Interactions of Pleiotrophin with a Structurally Defined Heparin Hexasaccharide
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of PTN WT and Truncation Mutants
2.2. Deuteration and Selective Protonation of PTN
2.3. Generation of GAG Fragments
2.4. Collection and Analysis of NMR Data
2.5. Computational Modeling of the PTN-Heparin Complex
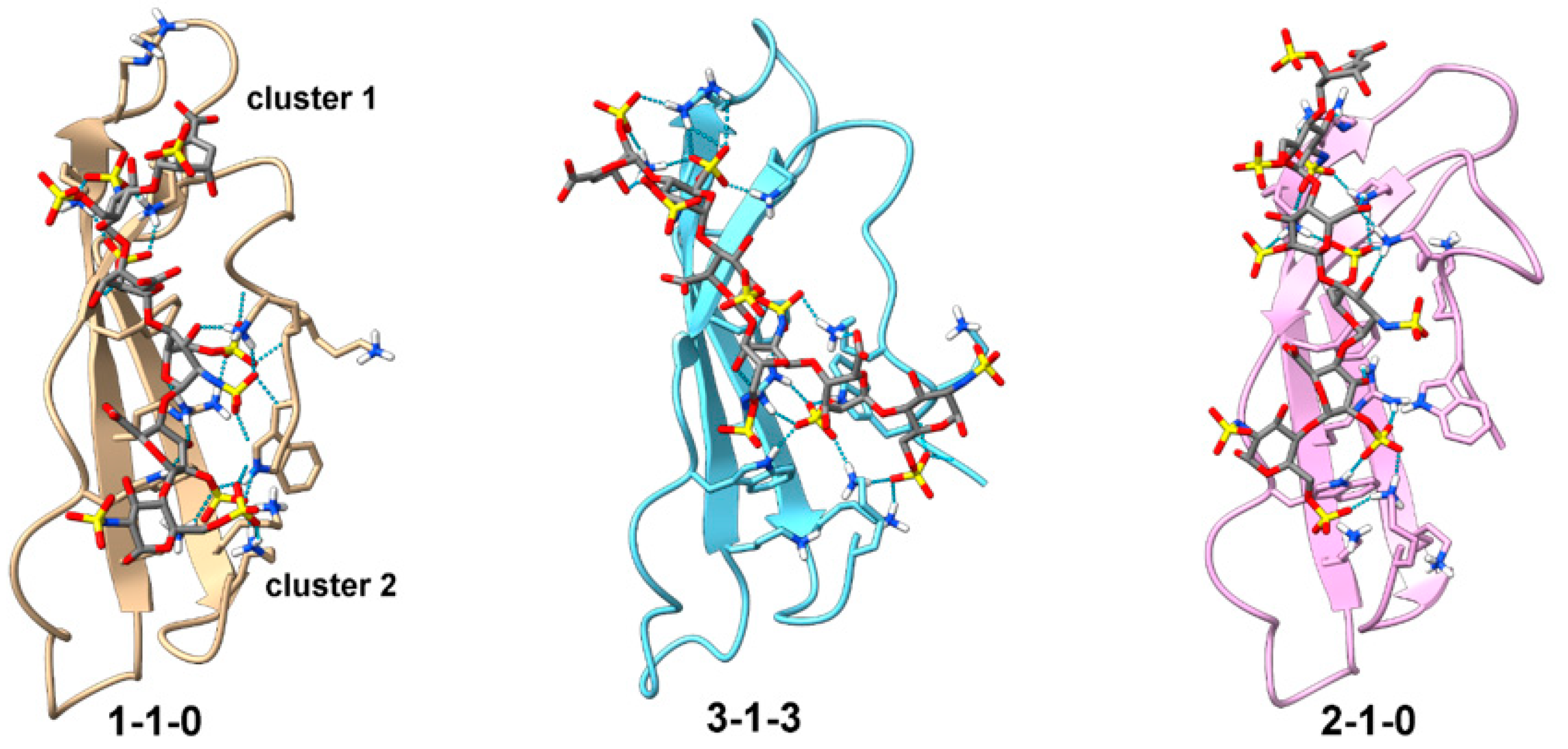
3. Results
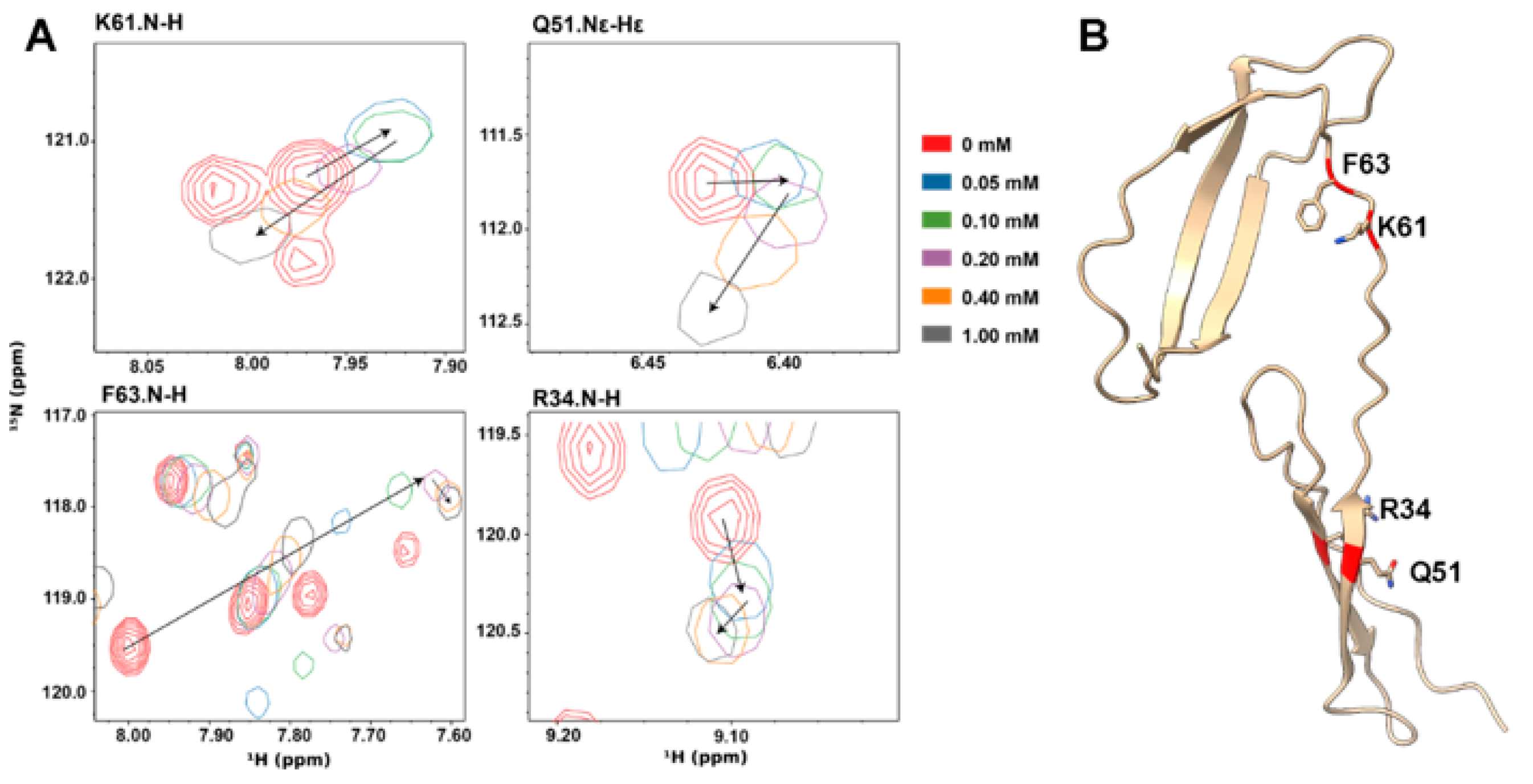
3.1. Stoichiometry of PTN Interactions with Heprin dp6
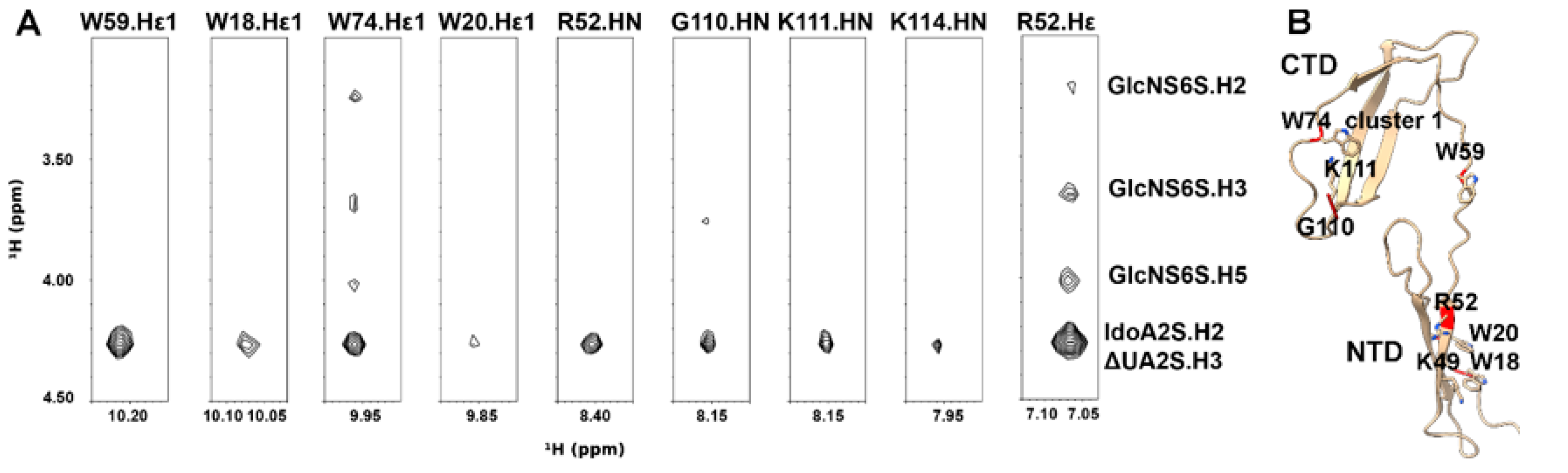
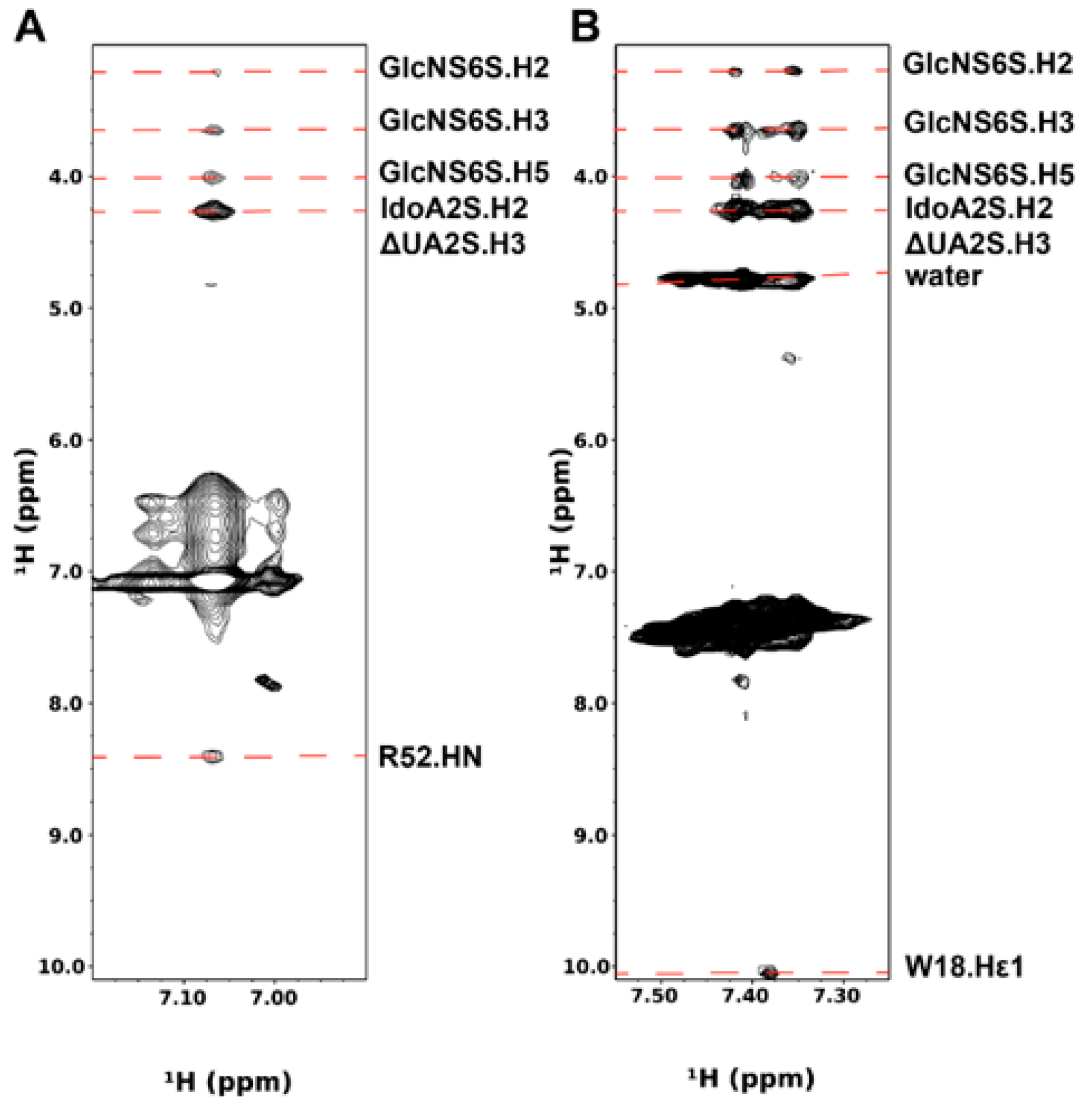
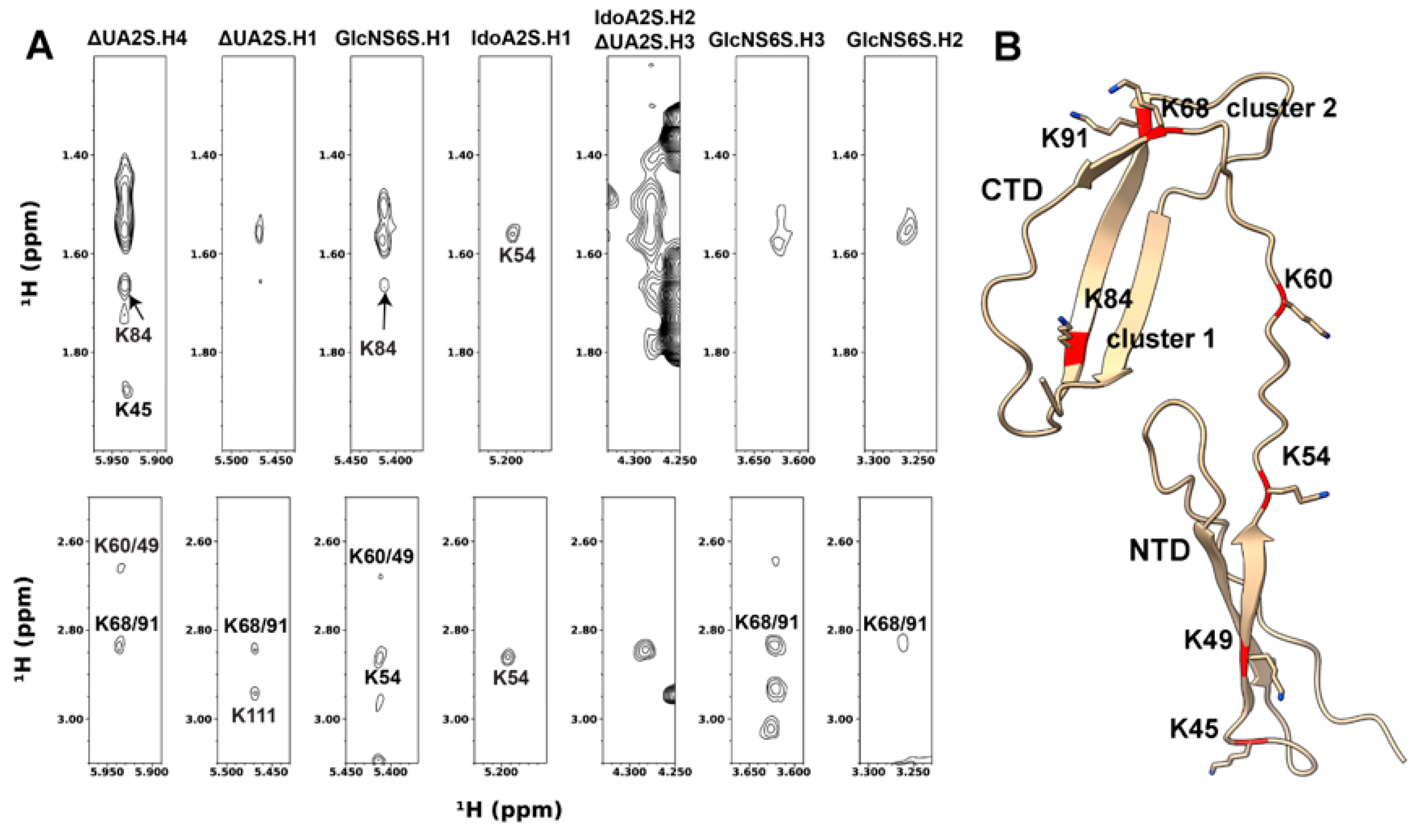
3.2. Intermolecular NOEs between PTN and DP6-C
3.3. Sulfate Specificity of PTN
3.4. Model of the PTN-GAG Complex
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Paveliev, M.; Fenrich, K.K.; Kislin, M.; Kuja-Panula, J.; Kulesskiy, E.; Varjosalo, M.; Kajander, T.; Mugantseva, E.; Ahonen-Bishopp, A.; Khiroug, L.; et al. HB-GAM (pleiotrophin) reverses inhibition of neural regeneration by the CNS extracellular matrix. Sci. Rep. 2016, 6, 33916. [Google Scholar] [CrossRef]
- Raulo, E.; Chernousov, M.A.; Carey, D.J.; Nolo, R.; Rauvala, H. Isolation of a neuronal cell surface receptor of heparin binding growth-associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3). J. Biol. Chem. 1994, 269, 12999–13004. [Google Scholar] [CrossRef]
