Impact of Tyrosine Kinase Inhibitors on the Expression Pattern of Epigenetic Regulators
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GEO Database
2.2. Data Visualization and Statistical Analysis
3. Results
3.1. Comparison of Expression Levels of Epigenetic Regulators Among Distinctive Subgroups of Malignant Diseases W/O Treatment
3.1.1. Distinctive Subgroups of a Solid Tumor: GSE37418. Novel Mutations Target Distinct Subgroups of Medulloblastoma
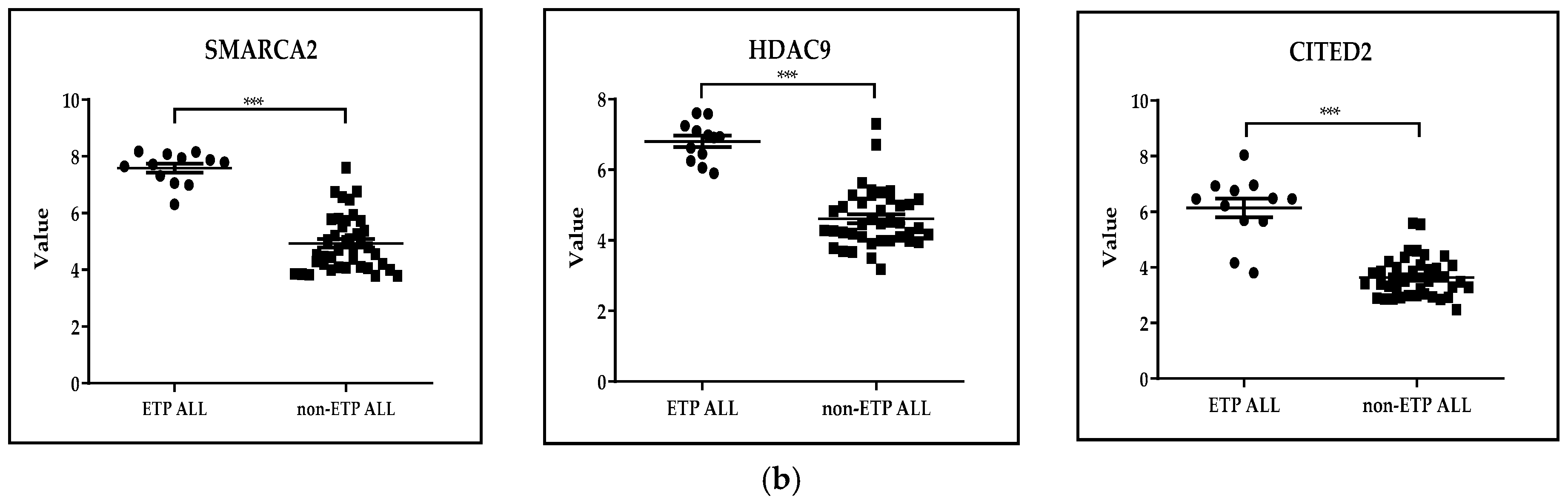
3.1.2. Distinctive Subgroups of a Hematological Malignant Disease: GSE28703. Discovery of Novel Recurrent Mutations and Rearrangements in Early T-Cell Precursor Acute Lymphoblastic Leukemia by Whole Genome Sequencing
3.2. Comparison of Expression Levels of Epigenetic Regulators Among Distinctive Subgroups TKI-Treated and Untreated Subgroups of Malignant Diseases
3.2.1. Solid Tumors
GSE66346: Expression Data from Renal Cancer Xenograft Tumor Treated with Sunitinib or Vehicle
GSE197555: Differential mRNA Expression Analysis of H460 Cells and A549 Cells After Trametinib Treatment
GSE59357: Gene Expression Profiles of Dasatinib-Resistant and Dasatinib-Sensitive Pancreatic Cancer Cell Lines
GSE42872: Expression Data from BRAFV600E A375 Melanoma Cells Treated with Vehicle or Vemurafenib
GSE98314: Melanoma Cell Lines Treated with Dabrafenib ± Trametinib
3.2.2. Leukemias and Lymphomas
GSE23743: Effect of Imatinib on Philadelphia Chromosome Positive Acute Lymphoblastic Leukemia
GSE24493: Effect of Imatinib on Chronic Myelogenous Leukemia
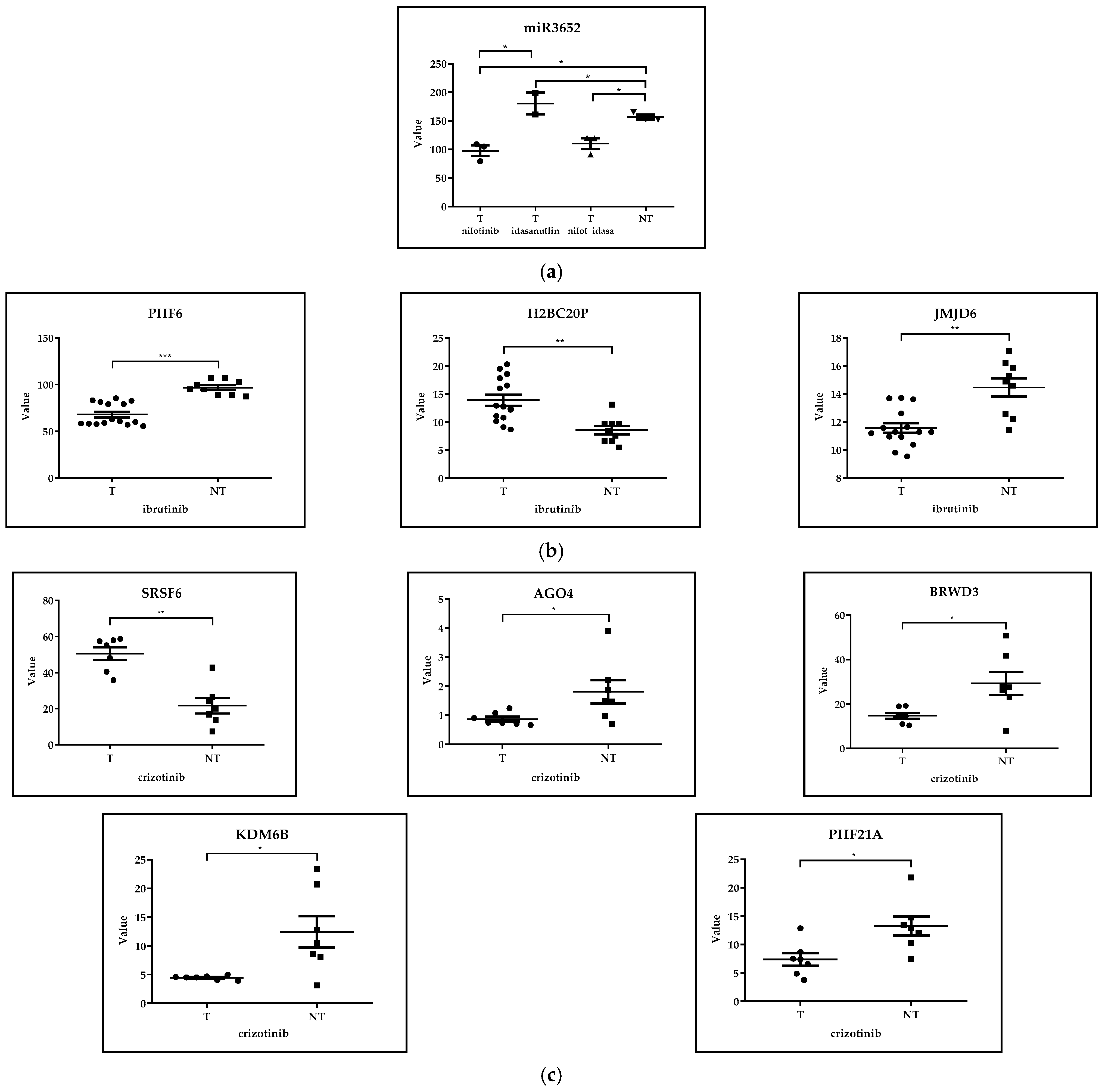
GSE218183: Bulk RNA-Seq Analysis of Primary CML CD34+ Cells (n = 3) Treated with Idasanutlin Alone or in Combination with Nilotinib In Vitro
GSE197811: Effect of the Janus Kinase Inhibitor Ruxolitinib on Gene Expression of Chronic Lymphocytic Leukemia Cells In Vivo
GSE171763: Inhibitors of Bcl-2 and Bruton’s Tyrosine Kinase Synergize to Abrogate Diffuse Large B-Cell Lymphoma (DLBCL) Growth
GSE173306: Transcription Profiling of ALK-Rearranged Cell Lines Resistant and Sensitive to Crizotinib
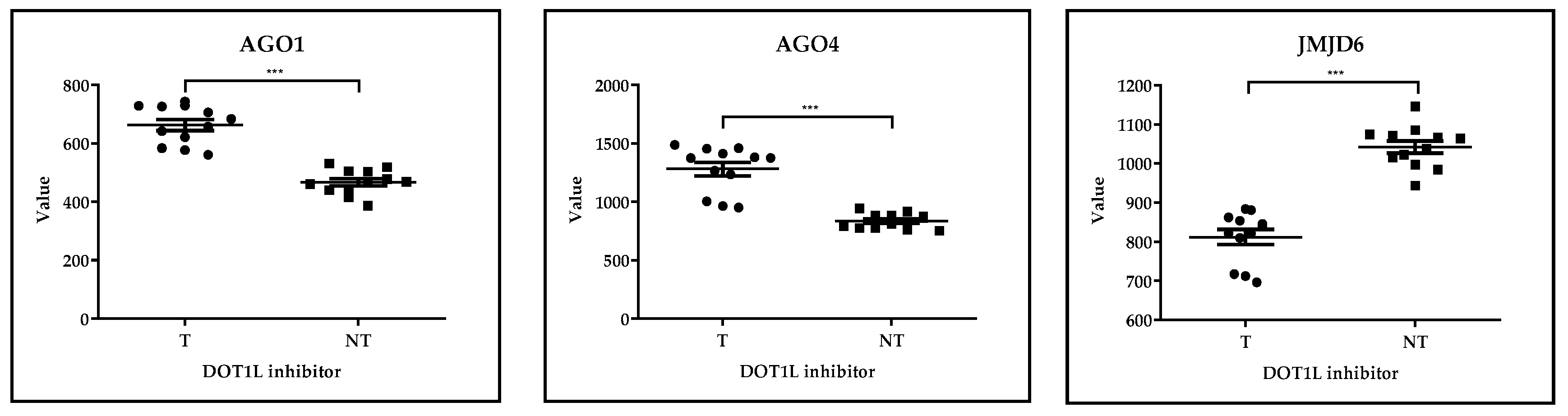
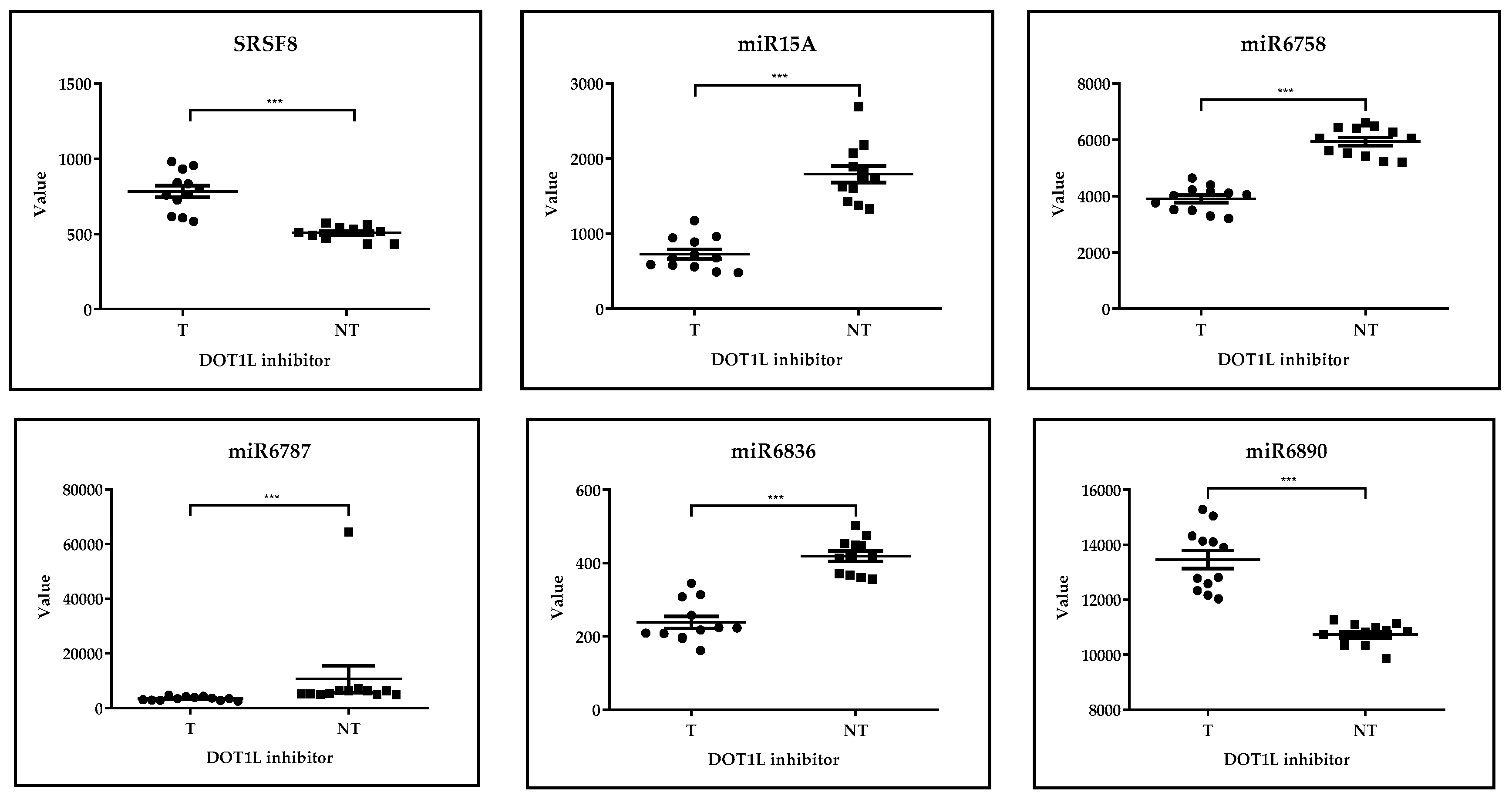
GSE29828: Expression Profiling of Mixed Lineage Leukemia Cells Treated with a Potent Small-Molecule DOT1L Inhibitor
3.3. Summary of Epigenetic Regulators Modulated by TKI Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALCL | anaplastic large cell lymphoma |
| AGO | argonaute protein |
| AIRE | autoimmune regulator |
| ALL | acute lymphoblastic leukemia |
| AML | acute myeloid leukemia |
| ASH/KMT | lysine-specific methyltransferase |
| ATRX | alpha-thalassemia/mental retardation, X-linked; X-linked nuclear protein |
| BAF | Barrier to Autointegration Factor |
| BAZ | bromodomain adjacent to zinc finger |
| BET | bromodomain and extra-terminal containing protein family |
| BHC80/PHF21A | PHD Finger Protein |
| Bmi1 | (ub)/PCGF4 polycomb group RING finger protein 4 |
| BRD | bromodomain containing family |
| BRG/SMARCA | Brahma Related Gene, SWI/SNF Related BAF Chromatin Remodeling Complex |
| BRM | bromodomains of the mammalian ATPases Brahma |
| BRPF1 | bromodomain- and PHD finger-containing protein 1 |
| BRWD | bromodomain And WD Repeat Domain |
| BTK | bruton Tyrosine Kinase |
| CBP | CREB Binding Protein |
| CDK | cyclin Dependent Kinase |
| CHAF1A | chromatin Assembly Factor 1 Subunit A |
| CHD | chromodomain helicase DNA-binding |
| CHRAC | chromatin accessibility complex |
| CITED | cbp/P300 Interacting Transactivator With Glu/Asp Rich Carboxy-Terminal Domain |
| CK2 | casein Kinase2 |
| CLL | chronic lymphoblastic leukemia |
| CML | chronic myeloid leukemia |
| Cps35/SWD2 | component of COMPASS (complex of proteins associated with Set1) |
| DICER | dicer 1: Ribonuclease III |
| DLBCL | diffuse large B-cell lymphoma |
| DNMT | DNA Methyltransferase |
| DNA | deoxyribonucleic acid |
| DOT1L | DOT1 Like Histone Lysine Methyltransferase |
| DPY30 | Dpy-30 Histone Methyltransferase Complex Regulatory Subunit |
