Connexons Coupling to Gap Junction Channel: Potential Role for Extracellular Protein Stabilization Centers
Abstract
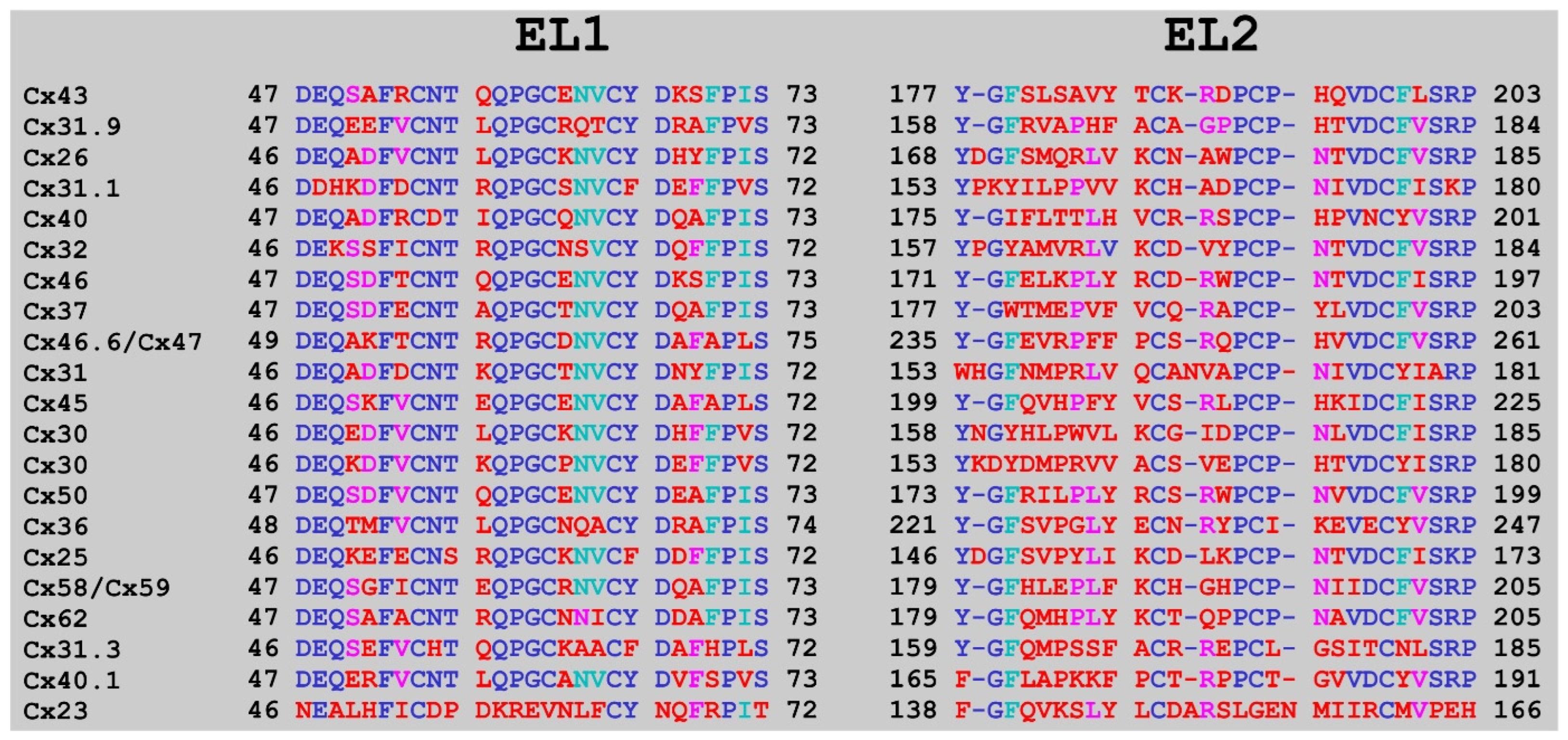
:1. Introduction
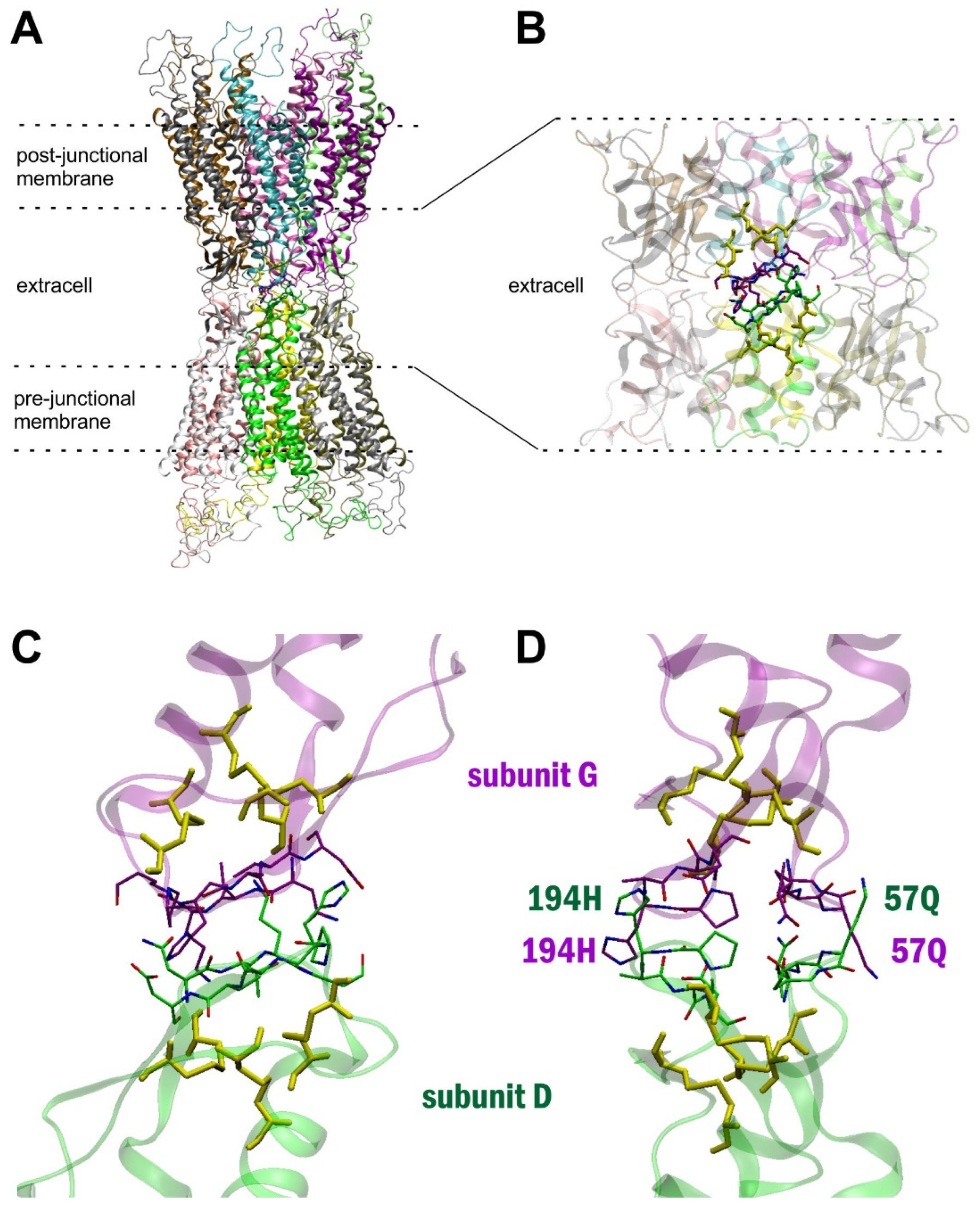
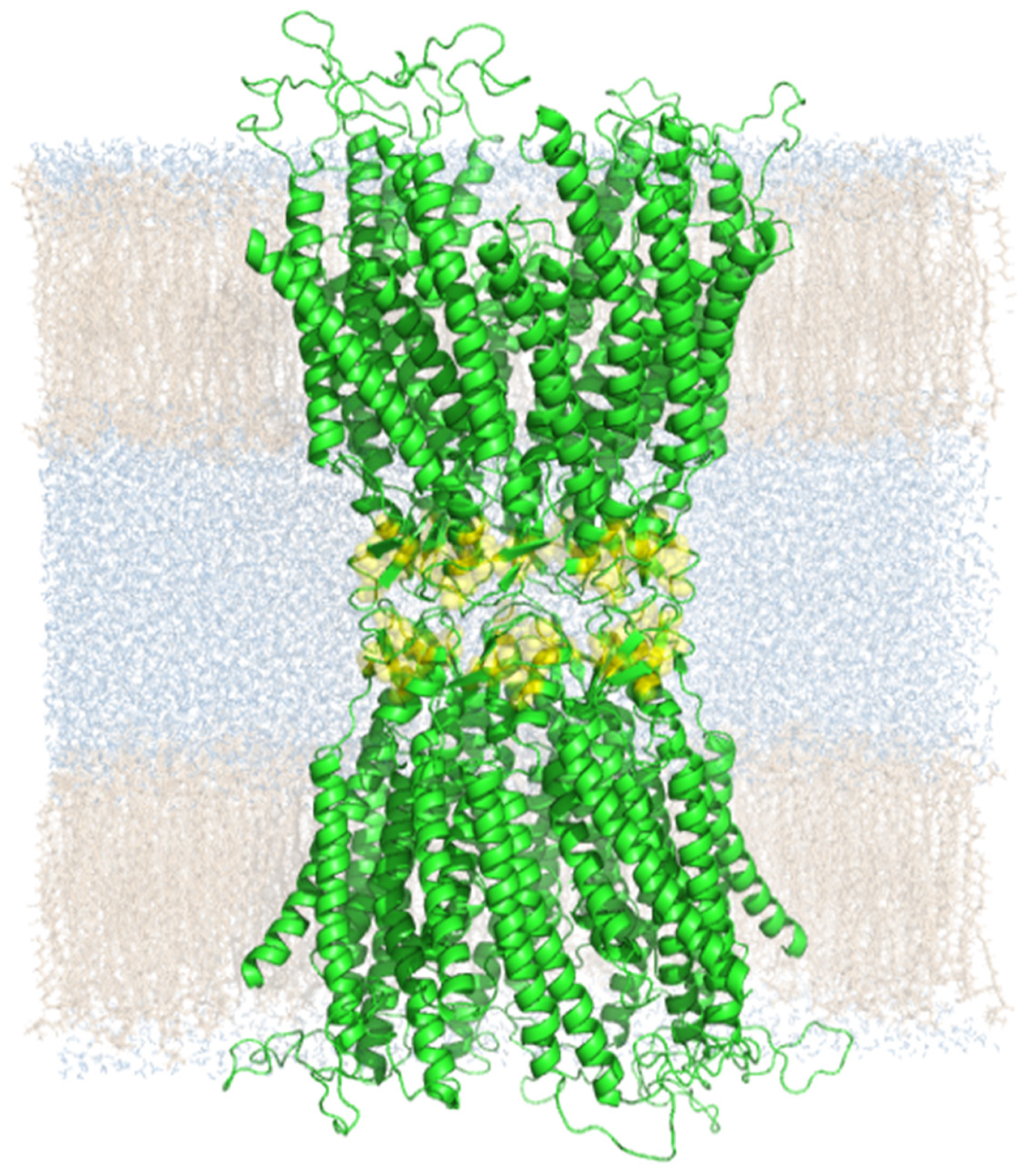
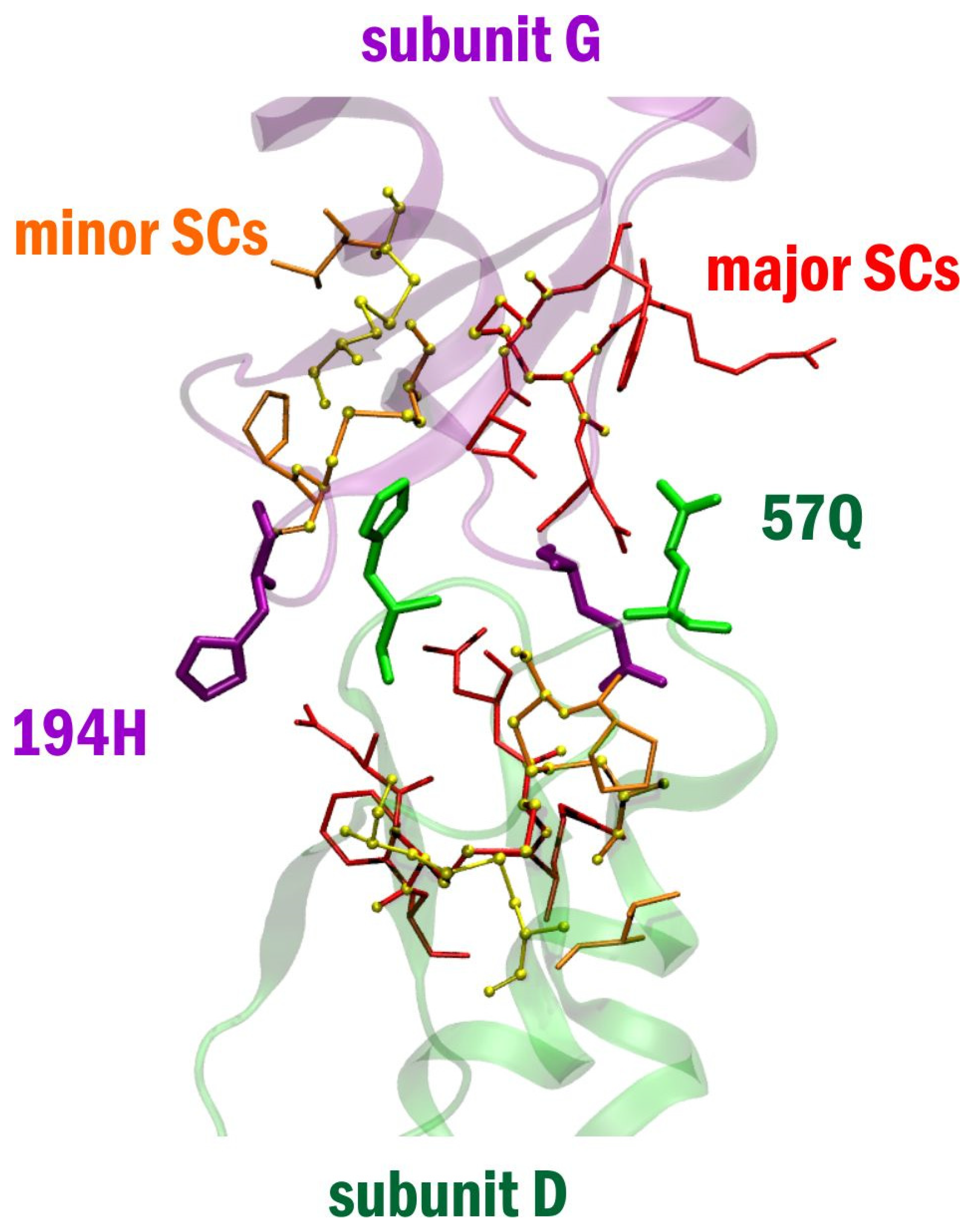
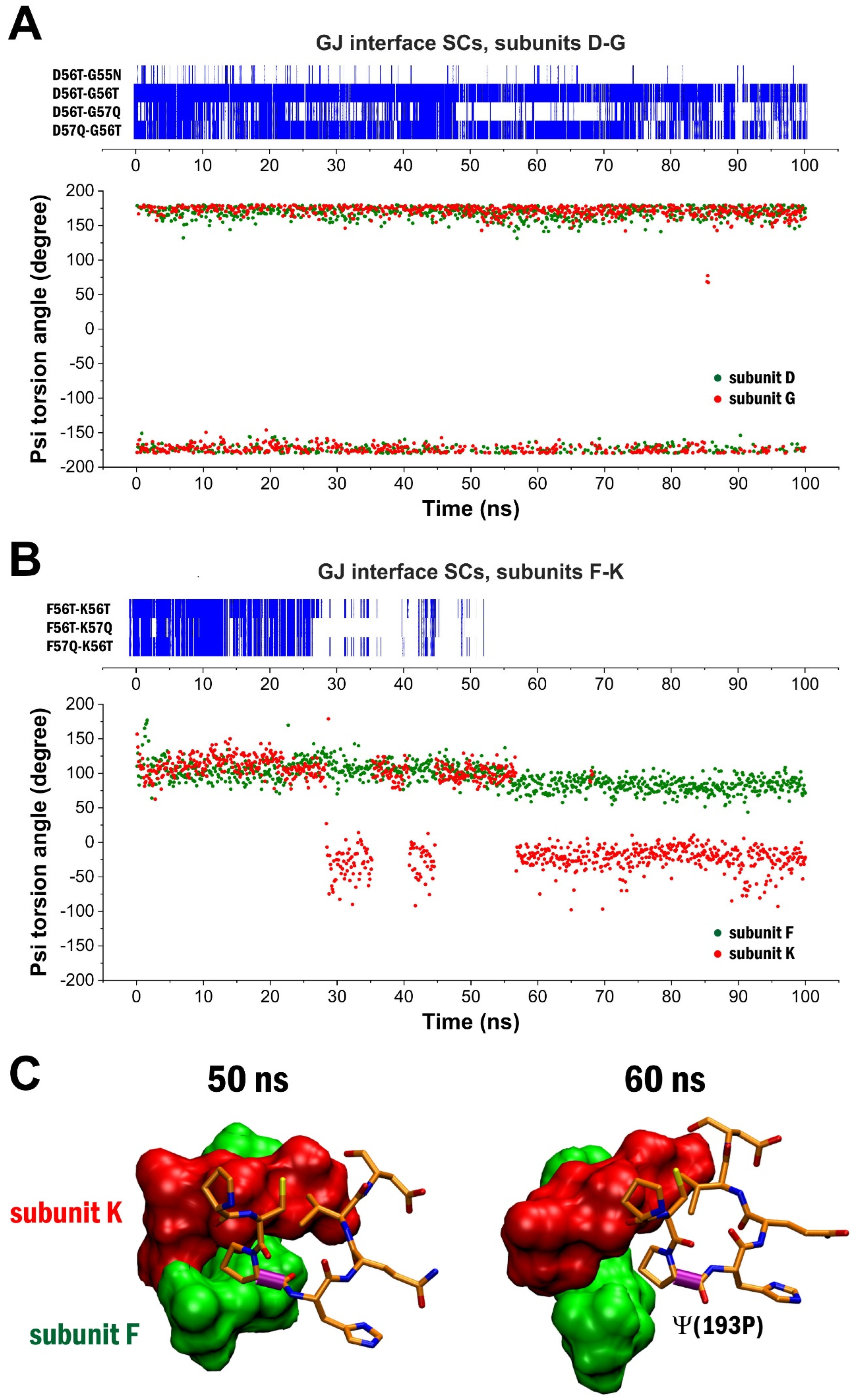
2. Results
3. Discussion
4. Methods
4.1. Preparation of the Cx43 GJC Homology Model
4.2. Preparation of the Cx43 GJC Homology Model with Two Membranes Using Virtual Molecular Dynamics (VMD)
4.3. SC Search and Analysis
4.4. Dihedral Angle Measurements
4.5. Alignment Calculations
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, D.; Yue, B.; Aoyama, H. Crucial motifs and residues in the extracellular loops influence the formation and specificity of connexin docking. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 9–21. [Google Scholar] [CrossRef]
- Laird, D.W.; Lampe, P.D. Therapeutic strategies targeting connexins. Nat. Rev. Drug Discov. 2018, 17, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.A.; Haddad, B.G.; Dolan, K.A.; Myers, J.B.; Yoshioka, C.C.; Copperman, J.; Zuckerman, D.M.; Reichow, S.L. Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 Å. Nat. Commun. 2020, 11, 4331. [Google Scholar] [CrossRef] [PubMed]
- Kotini, M.; Barriga, E.H.; Leslie, J.; Gentzel, M.; Rauschenberger, V.; Schambony, A.; Mayor, R. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat. Commun. 2018, 9, 3846. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Jeong, H.; Hyun, J.; Ryu, B.; Park, K.; Lim, H.-H.; Yoo, J.; Woo, J.-S. Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel. Sci. Adv. 2020, 6, eaba4996. [Google Scholar] [CrossRef] [PubMed]
- Oshima, A. Potential of cryo-EM for high-resolution structural analysis of gap junction channels. Curr. Opin. Struct. Biol. 2019, 54, 78–85. [Google Scholar] [CrossRef]
- Bennett, B.C.; Purdy, M.D.; Baker, K.A.; Acharya, C.; McIntire, W.E.; Stevens, R.C.; Zhang, Q.; Harris, A.L.; Abagyan, R.; Yeager, M. An electrostatic mechanism for Ca2+-mediated regulation of gap junction channels. Nat. Commun. 2016, 7, 8770. [Google Scholar] [CrossRef] [Green Version]
- Oshima, A.; Matsuzawa, T.; Murata, K.; Tani, K.; Fujiyoshi, Y. Hexadecameric structure of an invertebrate gap junction channel. J. Mol. Biol. 2016, 428, 1227–1236. [Google Scholar] [CrossRef] [Green Version]
- Iovine, M.K.; Gumpert, A.M.; Falk, M.M.; Mendelson, T.C. Cx23, a connexin with only four extracellular-loop cysteines, forms functional gap junction channels and hemichannels. FEBS Lett. 2007, 582, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosinsky, G.E.; Boassa, D.; Dermietzel, R.; Duffy, H.S.; Laird, D.W.; MacVicar, B.A.; Naus, C.C.; Penuela, S.; Scemes, E.; Spray, D.C.; et al. Pannexin channels are not gap junction hemichannels. Channels 2011, 5, 193–197. [Google Scholar] [CrossRef]