- Raulo, E.; Tumova, S.; Pavlov, I.; Pekkanen, M.; Hienola, A.; Klankki, E.; Kalkkinen, N.; Taira, T.; Kilpelainen, I.; Rauvala, H. The two thrombospondin type I repeat domains of the heparin-binding growth-associated molecule bind to heparin/heparan sulfate and regulate neurite extension and plasticity in hippocampal neurons. J. Biol. Chem. 2005, 280, 41576–41583. [Google Scholar] [CrossRef]
- Gramage, E.; Herradon, G. Genetic deletion of pleiotrophin leads to disruption of spinal nociceptive transmission: Evidence for pleiotrophin modulation of morphine-induced analgesia. Eur. J. Pharmacol. 2010, 647, 97–102. [Google Scholar] [CrossRef]
- Gramage, E.; Putelli, A.; Polanco, M.J.; Gonzalez-Martin, C.; Ezquerra, L.; Alguacil, L.F.; Perez-Pinera, P.; Deuel, T.F.; Herradon, G. The neurotrophic factor pleiotrophin modulates amphetamine-seeking behaviour and amphetamine-induced neurotoxic effects: Evidence from pleiotrophin knockout mice. Addict. Biol. 2010, 15, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Gramage, E.; Rossi, L.; Granado, N.; Moratalla, R.; Herradon, G. Genetic inactivation of pleiotrophin triggers amphetamine-induced cell loss in the substantia nigra and enhances amphetamine neurotoxicity in the striatum. Neuroscience 2010, 170, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Gramage, E.; Vicente-Rodriguez, M.; Herradon, G. Pleiotrophin modulates morphine withdrawal but has no effects on morphine-conditioned place preference. Neurosci. Lett. 2015, 604, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Herradon, G.; Ramos-Alvarez, M.P.; Gramage, E. Connecting Metainflammation and Neuroinflammation Through the PTN-MK-RPTPbeta/zeta Axis: Relevance in Therapeutic Development. Front. Pharmacol. 2019, 10, 377. [Google Scholar] [CrossRef]
