Microbial and Immune Landscape of Malignant Ascites: Insights from Gut, Bladder, and Ascitic Fluid Analyses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
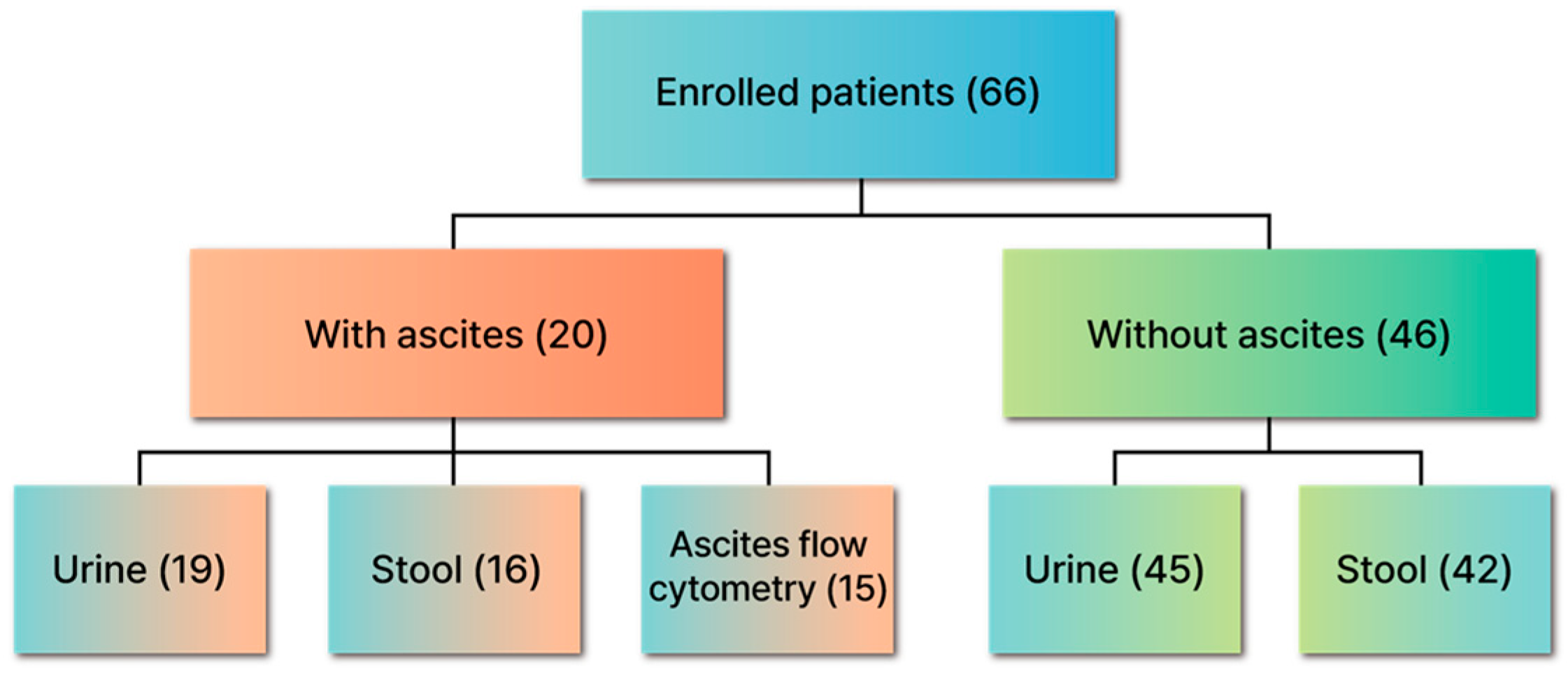
2.1. Patients and Study Protocol
2.2. DNA Extraction and 16S rDNA Sequencing
2.3. Bioinformatic Analysis of the Gut Microbiome and Statistical Analysis
2.4. Ethics Statement
3. Results
3.1. Baseline Characteristics of the Study Population
3.2. Malignant Ascites Typically Shows Minimal Bacterial Load
3.3. Gut Microbiome Shifts: Higher Diversity in Stage IV Colorectal Cancer
3.3.1. Overview and Subgroup Comparisons
3.3.2. Alpha Diversity: Stage IV vs. Stage I Colorectal Cancer
3.3.3. Beta Diversity: Subtle Changes Without Significant Clustering
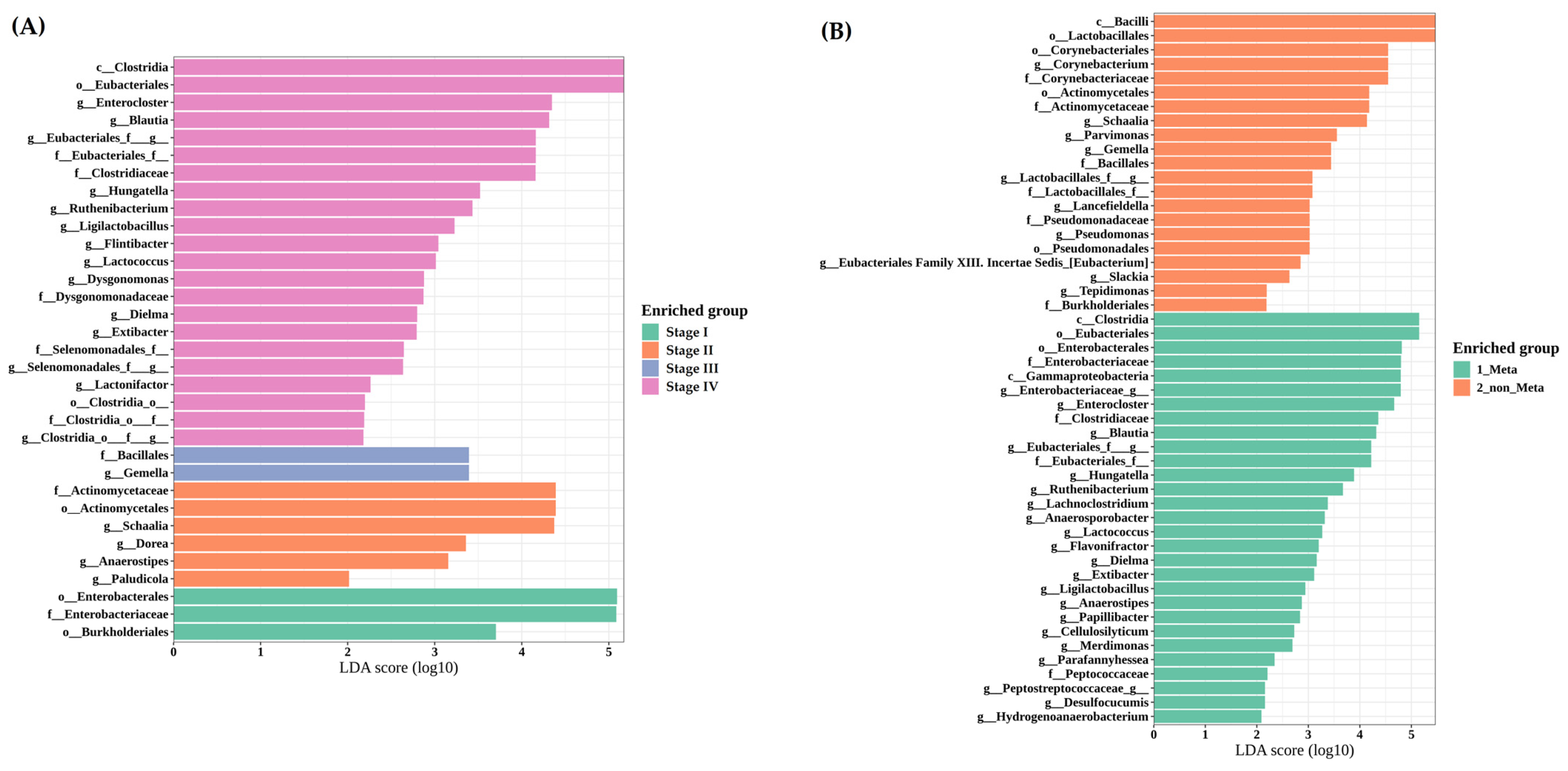
3.3.4. LEfSe Reveals Clostridia and Gammaproteobacteria Enrichment in Metastatic Cases
3.4. Urine Microbiome Remains Largely Unchanged Across Clinical Groups
3.4.1. Alpha Diversity: No Notable Variation by Cancer Type, Stage, or Ascites
3.4.2. Beta Diversity: Lack of Distinct Clusters Among Clinical Subgroups
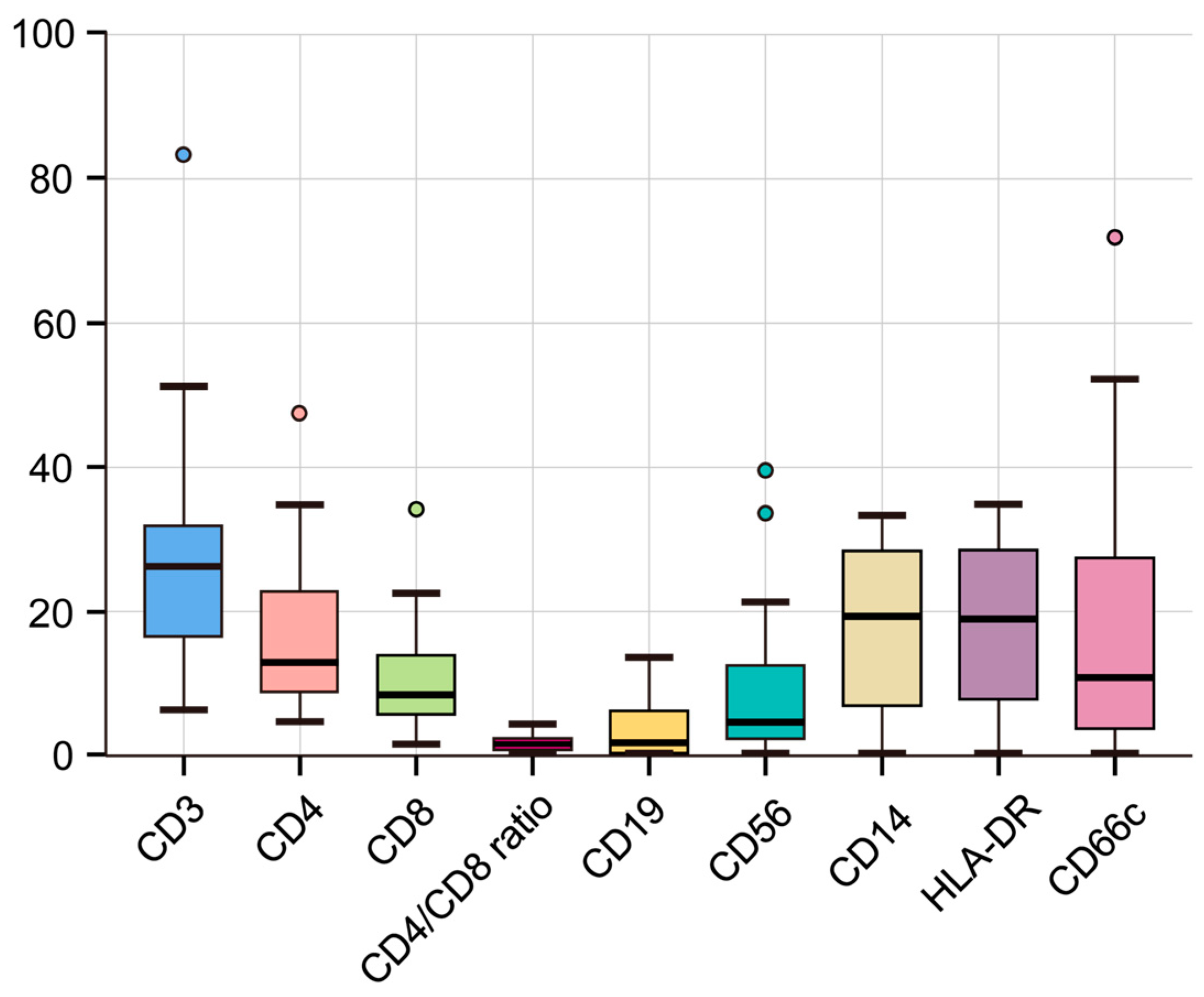
3.5. Flow Cytometric Analysis Reveals an Immunosuppressive Profile in Malignant Ascites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RAAS | renin–angiotensin–aldosterone system |
| VEGF | vascular endothelial growth factor |
| IL-2 | interleukin-2 |
| TNF-alpha | tumor necrosis factor-alpha |
| EMT | epithelial–mesenchymal transition |
| HLA-DR | human leukocyte antigen-DR |
| SBP | spontaneous bacterial peritonitis |
| LDA | linear discriminant analysis |