- Mersman, B.A.; Jolly, S.N.; Lin, Z.; Xu, F. Gap Junction Coding Innexin in Lymnaea stagnalis: Sequence Analysis and Characterization in Tissues and the Central Nervous System. Front. Synaptic Neurosci. 2020, 12, 1. [Google Scholar] [CrossRef]
- Jin, N.; Zhang, Z.; Keung, J.; Youn, S.B.; Ishibashi, M.; Tian, L.-M.; Marshak, D.W.; Solessio, E.; Umino, Y.; Fahrenfort, I.; et al. Molecular and functional architecture of the mouse photoreceptor network. Sci. Adv. 2020, 6, eaba7232. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Aghayeva, U.; Berghoff, E.G.; Hobert, O. Plasticity of the Electrical Connectome of C. elegans. Cell 2019, 176, 1174–1189. [Google Scholar] [CrossRef] [Green Version]
- Foote, C.I.; Zhou, L.; Zhu, X.; Nicholson, B.J. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J. Cell Biol. 1998, 140, 1187–1197. [Google Scholar] [CrossRef] [Green Version]
- Simon, Á.; Magyar, C.; Héja, L.; Kardos, J. Peptide Binding Sites of Connexin Proteins. Chemistry 2020, 2, 42. [Google Scholar] [CrossRef]
- Arkin, M.R.; Tang, Y.; Wells, J.A. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality. Chem. Biol. 2014, 21, 1102–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, X.; Gestwicki, J.E. Inhibitors of protein–protein interactions (PPIs): An analysis of scaffold choices and buried surface area. Curr. Opin. Chem. Biol. 2018, 44, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Shui, S.; Gainza, P.; Scheller, L.; Yang, C.; Kurumida, Y.; Rosset, S.; Georgeon, S.; Di Roberto, R.B.; Castellanos-Rueda, R.; Reddy, S.T.; et al. A rational blueprint for the design of chemically-controlled protein switches. Nat. Commun. 2021, 12, 5754. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, A.; Heuninck, J.; Pizzoccaro, A.; Moutin, E.; Koenen, J.; Séveno, M.; Durroux, T.; Junier, M.-P.; Schlecht-Louf, G.; Bachelerie, F.; et al. The atypical chemokine receptor 3 interacts with Connexin 43 inhibiting astrocytic gap junctional intercellular communication. Nat. Commun. 2020, 11, 4855. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Ghetti, A.; Pinto-Duarte, A.; Wang, X.; Dziewczapolski, G.; Galimi, F.; Huitron-Resendiz, S.; Piña-Crespo, J.; Roberts, A.J.; Verma, I.M.; et al. Astrocytes contribute to gamma oscillations and recognition memory. Proc. Natl. Acad. Sci. USA 2014, 111, E3343–E3352. [Google Scholar] [CrossRef] [Green Version]
- Reaume, A.G.; De Sousa, P.A.; Kulkarni, S.; Langille, B.L.; Zhu, D.; Davies, T.C.; Juneja, S.C.; Kidder, G.M.; Rossant, J. Cardiac malformation in neonatal mice lacking connexin43. Science 1995, 267, 1831–1834. [Google Scholar] [CrossRef]
- Price, G.W.; Chadjichristos, C.E.; Kavvadas, P.; Tang, S.C.W.; Yiu, W.H.; Green, C.R.; Potter, J.A.; Siamantouras, E.; Squires, P.E.; Hills, C.E. Blocking Connexin-43 mediated hemichannel activity protects against early tubular injury in experimental chronic kidney disease. Cell Commun. Signal. 2020, 18, 79. [Google Scholar] [CrossRef]