- Vicente-Rodriguez, M.; Perez-Garcia, C.; Ferrer-Alcon, M.; Uribarri, M.; Sanchez-Alonso, M.G.; Ramos, M.P.; Herradon, G. Pleiotrophin differentially regulates the rewarding and sedative effects of ethanol. J. Neurochem. 2014, 131, 688–695. [Google Scholar] [CrossRef]
- Vicente-Rodriguez, M.; Rojo Gonzalez, L.; Gramage, E.; Fernandez-Calle, R.; Chen, Y.; Perez-Garcia, C.; Ferrer-Alcon, M.; Uribarri, M.; Bailey, A.; Herradon, G. Pleiotrophin overexpression regulates amphetamine-induced reward and striatal dopaminergic denervation without changing the expression of dopamine D1 and D2 receptors: Implications for neuroinflammation. Eur. Neuropsychopharmacol. 2016, 26, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Klomp, H.J.; Czubayko, F.; Wellstein, A.; Juhl, H. Pleiotrophin can be rate-limiting for pancreatic cancer cell growth. Cancer Res. 2000, 60, 5284–5288. [Google Scholar]
- Yao, J.; Zhang, M.; Ma, Q.Y.; Wang, Z.; Wang, L.C.; Zhang, D. PAd-shRNA-PTN reduces pleiotrophin of pancreatic cancer cells and inhibits neurite outgrowth of DRG. World J. Gastroenterol. 2011, 17, 2667–2673. [Google Scholar] [CrossRef] [PubMed]
- Hamma-Kourbali, Y.; Bernard-Pierrot, I.; Heroult, M.; Dalle, S.; Caruelle, D.; Milhiet, P.E.; Fernig, D.G.; Delbe, J.; Courty, J. Inhibition of the mitogenic, angiogenic and tumorigenic activities of pleiotrophin by a synthetic peptide corresponding to its C-thrombospondin repeat-I domain. J. Cell. Physiol. 