| Drosha | drosha Ribonuclease III |
| EAF3 | essential Sas2-related acetyltransferase1-associated factor 3 |
| EED | embryonic Ectoderm Development |
| EGFR | epidermal Growth Factor Receptor |
| EHMT | euchromatic histone methyltransferase |
| ELP3 | Elongator Acetyltransferase Complex Subunit 3 |
| ESA1 | essential SAS2-related acetyltransferase |
| ETP | early T-cell precursor |
| EZH2 | Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit |
| FDA | Food and Drug Administration |
| FLT3 | Fms Related Receptor Tyrosine Kinase 3 |
| G9a/EHMT2 | euchromatin-localized histone methyltransferase 2 |
| Gcn/MTTA | Mitochondrially Encoded TRNA-Ala |
| GCNT1P1 | Glucosaminyl (N-Acetyl) Transferase 1 Pseudogene 1 |
| GEO | Gene Expression Omnibus |
| GLP | Glucagon Like Peptide |
| H2BC20P | H2B Clustered Histone 20, Pseudogene |
| H4C1 | H4 Clustered Histone 1 |
| hACF1 | ATP-utilizing chromatin assembly and remodeling factor 1 |
| HAT | Histone acetyltransferases |
| HBO1/KAT7 | Lysine Acetyltransferase 7 |
| HDAC | Histone Deacetylase |
| HIST | histone |
| HPA2 | heparanase-2 |
| HSC | hematopoietic stem cell |
| ING2 | Inhibitor Of Growth Family Member 2 |
| Ini1 | integrase interactor 1 |
| INO80 | INO80 Complex ATPase Subunit |
| ISWI | imitation SWItch |
| ITD | internal tandem duplications |
| JAK | Janus Kinase |
| JARID | Jumonji-AT Rich Interactive Domain |
| JHDM | Jumonji-C domain histone demethylase |
| JMJD | Jumonji C domain-containing protein |
| KAT2B | Lysine Acetyltransferase 2B. |
| KDM | Lysine Demethylase |
| KMT | Lysine Methyl Transferases |
| LncRNA | long non-coding RNA |
| LSD | lysergic acid diethylamide |
| MBD | methyl-CpG Binding Domain |
| MDC1 | Mediator Of DNA Damage Checkpoint 1 |
| MeCP | Methyl-CpG Binding Protein |
| Mi-2 | Menin-MLL inhibitor 2 |
| miR | micro RNA |
| miRNA | microRNA |
| MLL/KMT2A | mixed lineage leukemia, Lysine Methyltransferase 2A |
| MOF/MYST1/KAT8 | males absent on the first |
| MORF | monocytic leukemia zinc-finger protein-related factor |
| MOZ | monocytic leukemic zinc finger |
| NCoR | Nuclear Receptor Corepressor |
| NSCLC | non-small cell lung cancer |
| NSD | Nuclear Receptor Binding SET Domain Protein |
| NURD | Nucleosome Remodeling and Deacetylase |
| OS | overall survival |
| p300 | Histone acetyltransferase p300 |
| PB1 | protein polybromo 1 |
| PCAF | P300/CBP-associated factor |
| PHF | PHD Finger Protein |
| piRNA | PIWI-interacting RNAs |
| PRC | polycomb repressive complex |
| PRDM | PR/SET Domain |
| PRMT | Protein Arginine Methyltransferase |
| RCC | renal cell carcinoma |
| RNA | deoxyribonucleic acid |
| RTK | Receptor tyrosine kinase |
| Sas | something about silencing |
| SCLC | small cell lung cancer |
| SETD | SET Domain Containing |
| SIRT | sirtuin |
| SMARC | SWI/SNF Related BAF Chromatin Remodeling Complex |
| Snf | Sucrose Non-Fermenting |
| SP1 | Sp1 Transcription Factor |
| SRSF | Serine/arginine-rich splicing factor |
| SUV | Suppressor of variegation |
| SUZ | suppressor of zeste |
| Swi | SWItch |
| Swp | SWIRM domain PAO protein |
| SWR1 | SWI2/SNF2-Related 1 Chromatin Remodeling Complex |
| TAF | TATA-Box Binding Protein Associated Factor. |
| TDRD3 | Tudor Domain Containing 3 |
| TET | Tet Methylcytosine Dioxygenase |
| Tip60 | Tat interactive protein 60 |
| TKI | Tyrosine kinase inhibitor |
| TRIM | Tripartite motif proteins |
| UHRF | Ubiquitin Like With PHD And Ring Finger |
| UTF | Undifferentiated Embryonic Cell Transcription Factor |
| YBF | part of the MYST (histone acetyltransferases) complex |
References
- Quaresma, M.; Coleman, M.P.; Rachet, B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971-2011: A population-based study. Lancet 2015, 385, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Tonorezos, E.; Devasia, T.; Mariotto, A.B.; Mollica, M.A.; Gallicchio, L.; Green, P.; Doose, M.; Brick, R.; Streck, B.; Reed, C.; et al. Prevalence of cancer survivors in the United States. J. Natl. Cancer Inst. 2024, 116, 1784–1790. [Google Scholar] [CrossRef]