- Nouet, J.; Himelman, E.; Lahey, K.C.; Zhao, Q.; Fraidenraich, D. Connexin-43 reduction prevents muscle defects in a mouse model of manifesting Duchenne muscular dystrophy female carriers. Sci. Rep. 2020, 10, 5683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dermietzel, R.; Meier, C. Gap junction expression in brain tissues with focus on development. In Gap Junctions in Development and Disease; Springer: Berlin/Heidelberg, Germany, 2005; pp. 83–110. ISBN 3540261567. [Google Scholar]
- Kékesi, O.; Ioja, E.E.; Szabó, Z.; Kardos, J.; Héja, L. Recurrent seizure-like events are associated with coupled astroglial synchronization. Front. Cell. Neurosci. 2015, 9, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincze, R.; Péter, M.; Szabó, Z.; Kardos, J.; Héja, L.; Kovacs, Z. Connexin 43 differentially regulates epileptiform activity in models of convulsive and non-convulsive epilepsies. Front. Cell. Neurosci. 2019, 13, 173. [Google Scholar] [CrossRef]
- Szabó, Z.; Héja, L.; Szalay, G.; Kékesi, O.; Füredi, A.; Szebényi, K.; Dobolyi, Á.; Orbán, T.I.; Kolacsek, O.; Tompa, T.; et al. Extensive astrocyte synchronization advances neuronal coupling in slow wave activity in vivo. Sci. Rep. 2017, 7, 6018. [Google Scholar] [CrossRef]
- Maeda, S.; Nakagawa, S.; Suga, M.; Yamashita, E.; Oshima, A.; Fujiyoshi, Y.; Tsukihara, T. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 2009, 458, 597–602. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Fiser, A.; Simon, I. Stabilization centers in proteins:Identification, characterization and predictions. J. Mol. Biol. 1997, 272, 597–612. [Google Scholar] [CrossRef]
- Forbes, C.R.; Sinha, S.K.; Ganguly, H.K.; Bai, S.; Yap, G.P.A.; Patel, S.; Zondlo, N.J. Insights into Thiol–Aromatic Interactions: A Stereoelectronic Basis for S–H/π Interactions. J. Am. Chem. Soc. 2017, 139, 1842–1855. [Google Scholar] [CrossRef] [Green Version]
- Weber, D.S.; Warren, J.J. The interaction between methionine and two aromatic amino acids is an abundant and multifunctional motif in proteins. Arch. Biochem. Biophys. 2019, 672, 108053. [Google Scholar] [CrossRef] [PubMed]
- Yeung, P.S.-W.; Ing, C.E.; Yamashita, M.; Pomès, R.; Prakriya, M. A sulfur-aromatic gate latch is essential for opening of the Orai1 channel pore. eLife 2020, 9, e60751. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.I.; Shakhnovich, E.I. What can disulfide bonds tell us about protein energetics, function and folding: Simulations and bioninformatics analysis. J. Mol. Biol. 2000, 300, 975–985. [Google Scholar] [CrossRef]