| LEfSe | linear discriminant analysis effect size |
| PCoA | principal coordinates analysis |
| PERMANOVA | permutational multivariate analysis of variance |
| NK | natural killer |
| SCFA | short-chain fatty acid |
| MHC | major histocompatibility complex |
References
- Runyon, B.A.; Hoefs, J.C.; Morgan, T.R. Ascitic fluid analysis in malignancy-related ascites. Hepatology 1988, 8, 1104–1109. [Google Scholar] [CrossRef]
- Becker, G.; Galandi, D.; Blum, H.E. Malignant ascites: Systematic review and guideline for treatment. Eur. J. Cancer 2006, 42, 589–597. [Google Scholar] [CrossRef] [PubMed]
- A Adam, R.; Adam, Y.G. Malignant ascites: Past, present, and future. J. Am. Coll. Surg. 2004, 198, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, Z.; Cui, L.; Wen, Y.; Chen, X.; Gong, F.; Yi, H. Opportunities and Challenges of the Human Microbiome in Ovarian Cancer. Front. Oncol. 2020, 10, 163. [Google Scholar] [CrossRef]
- Patel, M.; McAllister, M.; Nagaraju, R.; Al Badran, S.S.F.; Edwards, J.; McBain, A.J.; Barriuso, J.; Aziz, O. The intestinal microbiota in colorectal cancer metastasis—Passive observer or key player? Crit. Rev. Oncol. 2022, 180, 103856. [Google Scholar] [CrossRef] [PubMed]
- Colella, M.; Topi, S.; Palmirotta, R.; D’agostino, D.; Charitos, I.A.; Lovero, R.; Santacroce, L. An Overview of the Microbiota of the Human Urinary Tract in Health and Disease: Current Issues and Perspectives. Life 2023, 13, 1486. [Google Scholar] [CrossRef]