- Botta, L.; Gatta, G.; Capocaccia, R.; Stiller, C.; Cañete, A.; Dal Maso, L.; Innos, K.; Mihor, A.; Erdmann, F.; Spix, C.; et al. Long-term survival and cure fraction estimates for childhood cancer in Europe (EUROCARE-6): Results from a population-based study. Lancet Oncol. 2022, 23, 1525–1536. [Google Scholar] [CrossRef]
- Casolino, R.; Beer, P.A.; Chakravarty, D.; Davis, M.B.; Malapelle, U.; Mazzarella, L.; Normanno, N.; Pauli, C.; Subbiah, V.; Turnbull, C.; et al. Interpreting and integrating genomic tests results in clinical cancer care: Overview and practical guidance. CA Cancer J. Clin. 2024, 74, 264–285. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, B.J. Imaging and Interventional Radiology for Cancer Management. Surg. Clin. N. Am. 2020, 100, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, N.; Fardi, E.; Ghaderi, H.; Palizdar, S.; Khorram, R.; Vafadar, R.; Ghanaatian, M.; Rezaei-Tazangi, F.; Baziyar, P.; Ahmadi, A.; et al. Receptor tyrosine kinase inhibitors in cancer. Cell Mol. Life Sci. 2023, 80, 104. [Google Scholar] [CrossRef]
- Pottier, C.; Fresnais, M.; Gilon, M.; Jérusalem, G.; Longuespée, R.; Sounni, N.E. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers 2020, 12, 731. [Google Scholar] [CrossRef]
- Hussain, S.; Mursal, M.; Verma, G.; Hasan, S.M.; Khan, M.F. Targeting oncogenic kinases: Insights on FDA approved tyrosine kinase inhibitors. Eur. J. Pharmacol. 2024, 970, 176484. [Google Scholar] [CrossRef]
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Müller, S.; Knapp, S. SH2 domains: Modulators of nonreceptor tyrosine kinase activity. Curr. Opin. Struct. Biol. 2009, 19, 643–649. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Shyam Sunder, S.; Sharma, U.C.; Pokharel, S. Adverse effects of tyrosine kinase inhibitors in cancer therapy: Pathophysiology, mechanisms and clinical management. Signal Transduct. Target. Ther. 2023, 8, 262. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Wang, Z.; Feng, Y.; Jia, Y.; Jiang, L.; Xia, Y.; Cao, J.; Liu, Y. Comparison of Hepatotoxicity Associated With New BCR-ABL Tyrosine Kinase Inhibitors vs Imatinib Among Patients With Chronic Myeloid Leukemia: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2120165. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hu, R.; Han, M.; Tan, Y.; Xie, M.; Gao, S.; Hu, J.F. Mechanisms underlying therapeutic resistance of tyrosine kinase inhibitors in chronic myeloid leukemia. Int. J. Biol. Sci. 2024, 20, 175–181. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef]
- Bouffet, E.; Hansford, J.R.; Garrè, M.L.; Hara, J.; Plant-Fox, A.; Aerts, I.; Locatelli, F.; van der Lugt, J.; Papusha, L.; Sahm, F.; et al. Dabrafenib plus Trametinib in Pediatric Glioma with BRAF V600 Mutations. N. Engl. J. Med. 2023, 389, 1108–1120. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wang, N.; Lam, W.; Guo, W.; Feng, Y.; Cheng, Y.C. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol. Cancer 2018, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Varlamova, E.G.; Goltyaev, M.V.; Simakin, A.V.; Gudkov, S.V.; Turovsky, E.A. Comparative Analysis of the Cytotoxic Effect of a Complex of Selenium Nanoparticles Doped with Sorafenib, “Naked” Selenium Nanoparticles, and Sorafenib on Human Hepatocyte Carcinoma HepG2 Cells. Int. J. Mol. Sci. 2022, 23, 6641. [Google Scholar] [CrossRef]
- Ladan, D.; Mahla, D.; Mahmoud, A.; Somayeh, T.; Azade, T. Preparation and evaluation of targeted albumin lipid nanoparticles with lactobionic acid for targeted drug delivery of sorafenib in hepatocellular carcinoma. J. Drug Deliv. Sci. Technol. 2022, 69, 103142. [Google Scholar] [CrossRef]
- Kurtze, I.; Sonnemann, J.; Beck, J.F. KRAS-mutated non-small cell lung cancer cells are responsive to either co-treatment with erlotinib or gefitinib and histone deacetylase inhibitors or single treatment with lapatinib. Oncol. Rep. 2011, 25, 1021–1029. [Google Scholar] [CrossRef][Green Version]