- Craveur, P.; Joseph, A.P.; Poulain, P.; De Brevern, A.G.; Rebehmed, J. Cis–trans isomerization of omega dihedrals in proteins. Amino Acids 2013, 45, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. Electrical synapses in mammalian CNS: Past eras, present focus and future directions. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 102–123. [Google Scholar] [CrossRef]
- O’Brien, J. Design principles of electrical synaptic plasticity. Neurosci. Lett. 2019, 695, 4–11. [Google Scholar] [CrossRef]
- Raškevičius, V.; Jotautis, V.; Rimkutė, L.; Marandykina, A.; Kazokaitė, M.; Kairys, V.; Skeberdis, V.A. Molecular basis for potentiation of Cx36 gap junction channel conductance by n-alcohols and general anesthetics. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, A.; Castro, C.; Flores, D.-L.; Gutierrez, E.; Baldi, P. Gap Junction Channels of Innexins and Connexins: Relations and Computational Perspectives. Int. J. Mol. Sci. 2019, 20, 2476. [Google Scholar] [CrossRef] [Green Version]
- Su, V.; Lau, A.F. Connexins: Mechanisms regulating protein levels and intercellular communication. FEBS Lett. 2014, 588, 1212–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unwin, P.N.; Ennis, P.D. Calcium-mediated changes in gap junction structure: Evidence from the low angle X-ray pattern. J. Cell Biol. 1983, 97, 1459–1466. [Google Scholar] [CrossRef] [Green Version]
- Verselis, V.K.; Trelles, M.P.; Rubinos, C.; Bargiello, T.A.; Srinivas, M. Loop Gating of Connexin Hemichannels Involves Movement of Pore-lining Residues in the First Extracellular Loop Domain. J. Biol. Chem. 2009, 284, 4484–4493. [Google Scholar] [CrossRef] [Green Version]
- Wiedemann, C.; Kumar, A.; Lang, A.; Ohlenschläger, O. Cysteines and Disulfide Bonds as Structure-Forming Units: Insights from Different Domains of Life and the Potential for Characterization by NMR. Front. Chem. 2020, 8, 280. [Google Scholar] [CrossRef]
- Wu, L.; Dong, A.; Dong, L.; Wang, S.-Q.; Li, Y. PARIS, an optogenetic method for functionally mapping gap junctions. eLife 2019, 8, e43366. [Google Scholar] [CrossRef]
- Zonta, F.; Polles, G.; Zanotti, G.; Mammano, F. Permeation Pathway of Homomeric Connexin 26 and Connexin 30 Channels Investigated by Molecular Dynamics. J. Biomol. Struct. Dyn. 2012, 29, 985–998. [Google Scholar] [CrossRef] [Green Version]
- Aasen, T.; Johnstone, S.; Vidal-Brime, L.; Lynn, K.S.; Koval, M. Connexins: Synthesis, post-translational modifications, and trafficking in health and disease. Int. J. Mol. Sci. 2018, 19, 1296. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Salarian, M.; Chen, Y.; Veenstra, R.; Louis, C.F.; Yang, J.J. Gap junction regulation by calmodulin. FEBS Lett. 2014, 588, 1430–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosinsky, G.E.; Solan, J.L.; Gaietta, G.M.; Ngan, L.; Lee, G.J.; Mackey, M.R.; Lampe, P.D. The C-terminus of connexin43 adopts different conformations in the Golgi and gap junction as detected with structure-specific antibodies. Biochem. J. 2007, 408, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.B.; Haddad, B.G.; O’Neill, S.E.; Chorev, D.S.; Yoshioka, C.C.; Robinson, C.V.; Zuckerman, D.M.; Reichow, S.L. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 2018, 564, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Bargiello, T.A.; Oh, S.; Tang, Q.; Bargiello, N.K.; Dowd, T.L.; Kwon, T. Gating of Connexin Channels by transjunctional-voltage: Conformations and models of open and closed states. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 22–39. [Google Scholar] [CrossRef]
- Batir, Y.; Bargiello, T.A.; Dowd, T.L. Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy. Arch. Biochem. Biophys. 2016, 608, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Beyer, E.C.; Berthoud, V.M. Gap junction structure: Unraveled, but not fully revealed. F1000Research 2017, 6, 568. [Google Scholar] [CrossRef] [Green Version]
- Flores, C.E.; Nannapaneni, S.; Davidson, K.G.V.; Yasumura, T.; Bennett, M.V.L.; Rash, J.E.; Pereda, A.E. Trafficking of gap junction channels at a vertebrate electrical synapse in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E573–E582. [Google Scholar] [CrossRef] [Green Version]
- Gaietta, G.; Deerinck, T.J.; Adams, S.R.; Bouwer, J.; Tour, O.; Laird, D.W.; Sosinsky, G.E.; Tsien, R.Y.; Ellisman, M.H. Multicolor and Electron Microscopic Imaging of Connexin Trafficking. Science 2002, 296, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.L. Electrical coupling and its channels. J. Gen. Physiol. 2018, 150, 1606–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, W.; Ramachandran, J.; Alsamarah, A.; Luo, Y.; Harris, A.L.; Contreras, J.E. Mechanism of gating by calcium in connexin hemichannels. Proc. Natl. Acad. Sci. USA 2016, 113, E7986–E7995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, S.R.; Williams, Z.J.; Pridham, K.J.; Gourdie, R.G. Peptidic connexin43 therapeutics in cardiac reparative medicine. J. Cardiovasc. Dev. Dis. 2021, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- King, D.R.; Sedovy, M.W.; Leng, X.; Xue, J.; Lamouille, S.; Koval, M.; Isakson, B.E.; Johnstone, S.R. Mechanisms of connexin regulating peptides. Int. J. Mol. Sci. 2021, 22, 10186. [Google Scholar] [CrossRef]
- Stout, R.F.; Spray, D.C. Cysteine residues in the cytoplasmic carboxy terminus of connexins dictate gap junction plaque stability. Mol. Biol. Cell 2017, 28, 2757–2764. [Google Scholar] [CrossRef]
- Fra, A.; Yoboue, E.D.; Sitia, R. Cysteines as redox molecular switches and targets of disease. Front. Mol. Neurosci. 2017, 10, 167. [Google Scholar] [CrossRef] [Green Version]
- Retamal, M.A.; García, I.E.; Pinto, B.I.; Pupo, A.; Báez, D.; Stehberg, J.; Del Rio, R.; González, C. Extracellular cysteine in connexins: Role as redox sensors. Front. Physiol. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.-A.; Wang, Y.; Zhang, Q.; Xia, Y.; Ge, W.; Guo, D. Prediction of reversible disulfide based on features from local structural signatures. BMC Genom. 2017, 18, 279. [Google Scholar] [CrossRef]