- Wagner, P.L.; Knotts, C.M.; Donneberg, V.S.; Dadgar, N.; Pico, C.X.C.; Xiao, K.; Zaidi, A.; Schiffman, S.C.; Allen, C.J.; Donnenberg, A.D.; et al. Characterizing the Immune Environment in Peritoneal Carcinomatosis: Insights for Novel Immunotherapy Strategies. Ann. Surg. Oncol. 2023, 31, 2069–2077. [Google Scholar] [CrossRef]
- Yang, C.J.; Song, J.S.; Yoo, J.-J.; Park, K.W.; Yun, J.; Kim, S.G.; Kim, Y.S. 16S rRNA Next-Generation Sequencing May Not Be Useful for Examining Suspected Cases of Spontaneous Bacterial Peritonitis. Medicina 2024, 60, 289. [Google Scholar] [CrossRef]
- Yoo, J.-J.; Song, J.S.; Bin Kim, W.; Yun, J.; Shin, H.B.; Jang, M.-A.; Ryu, C.B.; Kim, S.S.; Chung, J.C.; Kuk, J.C.; et al. Gardnerella vaginalis in Recurrent Urinary Tract Infection Is Associated with Dysbiosis of the Bladder Microbiome. J. Clin. Med. 2022, 11, 2295. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Ghorbani, E.; Avan, A.; Ryzhikov, M.; Ferns, G.; Khazaei, M.; Soleimanpour, S. Role of lactobacillus strains in the management of colorectal cancer: An overview of recent advances. Nutrition 2022, 103–104, 111828. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.; Zeng, T.; Zhang, Y.-H.; Liu, D.; Li, H.; Huang, T.; Cai, Y.-D. Identifying Robust Microbiota Signatures and Interpretable Rules to Distinguish Cancer Subtypes. Front. Mol. Biosci. 2020, 7, 604794. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Liu, G.; Tang, J.; Zhou, J.; Dong, M. Short-chain fatty acids play a positive role in colorectal cancer. Discov. Oncol. 2024, 15, 425. [Google Scholar] [CrossRef]
- Anderson, S.M.; Sears, C.L. The Role of the Gut Microbiome in Cancer: A Review, With Special Focus on Colorectal Neoplasia and Clostridioides difficile. Clin. Infect. Dis. 2023, 77, S471–S478. [Google Scholar] [CrossRef]