- Yang, Z.; Liu, C.; Hu, Y.; Liu, H.; Li, J.; Wu, L.; Liu, Q.; Zheng, Y.; Huang, P.; Wang, Y. Tyrosine kinase inhibitors combined with venetoclax and azacytidine as an effective therapy for de novo lymphoid blast phase-chronic myeloid leukemia. Leuk. Res. 2023, 127, 107039. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Morfouace, M.; Cheepala, S.; Jackson, S.; Fukuda, Y.; Patel, Y.T.; Fatima, S.; Kawauchi, D.; Shelat, A.A.; Stewart, C.F.; Sorrentino, B.P.; et al. ABCG2 Transporter Expression Impacts Group 3 Medulloblastoma Response to Chemotherapy. Cancer Res. 2015, 75, 3879–3889. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Gutierrez, A.; Kentsis, A.; Sanda, T.; Holmfeldt, L.; Chen, S.C.; Zhang, J.; Protopopov, A.; Chin, L.; Dahlberg, S.E.; Neuberg, D.S.; et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 2011, 118, 4169–4173. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, N.; Yamasaki, T.; Kanno, T.; Arakaki, R.; Sakamoto, H.; Utsunomiya, N.; Inoue, T.; Tsuruyama, T.; Nakamura, E.; Ogawa, O.; et al. Role of IL13RA2 in Sunitinib Resistance in Clear Cell Renal Cell Carcinoma. PLoS ONE 2015, 10, e0130980. [Google Scholar] [CrossRef]
- Feng, J.; Lian, Z.; Xia, X.; Lu, Y.; Hu, K.; Zhang, Y.; Liu, T.; Hu, L.; Yuan, K.; Sun, C.; et al. Targeting metabolic vulnerability in mitochondria conquers MEK inhibitor resistance in KRAS-mutant lung cancer. Acta Pharm. Sin. B 2023, 13, 1145–1163. [Google Scholar] [CrossRef]
- Chien, W.; Sun, Q.Y.; Lee, K.L.; Ding, L.W.; Wuensche, P.; Torres-Fernandez, L.A.; Tan, S.Z.; Tokatly, I.; Zaiden, N.; Poellinger, L.; et al. Activation of protein phosphatase 2A tumor suppressor as potential treatment of pancreatic cancer. Mol. Oncol. 2015, 9, 889–905. [Google Scholar] [CrossRef]
- Parmenter, T.J.; Kleinschmidt, M.; Kinross, K.M.; Bond, S.T.; Li, J.; Kaadige, M.R.; Rao, A.; Sheppard, K.E.; Hugo, W.; Pupo, G.M.; et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014, 4, 423–433. [Google Scholar] [CrossRef]
- GEO Database/GSE98314. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse98314 (accessed on 15 February 2025).
- Duy, C.; Hurtz, C.; Shojaee, S.; Cerchietti, L.; Geng, H.; Swaminathan, S.; Klemm, L.; Kweon, S.-M.; Nahar, R.; Braig, M.; et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 2011, 473, 384–388. [Google Scholar] [CrossRef]
- Hurtz, C.; Hatzi, K.; Cerchietti, L.; Braig, M.; Park, E.; Kim, Y.M.; Herzog, S.; Ramezani-Rad, P.; Jumaa, H.; Müller, M.C.; et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med. 2011, 208, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.T.; Liu, W.; Mitchell, R.; Clarke, C.J.; Kinstrie, R.; Warren, F.; Almasoudi, H.; Stevens, T.; Dunn, K.; Pritchard, J.; et al. Activating p53 abolishes self-renewal of quiescent leukaemic stem cells in residual CML disease. Nat. Commun. 2024, 15, 651. [Google Scholar] [CrossRef] [PubMed]
- Spaner, D.E.; Luo, T.Y.; Wang, G.; Schreiber, G.; Harari, D.; Shi, Y. Paradoxical activation of chronic lymphocytic leukemia cells by ruxolitinib in vitro and in vivo. Front. Oncol. 2023, 13, 1043694. [Google Scholar] [CrossRef]
- Bertram, K.; Leary, P.J.; Boudesco, C.; Fullin, J.; Stirm, K.; Dalal, V.; Zenz, T.; Tzankov, A.; Müller, A. Inhibitors of Bcl-2 and Bruton’s tyrosine kinase synergize to abrogate diffuse large B-cell lymphoma growth in vitro and in orthotopic xenotransplantation models. Leukemia 2022, 36, 1035–1047. [Google Scholar] [CrossRef]
- Mura, G.; Karaca Atabay, E.; Menotti, M.; Martinengo, C.; Ambrogio, C.; Giacomello, G.; Arigoni, M.; Olivero, M.; Calogero, R.A.; Chiarle, R.; et al. Regulation of CD45 phosphatase by oncogenic ALK in anaplastic large cell lymphoma. Front. Oncol. 2022, 12, 1085672. [Google Scholar] [CrossRef] [PubMed]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Majer, C.R.; Sneeringer, C.J.; Song, J.; Johnston, L.D.; Scott, M.P.; Smith, J.J.; Xiao, Y.; et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011, 20, 53–65. [Google Scholar] [CrossRef]