- Yi, M.C.; Khosla, C. Thiol–Disulfide Exchange Reactions in the Mammalian Extracellular Environment. Annu. Rev. Chem. Biomol. Eng. 2016, 7, 197–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Futera, Z.; Ali, M.E.; Gajdos, F.; Von Rudorff, G.F.; Carof, A.; Breuer, M.; Blumberger, J. Cysteine Linkages Accelerate Electron Flow through Tetra-Heme Protein STC. J. Am. Chem. Soc. 2017, 139, 17237–17240. [Google Scholar] [CrossRef] [Green Version]
- Moser, C.C.; Anderson, J.L.R.; Dutton, P.L. Guidelines for tunneling in enzymes. Biochim. Biophys. Acta (BBA)-Bioenerg. 2010, 1797, 1573–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuchebrukhov, A.A. Long-distance electron tunneling in proteins: A new challenge for time-resolved spectroscopy. Laser Phys. 2010, 20, 125–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delvaeye, T.; Vandenabeele, P.; Bultynck, G.; Leybaert, L.; Krysko, D.V. Therapeutic Targeting of Connexin Channels: New Views and Challenges. Trends Mol. Med. 2018, 24, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Kwak, B.R.; Jongsma, H.J. Selective inhibition of gap junction channel activity by synthetic peptides. J. Physiol. 1999, 516, 679–685. [Google Scholar] [CrossRef]
- Martin, P.E.M.; Wall, C.; Griffith, T.M. Effects of connexin-mimetic peptides on gap junction functionality and connexin expression in cultured vascular cells. Br. J. Pharmacol. 2005, 144, 617–627. [Google Scholar] [CrossRef] [Green Version]
- O’Carroll, S.J.; Alkadhi, M.; Nicholson, L.F.B.; Green, C.R. Connexin43 mimetic peptides reduce swelling, astrogliosis, and neuronal cell death after spinal cord injury. Cell Commun. Adhes. 2008, 15, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ma, M.; Locovei, S.; Keane, R.W.; Dahl, G. Modulation of membrane channel currents by gap junction protein mimetic peptides: Size matters. Am. J. Physiol.-Cell Physiol. 2007, 293, C1112–C1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schadzek, P.; Schlingmann, B.; Schaarschmidt, F.; Lindner, J.; Koval, M.; Heisterkamp, A.; Preller, M.; Ngezahayo, A. The cataract related mutation N188T in human connexin46 (hCx46) revealed a critical role for residue N188 in the docking process of gap junction channels. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Kopanic, J.L.; Al-Mugotir, M.H.; Kieken, F.; Zach, S.; Trease, A.J.; Sorgen, P.L. Characterization of the Connexin45 Carboxyl-Terminal Domain Structure and Interactions with Molecular Partners. Biophys. J. 2014, 106, 2184–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopanic, J.L.; Schlingmann, B.; Koval, M.; Lau, A.F.; Sorgen, P.L.; Su, V.F. Degradation of gap junction connexins is regulated by the interaction with Cx43-interacting protein of 75 kDa (CIP75). Biochem. J. 2015, 466, 571–585. [Google Scholar] [CrossRef] [Green Version]