2008, 214, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Garcia-Suarez, O.; Menendez-Rodriguez, P.; Mortimer, J.; Chang, Y.; Astudillo, A.; Deuel, T.F. The receptor protein tyrosine phosphatase (RPTP)beta/zeta is expressed in different subtypes of human breast cancer. Biochem. Biophys. Res. Commun. 2007, 362, 5–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wellstein, A.; Fang, W.J.; Khatri, A.; Lu, Y.; Swain, S.S.; Dickson, R.B.; Sasse, J.; Riegel, A.T.; Lippman, M.E. A heparin-binding growth factor secreted from breast cancer cells homologous to a developmentally regulated cytokine. J. Biol. Chem. 1992, 267, 2582–2587. [Google Scholar] [CrossRef]
- Zhang, N.; Zhong, R.; Wang, Z.Y.; Deuel, T.F. Human breast cancer growth inhibited in vivo by a dominant negative pleiotrophin mutant. J. Biol. Chem. 1997, 272, 16733–16736. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Yan, H.; Kurahashi, Y.; Ito, Y.; Maeda, H.; Tada, T.; Hongo, K.; Nakayama, J. Role of GalNAc4S-6ST in astrocytic tumor progression. PLoS ONE 2013, 8, e54278. [Google Scholar] [CrossRef] [PubMed]
- Koutsioumpa, M.; Polytarchou, C.; Courty, J.; Zhang, Y.; Kieffer, N.; Mikelis, C.; Skandalis, S.S.; Hellman, U.; Iliopoulos, D.; Papadimitriou, E. Interplay between alphavbeta3 integrin and nucleolin regulates human endothelial and glioma cell migration. J. Biol. Chem. 2013, 288, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Qin, E.Y.; Cooper, D.D.; Abbott, K.L.; Lennon, J.; Nagaraja, S.; Mackay, A.; Jones, C.; Vogel, H.; Jackson, P.K.; Monje, M. Neural Precursor-Derived Pleiotrophin Mediates Subventricular Zone Invasion by Glioma. Cell 2017, 170, 845–859.e19. [Google Scholar] [CrossRef]