- Tsuboyama, N.; Szczepanski, A.P.; Zhao, Z.; Wang, L. MBD5 and MBD6 stabilize the BAP1 complex and promote BAP1-dependent cancer. Genome Biol. 2022, 23, 206. [Google Scholar] [CrossRef]
- Dasari, S.; Pandhiri, T.; Grassi, T.; Visscher, D.W.; Multinu, F.; Agarwal, K.; Mariani, A.; Shridhar, V.; Mitra, A.K. Signals from the Metastatic Niche Regulate Early and Advanced Ovarian Cancer Metastasis through miR-4454 Downregulation. Mol. Cancer Res. 2020, 18, 1202–1217. [Google Scholar] [CrossRef]
- Jain, N.; Lamb, A.V.; O’Brien, S.; Ravandi, F.; Konopleva, M.; Jabbour, E.; Zuo, Z.; Jorgensen, J.; Lin, P.; Pierce, S.; et al. Early T-cell precursor acute lymphoblastic leukemia/lymphoma (ETP-ALL/LBL) in adolescents and adults: A high-risk subtype. Blood 2016, 127, 1863–1869. [Google Scholar] [CrossRef]
- Coustan-Smith, E.; Mullighan, C.G.; Onciu, M.; Behm, F.G.; Raimondi, S.C.; Pei, D.; Cheng, C.; Su, X.; Rubnitz, J.E.; Basso, G.; et al. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009, 10, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Moreno, D.A.; Scrideli, C.A.; Cortez, M.A.; de Paula Queiroz, R.; Valera, E.T.; da Silva Silveira, V.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2010, 150, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Andrades, A.; Peinado, P.; Alvarez-Perez, J.C.; Sanjuan-Hidalgo, J.; García, D.J.; Arenas, A.M.; Matia-González, A.M.; Medina, P.P. SWI/SNF complexes in hematological malignancies: Biological implications and therapeutic opportunities. Mol. Cancer. 2023, 22, 39. [Google Scholar] [CrossRef]
- Korthuis, P.M.; Berger, G.; Bakker, B.; Rozenveld-Geugien, M.; Jaques, J.; de Haan, G.; Schuringa, J.J.; Vellenga, E.; Schepers, H. CITED2-mediated human hematopoietic stem cell maintenance is critical for acute myeloid leukemia. Leukemia 2015, 29, 625–635. [Google Scholar] [CrossRef]
- Saleeb, R.; Kim, S.S.; Ding, Q.; Scorilas, A.; Lin, S.; Khella, H.W.; Boulos, C.; Ibrahim, G.; Yousef, G.M. The miR-200 family as prognostic markers in clear cell renal cell carcinoma. Urol. Oncol. 2019, 37, 955–963. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Z.; Xu, W.; Liu, H.; Chang, J.; Xu, W.; Li, S.; Cao, S.; Hou, J. Circ-SAR1A Promotes Renal Cell Carcinoma Progression Through miR-382/YBX1 Axis. Cancer Manag. Res. 2020, 12, 7353–7361. [Google Scholar] [CrossRef]
- Bertulli, C.; Marzollo, A.; Doria, M.; Di Cesare, S.; La Scola, C.; Mencarelli, F.; Pasini, A.; Affinita, M.C.; Vidal, E.; Magini, P.; et al. Expanding Phenotype of Schimke Immuno-Osseous Dysplasia: Congenital Anomalies of the Kidneys and of the Urinary Tract and Alteration of NK Cells. Int. J. Mol. Sci. 2020, 21, 8604. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Cheng, Z.; Cui, S.; Mao, X.; Li, B.; Fu, Y.; Wang, H.; Jin, H.; Ye, Q.; Zhao, X.; et al. circFOXM1 promotes proliferation of non-small cell lung carcinoma cells by acting as a ceRNA to upregulate FAM83D. J. Exp. Clin. Cancer Res. 2020, 39, 55. [Google Scholar] [CrossRef]
- Xu, N.; Liu, F.; Wu, S.; Ye, M.; Ge, H.; Zhang, M.; Song, Y.; Tong, L.; Zhou, J.; Bai, C. CHD4 mediates proliferation and migration of non-small cell lung cancer via the RhoA/ROCK pathway by regulating PHF5A. BMC Cancer 2020, 20, 262. [Google Scholar] [CrossRef]
- Yavas, A.; Ozcan, K.; Adsay, N.V.; Balci, S.; Tarcan, Z.C.; Hechtman, J.F.; Luchini, C.; Scarpa, A.; Lawlor, R.T.; Mafficini, A.; et al. SWI/SNF Complex-Deficient Undifferentiated Carcinoma of the Pancreas: Clinicopathologic and Genomic Analysis. Mod. Pathol. 2024, 37, 100585. [Google Scholar] [CrossRef]
- Mziaut, H.; Henniger, G.; Ganss, K.; Hempel, S.; Wolk, S.; McChord, J.; Chowdhury, K.; Ravassard, P.; Knoch, K.P.; Krautz, C.; et al. MiR-132 controls pancreatic beta cell proliferation and survival through Pten/Akt/Foxo3 signaling. Mol. Metab. 2020, 31, 150–162. [Google Scholar] [CrossRef] [PubMed]