- Kiyici, M.; Nak, S.G.; Budak, F.; Gurel, S.; Oral, B.; Dolar, E.; Gulten, M. Lymphocyte subsets and cytokines in ascitic fluid of decompensated cirrhotic patients with and without spontaneous ascites infection. J. Gastroenterol. Hepatol. 2006, 21, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Nunes, D.L.; Mendes-Frias, A.; Silvestre, R.; Dinis-Oliveira, R.J.; Ricardo, S. Immune Tumor Microenvironment in Ovarian Cancer Ascites. Int. J. Mol. Sci. 2022, 23, 10692. [Google Scholar] [CrossRef] [PubMed]
- Rickard, B.P.; Conrad, C.; Sorrin, A.J.; Ruhi, M.K.; Reader, J.C.; Huang, S.A.; Franco, W.; Scarcelli, G.; Polacheck, W.J.; Roque, D.M.; et al. Malignant Ascites in Ovarian Cancer: Cellular, Acellular, and Biophysical Determinants of Molecular Characteristics and Therapy Response. Cancers 2021, 13, 4318. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, N. The Gut-Peritoneum Axis in Peritoneal Dialysis and Peritoneal Fibrosis. Kidney Med. 2023, 5, 100645. [Google Scholar] [CrossRef]


| Characteristic | n = 66 |
|---|---|
| Age, years | |
| Mean ± SD (range) | 64.79 ± 10.84 (31–87) |
| Sex | |
| Male | 33 (50.0%) |
| Female | 33 (50.0%) |
| Solid malignancies | |
| Colorectal cancer | 48 (72.7%) |
| Gastric cancer | 6 (9.1%) |
| Ovary cancer | 10 (15.2%) |
| Others | 2 (3.0%) |
| Stage | |
| I/II | 18 (27.3%) |
| III | 19 (28.8%) |
| IV | 29 (43.9%) |
| Group | |
| With ascites | 20 (30.3%) |
| Without ascites | 46 (69.7%) |
| Peritoneal metastases | 27 (40.9%) |
| Pathologic confirmed | 12 (60.0%) |
| Atypical cell | 6 (30.0%) |