- Yokoi, H.; Kasahara, M.; Mori, K.; Ogawa, Y.; Kuwabara, T.; Imamaki, H.; Kawanishi, T.; Koga, K.; Ishii, A.; Kato, Y.; et al. Pleiotrophin triggers inflammation and increased peritoneal permeability leading to peritoneal fibrosis. Kidney Int. 2012, 81, 160–169. [Google Scholar] [CrossRef]
- Sevillano, J.; Sanchez-Alonso, M.G.; Zapateria, B.; Calderon, M.; Alcala, M.; Limones, M.; Pita, J.; Gramage, E.; Vicente-Rodriguez, M.; Horrillo, D.; et al. Pleiotrophin deletion alters glucose homeostasis, energy metabolism and brown fat thermogenic function in mice. Diabetologia 2019, 62, 123–135. [Google Scholar] [CrossRef]
- Zapateria, B.; Sevillano, J.; Sanchez-Alonso, M.G.; Limones, M.; Pizarro-Delgado, J.; Zuccaro, A.; Herradon, G.; Medina-Gomez, G.; Ramos-Alvarez, M.P. Deletion of pleiotrophin impairs glucose tolerance and liver metabolism in pregnant mice: Moonlighting role of glycerol kinase. FASEB J. 2021, 35, e21911. [Google Scholar] [CrossRef]
- Gu, D.; Yu, B.; Zhao, C.; Ye, W.; Lv, Q.; Hua, Z.; Ma, J.; Zhang, Y. The effect of pleiotrophin signaling on adipogenesis. FEBS Lett. 2007, 581, 382–388. [Google Scholar] [CrossRef]
- Yi, C.; Xie, W.D.; Li, F.; Lv, Q.; He, J.; Wu, J.; Gu, D.; Xu, N.; Zhang, Y. MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin. FEBS Lett. 2011, 585, 3303–3309. [Google Scholar] [CrossRef]
