Déjean, S.; Martin, P.; Baccini, A. CCA: An R Package to Extend Canonical Correlation Analysis. J. Stat. Softw. 2007, 23. [Google Scholar] [CrossRef]
- Gonzalez, I.; Déjean, S.; Martin, P.; Gonçalves, O.; Besse, P.; Baccini, A. Highlighting Relationships Between Heteregeneous Biological Data Through Graphical Displays Based On Regularized Canonical Correlation Analysis. J. Biol. Syst. 2009, 17, 173–199. [Google Scholar] [CrossRef]
- González, I.; Lê Cao, K.-A.; Davis, M.J.; Déjean, S. network: Relevance Network for (r)CCA and (s)PLS regression. Available online: https://rdrr.io/cran/mixOmics/man/network.html (accessed on 17 November 2020).
- Gonzalez, I.; Cao, K.A.; Davis, M.J.; Dejean, S. Visualising associations between paired ‘omics’ data sets. BioData Min. 2012, 5, 19. [Google Scholar] [CrossRef]
- Witkamp, R.F. The role of fatty acids and their endocannabinoid-like derivatives in the molecular regulation of appetite. Mol. Asp. Med. 2018, 64, 45–67. [Google Scholar] [CrossRef]
- Lefort, C.; Roumain, M.; Van Hul, M.; Rastelli, M.; Manco, R.; Leclercq, I.; Delzenne, N.M.; Marzo, V.D.; Flamand, N.; Luquet, S.; et al. Hepatic NAPE-PLD Is a Key Regulator of Liver Lipid Metabolism. Cells 2020, 9, 1247. [Google Scholar] [CrossRef]
- Gruden, G.; Barutta, F.; Kunos, G.; Pacher, P. Role of the endocannabinoid system in diabetes and diabetic complications. Br. J. Pharmacol. 2016, 173, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Bazwinsky-Wutschke, I.; Zipprich, A.; Dehghani, F. Endocannabinoid System in Hepatic Glucose Metabolism, Fatty Liver Disease, and Cirrhosis. Int. J. Mol. Sci. 2019, 20, 2516. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.A.T.; Black, W.C.; Hai, J.F.R. Multivariate Data Analysis; Prentice hall: Upper Saddle River, NJ, USA, 1998; Volume 5. [Google Scholar]
- Fanelli, F.; Mezzullo, M.; Repaci, A.; Belluomo, I.; Ibarra Gasparini, D.; Di Dalmazi, G.; Mastroroberto, M.; Vicennati, V.; Gambineri, A.; Morselli-Labate, A.M.; et al. Profiling plasma N-Acylethanolamine levels and their ratios as a biomarker of obesity and dysmetabolism. Mol. Metab. 2018, 14, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Côté, M.; Matias, I.; Lemieux, I.; Petrosino, S.; Alméras, N.; Després, J.P.; Di Marzo, V. Circulating endocannabinoid levels, abdominal adiposity and related cardiometabolic risk factors in obese men. Int. J. Obes. 2007, 31, 692–699. [Google Scholar] [CrossRef]
- Blüher, M.; Engeli, S.; Klöting, N.; Berndt, J.; Fasshauer, M.; Bátkai, S.; Pacher, P.; Schön, M.R.; Jordan, J.; Stumvoll, M. Dysregulation of the peripheral and adipose tissue endocannabinoid system in human abdominal obesity. Diabetes 2006, 55, 3053–3060. [Google Scholar] [CrossRef]
- Mahesh, S.; Jayas, D.; Paliwal, J.; White, N. Comparison of Partial Least Squares Regression (PLSR) and Principal Components Regression (PCR) Methods for Protein and Hardness Predictions using the Near-Infrared (NIR) Hyperspectral Images of Bulk Samples of Canadian Wheat. Food Bioprocess Technol. 2014, 8, 31–40. [Google Scholar] [CrossRef]