| Pathological negative | 2 (10.0%) |
| No. | Sex | Age | Cancer Type | Ascites 16sR | Ascites Culture | Ascites WBC | Ascites PMN | Ascites CEA |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 72 | Urachal cancer | Too low bac load | 648 | 45 | 4438 | |
| 2 | F | 68 | Ovary cancer | Too low bac load | 6768 | 5617 | 495,370 | |
| 3 | F | 53 | Ovary cancer | Too low bac load | 1040 | 21 | 0.33 | |
| 4 | F | 78 | Endometrial cancer | Too low bac load | 140 | 1 | 0.36 | |
| 5 | F | 65 | Colon cancer | Enterococcus (50.9%), Bacteroides (18.1%) | Enterococcus faecalis | 680 | 265 | 5896.0 |
| 6 | F | 56 | Cervical cancer | Too low bac load | 290 | 0 | NA | |
| 7 | M | 35 | AGC | Too low bac load | 3300 | 0 | 2049 | |
| 8 | F | 65 | Primary peritoneal cancer | Too low bac load | 1120 | 22 | 2.42 | |
| 9 | F | 82 | Extrapulmonary NET | Too low bac load | 1080 | 259 | 0.86 | |
| 10 | M | 81 | Cecal cancer | Too low bac load | 900 | 18 | 4954 | |
| 11 | F | 70 | Ovary cancer | Too low bac load | 396 | 4 | 1.35 | |
| 12 | F | 46 | Colon cancer | Too low bac load | 3600 | 3348 | 27.3 | |
| 13 | F | 58 | Colon cancer | Too low bac load | 190 | 6 | 217 | |
| 14 | M | 63 | Appendiceal cancer | Too low bac load | 980 | 59 | 114 | |
| 15 | M | 62 | Colon cancer | Too low bac load | 310 | 0 | 105 | |
| 16 | M | 65 | AGC | Too low bac load | 360 | 4 | 3937 | |
| 17 | M | 70 | AGC | Too low bac load | 3816 | 2519 | 271 | |
| 18 | F | 57 | AGC | Too low bac load | 8 | 0 | 68.5 | |
| 19 | F | 59 | AGC | Too low bac load | 48 | 0 | 1268 | |
| 20 | M | 56 | AGC | Too low bac load | 369 | 4 | 301 |
| n = 15 | |