- De Martino, E.; Gandin, I.; Azzalini, E.; Massone, C.; Pizzichetta, M.A.; Giulioni, E.; Javor, S.; Pinzani, C.; Conforti, C.; Zalaudek, I.; et al. A group of three miRNAs can act as candidate circulating biomarkers in liquid biopsies from melanoma patients. Front Med. 2023, 10, 1180799. [Google Scholar] [CrossRef]
- Kuser-Abali, G.; Zhang, Y.; Szeto, P.; Zhao, P.; Masoumi-Moghaddam, S.; Fedele, C.G.; Leece, I.; Huang, C.; Cheung, J.G.; Ameratunga, M.; et al. UHRF1/UBE2L6/UBR4-mediated ubiquitination regulates EZH2 abundance and thereby melanocytic differentiation phenotypes in melanoma. Oncogene 2023, 42, 1360–1373. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Silva, J.M.; Oliveira, L.S.; Chiminazo, C.B.; Fonseca, R.; de Souza, C.V.E.; Aissa, A.F.; de Almeida Lima, G.D.; Ionta, M.; Castro-Gamero, A.M. WT161, a selective HDAC6 inhibitor, decreases growth, enhances chemosensitivity, promotes apoptosis, and suppresses motility of melanoma cells. Cancer Chemother. Pharmacol. 2025, 95, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Song, C.; Ding, Y.; Pan, X.; Ge, Z.; Tan, B.H.; Gowda, C.; Sachdev, M.; Muthusami, S.; Ouyang, H.; et al. Transcriptional Regulation of JARID1B/KDM5B Histone Demethylase by Ikaros, Histone Deacetylase 1 (HDAC1), and Casein Kinase 2 (CK2) in B-cell Acute Lymphoblastic Leukemia. J. Biol. Chem. 2016, 291, 4004–4018. [Google Scholar] [CrossRef]
- Mar, B.G.; Chu, S.H.; Kahn, J.D.; Krivtsov, A.V.; Koche, R.; Castellano, C.A.; Kotlier, J.L.; Zon, R.L.; McConkey, M.E.; Chabon, J.; et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 2017, 130, 2631–2641. [Google Scholar] [CrossRef]
- McLean, C.M.; Karemaker, I.D.; van Leeuwen, F. The emerging roles of DOT1L in leukemia and normal development. Leukemia 2014, 28, 2131–2138. [Google Scholar] [CrossRef]
- Adriaanse, F.R.S.; Schneider, P.; Arentsen-Peters, S.T.C.J.M.; Fonseca, A.M.N.D.; Stutterheim, J.; Pieters, R.; Zwaan, C.M.; Stam, R.W. Distinct Responses to Menin Inhibition and Synergy with DOT1L Inhibition in KMT2A-Rearranged Acute Lymphoblastic and Myeloid Leukemia. Int. J. Mol. Sci. 2024, 25, 6020. [Google Scholar] [CrossRef]
- Izaguirre-Carbonell, J.; Christiansen, L.; Burns, R.; Schmitz, J.; Li, C.; Mokry, R.L.; Bluemn, T.; Zheng, Y.; Shen, J.; Carlson, K.S.; et al. Critical role of Jumonji domain of JMJD1C in MLL-rearranged leukemia. Blood Adv. 2019, 3, 1499–1511. [Google Scholar] [CrossRef]
- Staehle, H.F.; Heinemann, J.; Gruender, A.; Omlor, A.M.; Pahl, H.L.; Jutzi, J.S. Jmjd1c is dispensable for healthy adult hematopoiesis and Jak2V617F-driven myeloproliferative disease initiation in mice. PLoS ONE 2020, 15, e0228362. [Google Scholar] [CrossRef]
- Chen, S.H.; Chow, J.M.; Hsieh, Y.Y.; Lin, C.Y.; Hsu, K.W.; Hsieh, W.S.; Chi, W.M.; Shabangu, B.M.; Lee, C.H. HDAC1,2 Knock-Out and HDACi Induced Cell Apoptosis in Imatinib-Resistant K562 Cells. Int. J. Mol. Sci. 2019, 20, 2271. [Google Scholar] [CrossRef]
- Jiang, Q.; Stachelscheid, J.; Bloehdorn, J.; Pacholewska, A.; Aszyk, C.; Grotenhuijs, F.; Müller, T.; Onder, O.; Wagle, P.; Herling, C.D.; et al. Oncogenic role and target properties of the lysine-specific demethylase KDM1A in chronic lymphocytic leukemia. Blood 2023, 142, 44–61. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, D.; Liu, H.; Huang, Y.; Meng, F.; Tang, J.; Li, Z.; Xie, W. PHF13 epigenetically activates TGFβ driven epithelial to mesenchymal transition. Cell Death Dis. 2022, 13, 487. [Google Scholar] [CrossRef]
- Li, S.; Young, K.H.; Medeiros, L.J. Diffuse large B-cell lymphoma. Pathology 2018, 50, 74–87. [Google Scholar] [CrossRef]
- Lawson, H.; Sepulveda, C.; van de Lagemaat, L.N.; Durko, J.; Barile, M.; Tavosanis, A.; Georges, E.; Shmakova, A.; Timms, P.; Carter, R.N.; et al. JMJD6 promotes self-renewal and regenerative capacity of hematopoietic stem cells. Blood Adv. 2021, 5, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Eisa, Y.A.; Guo, Y.; Yang, F.C. The Role of PHF6 in Hematopoiesis and Hematologic Malignancies. Stem Cell Rev. Rep. 2023, 19, 67–75. [Google Scholar] [CrossRef]