- Kabir, A.; Rahman, M.J.; Shamim, A.A.; Klemm, R.D.W.; Labrique, A.B.; Rashid, M.; Christian, P.; West, K.P., Jr. Identifying maternal and infant factors associated with newborn size in rural Bangladesh by partial least squares (PLS) regression analysis. PLoS ONE 2017, 12, e0189677. [Google Scholar] [CrossRef]
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2019, 100, 171–210. [Google Scholar] [CrossRef]
- Faraj, M.; Lu, H.; Cianflone, K. Diabetes, lipids, and adipocyte secretagogues. Biochem. Cell Biol. 2004, 82, 170–190. [Google Scholar] [CrossRef]
- Blaak, E.E. Fatty acid metabolism in obesity and type 2 diabetes mellitus. Proc. Nutr. Soc. 2003, 62, 753–760. [Google Scholar] [CrossRef]
- Guzmán, M.; Lo Verme, J.; Fu, J.; Oveisi, F.; Blázquez, C.; Piomelli, D. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J. Biol. Chem. 2004, 279, 27849–27854. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Oveisi, F.; Gaetani, S.; Lin, E.; Piomelli, D. Oleoylethanolamide, an endogenous PPAR-alpha agonist, lowers body weight and hyperlipidemia in obese rats. Neuropharmacology 2005, 48, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Terrazzino, S.; Berto, F.; Dalle Carbonare, M.; Fabris, M.; Guiotto, A.; Bernardini, D.; Leon, A. Stearoylethanolamide exerts anorexic effects in mice via down-regulation of liver stearoyl-coenzyme A desaturase-1 mRNA expression. FASEB J. 2004, 18, 1580–1582. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Zhan, X.; Yu, X.; Jia, M.; Zhang, Y.; Yao, J.; Hu, X.; Bao, Z. Inhibiting CB1 receptors improves lipogenesis in an in vitro non-alcoholic fatty liver disease model. Lipids Health Dis. 2014, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R. Endocannabinoids promote hepatic lipogenesis and steatosis through CB1 receptors. Hepatology 2005, 42, 959–961. [Google Scholar] [CrossRef]
- Buettner, C.; Muse, E.D.; Cheng, A.; Chen, L.; Scherer, T.; Pocai, A.; Su, K.; Cheng, B.; Li, X.; Harvey-White, J.; et al. Leptin controls adipose tissue lipogenesis via central, STAT3–independent mechanisms. Nat. Med. 2008, 14, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Joosten, M.M.; Balvers, M.G.; Verhoeckx, K.C.; Hendriks, H.F.; Witkamp, R.F. Plasma anandamide and other N-acylethanolamines are correlated with their corresponding free fatty acid levels under both fasting and non-fasting conditions in women. Nutr. Metab. 2010, 7, 49. [Google Scholar] [CrossRef]
- Pérez-Pérez, A.; Sánchez-Jiménez, F.; Vilariño-García, T.; Sánchez-Margalet, V. Role of Leptin in Inflammation and Vice Versa. Int. J. Mol. Sci. 2020, 21, 5887. [Google Scholar] [CrossRef]
- Balvers, M.G.; Verhoeckx, K.C.; Plastina, P.; Wortelboer, H.M.; Meijerink, J.; Witkamp, R.F. Docosahexaenoic acid and eicosapentaenoic acid are converted by 3T3-L1 adipocytes to N-acyl ethanolamines with anti-inflammatory properties. Biochim. Biophys. Acta 2010, 1801, 1107–1114. [Google Scholar] [CrossRef]
- Gdula-Argasińska, J.; Bystrowska, B. Docosahexaenoic acid attenuates in endocannabinoid synthesis in RAW 264.7 macrophages activated with benzo(a)pyrene and lipopolysaccharide. Toxicol. Lett. 2016, 258, 93–100. [Google Scholar] [CrossRef]