|---|---|
| Age, years | |
| Mean ± SD (range) | 61.87 ± 12.74 (35–82) |
| Sex | |
| Male | 7 (46.67%) |
| Female | 8 (53.33%) |
| Flow cytometry | Median (IQR) |
| CD3 | 26.03 (17.21, 31.67) |
| CD4 | 12.81 (9.54, 21.39) |
| CD8 | 8.37 (5.87, 13.12) |
| CD4/CD8 ratio | 1.63 (0.98, 2.4) |
| CD19 | 2.23 (0.52, 6.1) |
| CD56 | 4.64 (3, 11.13) |
| HLA-DR | 18.86 (10.38, 26.74) |
| CD66c | 11.07 (4, 26.55) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, J.; Song, J.-S.; Yoo, J.-J.; Kweon, S.; Choi, Y.-Y.; Lim, D.; Kuk, J.-C.; Kim, H.-J.; Park, S.-K. Microbial and Immune Landscape of Malignant Ascites: Insights from Gut, Bladder, and Ascitic Fluid Analyses. Cancers 2025, 17, 1280. https://doi.org/10.3390/cancers17081280
Yun J, Song J-S, Yoo J-J, Kweon S, Choi Y-Y, Lim D, Kuk J-C, Kim H-J, Park S-K. Microbial and Immune Landscape of Malignant Ascites: Insights from Gut, Bladder, and Ascitic Fluid Analyses. Cancers. 2025; 17(8):1280. https://doi.org/10.3390/cancers17081280
Chicago/Turabian StyleYun, Jina, Ju-Sun Song, Jeong-Ju Yoo, Solbi Kweon, Yoon-Young Choi, Daero Lim, Jung-Cheol Kuk, Hyun-Jung Kim, and Seong-Kyu Park. 2025. "Microbial and Immune Landscape of Malignant Ascites: Insights from Gut, Bladder, and Ascitic Fluid Analyses" Cancers 17, no. 8: 1280. https://doi.org/10.3390/cancers17081280
APA StyleYun, J., Song, J.-S., Yoo, J.-J., Kweon, S., Choi, Y.-Y., Lim, D., Kuk, J.-C., Kim, H.-J., & Park, S.-K. (2025). Microbial and Immune Landscape of Malignant Ascites: Insights from Gut, Bladder, and Ascitic Fluid Analyses. Cancers, 17(8), 1280. https://doi.org/10.3390/cancers17081280