- Mathur, R.; Sehgal, L.; Havranek, O.; Köhrer, S.; Khashab, T.; Jain, N.; Burger, J.A.; Neelapu, S.S.; Davis, R.E.; Samaniego, F. Inhibition of demethylase KDM6B sensitizes diffuse large B-cell lymphoma to chemotherapeutic drugs. Haematologica 2017, 102, 373–380. [Google Scholar] [CrossRef]
- Issa, N.; Bjeije, H.; Wilson, E.R.; Krishnan, A.; Dunuwille, W.M.B.; Parsons, T.M.; Zhang, C.R.; Han, W.; Young, A.L.; Ren, Z.; et al. KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress. Leukemia 2023, 37, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Maienschein-Cline, M.; Maffucci, P.; Veselits, M.; Kennedy, D.E.; McLean, K.C.; Okoreeh, M.K.; Karki, S.; Cunningham-Rundles, C.; Clark, M.R. BRWD1 orchestrates epigenetic landscape of late B lymphopoiesis. Nat. Commun. 2018, 9, 3888. [Google Scholar] [CrossRef]
- Liao, Q.; Wang, B.; Li, X.; Jiang, G. miRNAs in acute myeloid leukemia. Oncotarget 2017, 8, 3666–3682. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Saenz, D.T.; Fiskus, W.; Mill, C.P.; Perera, D.; Manshouri, T.; Lara, B.H.; Karkhanis, V.; Sharma, S.; Horrigan, S.K.; Bose, P.; et al. Mechanistic basis and efficacy of targeting the β-catenin-TCF7L2-JMJD6-c-Myc axis to overcome resistance to BET inhibitors. Blood 2020, 135, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Bollaert, E.; Claus, M.; Vandewalle, V.; Lenglez, S.; Essaghir, A.; Demoulin, J.B.; Havelange, V. MiR-15a-5p Confers Chemoresistance in Acute Myeloid Leukemia by Inhibiting Autophagy Induced by Daunorubicin. Int. J. Mol. Sci. 2021, 22, 5153. [Google Scholar] [CrossRef]
- Li, F.; Zhao, C.; Wang, L. Molecular-targeted agents combination therapy for cancer: Developments and potentials. Int. J. Cancer 2014, 134, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Petrazzuolo, A.; Maiuri, M.C.; Zitvogel, L.; Kroemer, G.; Kepp, O. Trial Watch: Combination of tyrosine kinase inhibitors (TKIs) and immunotherapy. Oncoimmunology 2022, 11, 2077898. [Google Scholar] [CrossRef]


| Writer | Eraser | Reader | ncRNA | Chromatin Remodeling |
| DNA methyltransferase DNMT DNA hydroxymethylase TET Histone acetyltransferase CBP ELP3 ESA1 Gcn HAT HBO1 HPA2 MOF MORF MOZ p300 PCAF Sas Tip60 YBF Histone methyltransferase ASH1L Bmi1 (ub) DOT1L EHMT EZH2 G9a GLP KMT MLL NSD PRDM PRMT SETD SUV39H SUV420H | Histone demethylase JARID JHDM JMJD KDM LSD PHF UTF Histone deacetylase HDAC SIRT | AIRE ASH ATRX BAZ BHC80 BRD BRPF1 CHD Cps35 EAF3 EED ING2 MBD MDC1 MeCP PB1 SP1 SUZ TAF TDRD3 TRIM | LncRNA microRNA miRNA piRNA Components of RISC complexes AGO DICER Drosha | BAF BRG BRM CHRAC hACF1 Ini1 INO80 ISWI Mi-2 NCoR NURD PRC SMARC Snf Swi Swp SWR1 |
| Dataset Number | Title | Citations |
|---|---|---|
| GSE37418 | Novel mutations target distinct subgroups of medulloblastoma. | [23,24] |
| GSE28703 | Discovery of novel recurrent mutations and rearrangements in early T-cell precursor acute lymphoblastic leukemia by whole genome sequencing. | [25,26] |
| GSE66346 | Expression data from renal cancer xenograft tumor treated with sunitinib or vehicle. | [27] |
| GSE197555 | Differential mRNA expression analysis of H460 cells and A549 cells after trametinib treatment. | [28] |
| GSE59357 | Gene expression profiles of dasatinib-resistant and dasatinib-sensitive pancreatic cancer cell lines. | [29] |
| GSE42872 | Expression data from BRAFV600E A375 melanoma cells treated with vehicle or vemurafenib. | [30] |
| GSE98314 | Melanoma cell lines treated with dabrafenib ± trametinib. | [31] |
| GSE23743 | Effect of imatinib on philadelphia chromosome positive acute lymphoblastic leukemia. | [32] |
| GSE24493 | Effect of Imatinib on chronic myelogenous leukemia. | [32,33] |
| GSE218183 | Bulk RNA-seq analysis of primary CML CD34+ cells (n = 3) treated with idasanutlin alone or in combination with nilotinib in vitro. | [34] |
| GSE197811 | Effect of the janus kinase inhibitor ruxolitinib on gene expression of chronic lymphocytic leukemia cells in vivo. | [35] |