- McDougle, D.R.; Watson, J.E.; Abdeen, A.A.; Adili, R.; Caputo, M.P.; Krapf, J.E.; Johnson, R.W.; Kilian, K.A.; Holinstat, M.; Das, A. Anti-inflammatory ω-3 endocannabinoid epoxides. Proc. Natl. Acad. Sci. USA 2017, 114, E6034. [Google Scholar] [CrossRef] [PubMed]
- Valassi, E.; Scacchi, M.; Cavagnini, F. Neuroendocrine control of food intake. Nutr. Metab. Cardiovasc. Dis. 2008, 18, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Goparaju, S.K.; Wang, L.; Liu, J.; Batkai, S.; Jarai, Z.; Fezza, F.; Miura, G.I.; Palmiter, R.D.; Sugiura, T.; et al. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 2001, 410, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Balsevich, G.; Petrie, G.N.; Hill, M.N. Endocannabinoids: Effectors of glucocorticoid signaling. Front. Neuroendocrinol. 2017, 47, 86–108. [Google Scholar] [CrossRef]
- Ruiz, A.G.; Casafont, F.; Crespo, J.; Cayón, A.; Mayorga, M.; Estebanez, A.; Fernadez-Escalante, J.C.; Pons-Romero, F. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: Evidence for the potential role of endotoxin in the pathogenesis of non-alcoholic steatohepatitis. Obes. Surg. 2007, 17, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, Z.; Ye, X.; Zou, S.; Li, H.; Yu, D.; Wu, H.; Chen, Y.; Dore, J.; Clement, K.; et al. A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese. Diabetes Care 2010, 33, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Navarrete, J.M.; Ortega, F.; Serino, M.; Luche, E.; Waget, A.; Pardo, G.; Salvador, J.; Ricart, W.; Frühbeck, G.; Burcelin, R.; et al. Circulating lipopolysaccharide-binding protein (LBP) as a marker of obesity-related insulin resistance. Int. J. Obes. 2012, 36, 1442–1449. [Google Scholar] [CrossRef]
- Genser, L.; Aguanno, D.; Soula, H.A.; Dong, L.; Trystram, L.; Assmann, K.; Salem, J.E.; Vaillant, J.C.; Oppert, J.M.; Laugerette, F.; et al. Increased jejunal permeability in human obesity is revealed by a lipid challenge and is linked to inflammation and type 2 diabetes. J. Pathol. 2018, 246, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Chouinard, F.; Lefebvre, J.S.; Flamand, N. Regulation of inflammation by cannabinoids, the endocannabinoids 2-arachidonoyl-glycerol and arachidonoyl-ethanolamide, and their metabolites. J. Leukoc. Biol. 2015, 97, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Fredman, G.; Krishnamoorthy, S.; Agrawal, N.; Irimia, D.; Piomelli, D.; Serhan, C.N. Decoding functional metabolomics with docosahexaenoyl ethanolamide (DHEA) identifies novel bioactive signals. J. Biol. Chem. 2011, 286, 31532–31541. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Blanchet, M.R.; Laviolette, M.; Flamand, N. The CB(2) receptor and its role as a regulator of inflammation. Cell. Mol. Life Sci. 2016, 73, 4449–4470. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Izzo, A.A. Endocannabinoid overactivity and intestinal inflammation. Gut 2006, 55, 1373–1376. [Google Scholar] [CrossRef] [PubMed]
- Karwad, M.A.; Couch, D.G.; Theophilidou, E.; Sarmad, S.; Barrett, D.A.; Larvin, M.; Wright, K.L.; Lund, J.N.; O’Sullivan, S.E. The role of CB(1) in intestinal permeability and inflammation. FASEB J. 2017, 31, 3267–3277. [Google Scholar] [CrossRef] [PubMed]