- Héja, L.; Nyitrai, G.; Kékesi, O.; Dobolyi, A.; Szabó, P.; Fiáth, R.; Ulbert, I.; Pál-Szenthe, B.; Palkovits, M.; Kardos, J. Astrocytes convert network excitation to tonic inhibition of neurons. BMC Biol. 2012, 10, 26. [Google Scholar] [CrossRef] [Green Version]
- Rouach, N.; Koulakoff, A.; Abudara, V.; Willecke, K.; Giaume, C. Astroglial metabolic networks sustain hippocampal synaptic transmission. Science 2008, 322, 1551–1555. [Google Scholar] [CrossRef]
- Timofeev, I.; Grenier, F.; Steriade, M. Impact of intrinsic properties and synaptic factors on the activity of neocortical networks in vivo. J. Physiol.-Paris 2000, 94, 343–355. [Google Scholar] [CrossRef]
- Timofeev, I.; Bazhenov, M.; Sejnowski, T.; Steriade, M. Cortical hyperpolarization-activated depolarizing current takes part in the generation of focal paroxysmal activities. Proc. Natl. Acad. Sci. USA 2002, 99, 9533–9537. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Gonzalo, M.; Losi, G.; Brondi, M.; Uva, L.; Sato, S.S.; de Curtis, M.; Ratto, G.M.; Carmignoto, G. Ictal but not interictal epileptic discharges activate astrocyte endfeet and elicit cerebral arteriole responses. Front. Cell. Neurosci. 2011, 5, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs, R.; Heinemann, U.; Steinhäuser, C. Mechanisms underlying blood-brain barrier dysfunction in brain pathology and epileptogenesis: Role of astroglia. Epilepsia 2012, 53 (Suppl. 6), 53–59. [Google Scholar] [CrossRef]
- Nadarajah, B.; Thomaidou, D.; Evans, W.H.; Parnavelas, J.G. Gap junctions in the adult cerebral cortex: Regional differences in their distribution and cellular expression of connexins. J. Comp. Neurol. 1996, 376, 326–342. [Google Scholar] [CrossRef]
- Amzica, F.; Massimini, M. Glial and neuronal interactions during slow wave and paroxysmal activities in the neocortex. Cereb. cortex 2002, 12, 1101–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samoilova, M.; Li, J.; Pelletier, M.R.; Wentlandt, K.; Adamchik, Y.; Naus, C.C.; Carlen, P.L. Epileptiform activity in hippocampal slice cultures exposed chronically to bicuculline: Increased gap junctional function and expression. J. Neurochem. 2003, 86, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Nyikos, L.; Lasztóczi, B.; Antal, K.; Kovács, R.; Kardos, J. Desynchronisation of spontaneously recurrent experimental seizures proceeds with a single rhythm. Neuroscience 2003, 121, 705–717. [Google Scholar] [CrossRef]
- Kardos, J.; Szabó, Z.; Héja, L. Framing Neuro-Glia Coupling in Antiepileptic Drug Design. J. Med. Chem. 2016, 59, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, S.S.; Wentlandt, K.; Piran, S.; Carlen, P.L. Anticonvulsant actions of gap junctional blockers in an in vitro seizure model. J. Neurophysiol. 2002, 88, 1893–1902. [Google Scholar] [CrossRef] [Green Version]