| GSE171763 | Inhibitors of Bcl-2 and Bruton’s tyrosine kinase synergize to abrogate diffuse large B-cell lymphoma (DLBCL) growth. | [36] |
| GSE173306 | Transcription profiling of ALK-rearranged cell lines resistent and sensitive to crizotinib. | [37] |
| GSE29828 | Expression Profiling of Mixed Lineage Leukemia Cells Treated with a Potent Small-Molecule DOT1L Inhibitor. | [38] |
| Dataset Number | Diagnosis | TKI | In Vivo/ In Vitro | Epigenetic Regulator | Expression Level After TKI Treatment | Statistical Correlation | Clinical Trial Number of the Drug Against the Epigenetic Regulator |
|---|---|---|---|---|---|---|---|
| GSE66346 | renal cancer | sunitinib | in vivo | SMARCAL1 |  | p< 0.05 | - |
| miR200C |  | NCT02579187 | |||||
| miR323A |  | - | |||||
| miR382 |  | - | |||||
| miR516B1 |  | - | |||||
| miR664B |  | - | |||||
| GSE197555 | NSCLC | trametinib | in vitro | CHD4 |  | p< 0.01 | - |
| DPY30 |  | - | |||||
| miR614 |  | - | |||||
| GSE59357 | pancreatic cancer | dasatinib | in vitro | CITED2 |  | p< 0.001 | - |
| SMARCD3 |  | - | |||||
| miR132 |  | - | |||||
| GSE42872 | melanoma | vemurafenib | in vitro | HIST1H1A |  | p = 0.100 | - |
| HIST1H1B |  | - | |||||
| HIST1H2AB |  | - | |||||
| HIST1H2BB |  | - | |||||
| HIST1H2BF |  | - | |||||
| HIST1H3A |  | - | |||||
| KAT2B |  | - | |||||
| UHRF1 |  | - | |||||
| miR17HG |  | - | |||||
| miR221 |  | LNA-i-miR-221 /NCT04811898/ | |||||
| GSE98314 | melanoma | dabrafenib, trametinib | in vitro | HIST1H4C |  | p< 0.001 | - |
| UHRF1 |  | p< 0.01 | - | ||||
| GSE23743 | Philadelphia positive ALL | imatinib | in vitro | GCNT1P1 |  | p = 0.125 | - |
| KDM5B |  | - | |||||
| MECP2 |  | trofinetide/NCT04181723/ | |||||
| PHF21A |  | - | |||||
| SETD2 |  | AZD1775/NCT03284385/ axitinib/NCT05941637/ | |||||
| miR3652 |  | - | |||||
| miR6733 |  | - | |||||
| GSE24493 | CML | imatinib | in vitro | BAZ1A |  | p = 0.100 | - |
| BAZ2B |  | - | |||||
| CHAF1A |  | - | |||||
| DOT1L |  | EPZ5676/NCT02141828/ pinometostat/NCT03701295/ | |||||
| JMJD1C |  | - | |||||
| miR612 |  | - | |||||
| GSE218183 | CML | idasanutlin, nilotinib | in vivo | miR3652 |  (n) (n) (i) (i) (n&i) (n&i) | p< 0.05 | - |
| H4C1 |  (n) (n) (i) (i) (n&i) (n&i) | p = 0.1087 | - | ||||
| HDAC1 |  (n) (n) (i) (i)  (n&i) (n&i) | p = 0.0764 | tucidinostat/NCT05320640/ chidamide/NCT04514081 NCT04233294 NCT06393361 NCT06563778/ | ||||
| GSE197811 | CLL | ruxolitinib | in vivo | KDM1A |  | p = 0.4225 | IMG-7289/NCT04081220/ |
| PHF13 |  | p = 0.4848 | - | ||||
| miR6859 |  | p = 0.6991 | - | ||||
| GSE171763 | DLBCL | ibrutinib | in vitro et in vivo | PHF6 |  | p< 0.0001 | - |
| H2BC20P |  | p< 0.001 | - | ||||
| JMJD6 |  | p< 0.001 | - | ||||
| GSE173306 | ALCL | crizotinib | in vitro | SRSF6 |  | p< 0.01 | - |
| AGO4 |  | p< 0.05 | - | ||||
| BRWD3 |  | p< 0.05 | - | ||||
| KDM6B |  | p< 0.05 | - | ||||
| PHF21A |  | p< 0.05 | - | ||||
| BRWD1 |  | p = 0.0842 | - | ||||
| GSE29828 | AML | DOT1L inhibitor | in vitro | AGO1 |  | p< 0.001 | - |
| AGO4 |  | - | |||||
| JMJD6 |  | - | |||||
| SRSF8 |  | - | |||||
| miR15A |  | - | |||||
| miR6758 |  | - | |||||
| miR6787 |  | - | |||||
| miR6836 |  | - | |||||
| miR6890 |  | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tóth, K.; Gaál, Z. Impact of Tyrosine Kinase Inhibitors on the Expression Pattern of Epigenetic Regulators. Cancers 2025, 17, 1282. https://doi.org/10.3390/cancers17081282
Tóth K, Gaál Z. Impact of Tyrosine Kinase Inhibitors on the Expression Pattern of Epigenetic Regulators. Cancers. 2025; 17(8):1282. https://doi.org/10.3390/cancers17081282
Chicago/Turabian StyleTóth, Klaudia, and Zsuzsanna Gaál. 2025. "Impact of Tyrosine Kinase Inhibitors on the Expression Pattern of Epigenetic Regulators" Cancers 17, no. 8: 1282. https://doi.org/10.3390/cancers17081282
APA StyleTóth, K., & Gaál, Z. (2025). Impact of Tyrosine Kinase Inhibitors on the Expression Pattern of Epigenetic Regulators. Cancers, 17(8), 1282. https://doi.org/10.3390/cancers17081282