| Endocannabinoids | Metabolic Parameters | ||
|---|---|---|---|
| AEA | N-arachidonoyl-ethanolamine | FM | fat mass |
| 2-AG (AG12) | 1(3)- and 2-arachidonoyl-glycerol | HbA1c | glycated hemoglobin A1c |
| DHEA | N-docosahexanoyl-ethanolamine | LBP | lipopolysaccharide-binding protein |
| DHA | docosahexaenoic acid | LDL | low density lipoprotein |
| 2-DHG (DHA12) | 1(3)- and 2-docosahexaenoyl-glycerol | NEFA | non-esterified fatty acids |
| 2-DPG (DPAG12) | 1(3)- and 2-docospentaenoyl-glycerol(n-3) | TC | total cholesterol |
| DPEA | N-docosapentaenoyl-ethanolamine(n-3) | T.H | total/HDL cholesterol |
| DPA | docosapentaenoic acid (n-3) | ||
| EPA | eicosapentaenoic acid | ||
| EPEA | N-eicosapentaenoyl-ethanolamine | ||
| LEA | N-linoleoyl-ethanolamine | ||
| 2-LG (LG12) | 1(3)- and 2-linoleoyl-glycerol | ||
| OEA | N-oleoyl-ethanolamine | ||
| 2-OG (OG12) | 1(3)- and 2-oleoylglycerol | ||
| PEA | N-palmitoyl-ethanolamine | ||
| SEA | N-stearoyl-ethanolamine | ||
| Predictors | Estimate | Std Error | T Value | Pr(>|t|) | * |
|---|---|---|---|---|---|
| PEA | 0.049287 | 0.203249 | 0.2425 | 0.809993 | |
| SEA | −0.575454 | 0.274860 | −2.0936 | 0.044567 | * |
| OEA | 0.597778 | 0.267343 | 2.2360 | 0.032689 | * |
| LEA | −0.139934 | 0.267431 | −0.5233 | 0.604519 | |
| AEA | 0.898707 | 0.201773 | 4.4541 | 0.000102 | *** |
| EPEA | 0.494766 | 0.264972 | 1.8672 | 0.071350 | |
| DPEA | −0.625028 | 0.221271 | −2.8247 | 0.008202 | ** |
| DHEA | −0.371274 | 0.177796 | −2.1687 | 0.037895 | * |
| DPA | −0.020114 | 0.206108 | −0.0976 | 0.922886 | |
| EPA | 0.271866 | 0.217275 | 1.2513 | 0.220202 | |
| DHA | 0.200733 | 0.182672 | 1.0989 | 0.280285 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Depommier, C.; Flamand, N.; Pelicaen, R.; Maiter, D.; Thissen, J.-P.; Loumaye, A.; Hermans, M.P.; Everard, A.; Delzenne, N.M.; Di Marzo, V.; et al. Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models. Cells 2021, 10, 71. https://doi.org/10.3390/cells10010071
Depommier C, Flamand N, Pelicaen R, Maiter D, Thissen J-P, Loumaye A, Hermans MP, Everard A, Delzenne NM, Di Marzo V, et al. Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models. Cells. 2021; 10(1):71. https://doi.org/10.3390/cells10010071
Chicago/Turabian StyleDepommier, Clara, Nicolas Flamand, Rudy Pelicaen, Dominique Maiter, Jean-Paul Thissen, Audrey Loumaye, Michel P. Hermans, Amandine Everard, Nathalie M. Delzenne, Vincenzo Di Marzo, and et al. 2021. "Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models" Cells 10, no. 1: 71. https://doi.org/10.3390/cells10010071
APA StyleDepommier, C., Flamand, N., Pelicaen, R., Maiter, D., Thissen, J.-P., Loumaye, A., Hermans, M. P., Everard, A., Delzenne, N. M., Di Marzo, V., & Cani, P. D. (2021). Linking the Endocannabinoidome with Specific Metabolic Parameters in an Overweight and Insulin-Resistant Population: From Multivariate Exploratory Analysis to Univariate Analysis and Construction of Predictive Models. Cells, 10(1), 71. https://doi.org/10.3390/cells10010071