- mer Bostanci, M.; Baǧirici, F. Anticonvulsive effects of carbenoxolone on penicillin-induced epileptiform activity: An in vivo study. Neuropharmacology 2007, 52, 362–367. [Google Scholar] [CrossRef]
- Elisevich, K.; Rempel, S.A.; Smith, B.; Hirst, K. Temporal profile of connexin 43 mRNA expression in a tetanus toxin-induced seizure disorder. Mol. Chem. Neuropathol. 1998, 35, 23–37. [Google Scholar] [CrossRef]
- Santos-Miranda, A.; Chen, H.; Chen, R.C.; Odoko-Ishimoto, M.; Aoyama, H.; Bai, D. The amino terminal domain plays an important role in transjunctional voltage-dependent gating kinetics of Cx45 gap junctions. J. Mol. Cell. Cardiol. 2020, 143, 71–84. [Google Scholar] [CrossRef]
- Tong, X.; Aoyama, H.; Sudhakar, S.; Chen, H.; Shilton, B.H.; Bai, D. The first extracellular domain plays an important role in unitary channel conductance of Cx50 gap junction channels. PLoS ONE 2015, 10, e0143876. [Google Scholar] [CrossRef]
- Tejada, M.G.; Sudhakar, S.; Kim, N.K.; Aoyama, H.; Shilton, B.H.; Bai, D. Variants with increased negative electrostatic potential in the Cx50 gap junction pore increased unitary channel conductance and magnesium modulation. Biochem. J. 2018, 475, 3315–3330. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Haddad, B.G.; Khan, U.; Chen, H.; Atalla, M.; Zhang, Z.; Zuckerman, D.M.; Reichow, S.L.; Bai, D. Connexin 46 and connexin 50 gap junction channel properties are shaped by structural and dynamic features of their N-terminal domains. J. Physiol. 2021, 599, 3313–3335. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Magyar, C.; Gromiha, M.M.; Pujadas, G.; Tusnády, G.E.; Simon, I. SRide: A server for identifying stabilizing residues in proteins. Nucleic Acids Res. 2005, 33, W303–W305. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Héja, L.; Simon, Á.; Szabó, Z.; Kardos, J. Connexons Coupling to Gap Junction Channel: Potential Role for Extracellular Protein Stabilization Centers. Biomolecules 2022, 12, 49. https://doi.org/10.3390/biom12010049
Héja L, Simon Á, Szabó Z, Kardos J. Connexons Coupling to Gap Junction Channel: Potential Role for Extracellular Protein Stabilization Centers. Biomolecules. 2022; 12(1):49. https://doi.org/10.3390/biom12010049
Chicago/Turabian StyleHéja, László, Ágnes Simon, Zsolt Szabó, and Julianna Kardos. 2022. "Connexons Coupling to Gap Junction Channel: Potential Role for Extracellular Protein Stabilization Centers" Biomolecules 12, no. 1: 49. https://doi.org/10.3390/biom12010049
APA StyleHéja, L., Simon, Á., Szabó, Z., & Kardos, J. (2022). Connexons Coupling to Gap Junction Channel: Potential Role for Extracellular Protein Stabilization Centers. Biomolecules, 12(1), 49. https://doi.org/10.3390/biom12010049