- Kuboyama, K.; Fujikawa, A.; Suzuki, R.; Tanga, N.; Noda, M. Role of Chondroitin Sulfate (CS) Modification in the Regulation of Protein-tyrosine Phosphatase Receptor Type Z (PTPRZ) Activity: Pleiotrophin-ptprz-a signaling is involved in oligodendrocyte differentiation. J. Biol. Chem. 2016, 291, 18117–18128. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Nishiwaki, T.; Shintani, T.; Hamanaka, H.; Noda, M. 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase zeta/RPTPbeta, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM). J. Biol. Chem. 1996, 271, 21446–21452. [Google Scholar] [CrossRef]
- Meng, K.; Rodriguez-Pena, A.; Dimitrov, T.; Chen, W.; Yamin, M.; Noda, M.; Deuel, T.F. Pleiotrophin signals increased tyrosine phosphorylation of beta beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. Proc. Natl. Acad. Sci. USA 2000, 97, 2603–2608. [Google Scholar] [CrossRef] [PubMed]
- Mourlevat, S.; Debeir, T.; Ferrario, J.E.; Delbe, J.; Caruelle, D.; Lejeune, O.; Depienne, C.; Courty, J.; Raisman-Vozari, R.; Ruberg, M. Pleiotrophin mediates the neurotrophic effect of cyclic AMP on dopaminergic neurons: Analysis of suppression-subtracted cDNA libraries and confirmation In Vitro. Exp. Neurol. 2005, 194, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Kilpelainen, I.; Kaksonen, M.; Kinnunen, T.; Avikainen, H.; Fath, M.; Linhardt, R.J.; Raulo, E.; Rauvala, H. Heparin-binding growth-associated molecule contains two heparin-binding beta -sheet domains that are homologous to the thrombospondin type I repeat. J. Biol. Chem. 2000, 275, 13564–13570. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Nandini, C.D.; Hattori, T.; Bao, X.; Murayama, D.; Nakamura, T.; Fukushima, N.; Sugahara, K. Structure of pleiotrophin- and hepatocyte growth factor-binding sulfated hexasaccharide determined by biochemical and computational approaches. J. Biol. Chem. 2010, 285, 27673–27685. [Google Scholar] [CrossRef]
- Mizumoto, S.; Fongmoon, D.; Sugahara, K. Interaction of chondroitin sulfate and dermatan sulfate from various biological sources with heparin-binding growth factors and cytokines. Glycoconj. J. 2013, 30, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.; Shen, D.; Wang, X. Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate. FEBS J. 2016, 283, 1488–1503. [Google Scholar] [CrossRef]
- Ori, A.; Free, P.; Courty, J.; Wilkinson, M.C.; Fernig, D.G. Identification of heparin-binding sites in proteins by selective labeling. Mol. Cell. Proteom. 2009, 8, 2256–2265. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.; Shen, D.; Wang, X. Pleiotrophin interacts with glycosaminoglycans in a highly flexible and adaptable manner. FEBS Lett. 2021, 595, 925–941. [Google Scholar] [CrossRef]
- Cesaretti, M.; Luppi, E.; Maccari, F.; Volpi, N. A 96-well assay for uronic acid carbazole reaction. Carbohydr. Polym. 2003, 54, 59–61. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. Nmrpipe—A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004, 278, 313–352. [Google Scholar]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; Gonzalez-Outeirino, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef]
- Huige, C.J.M.; Altona, C. Force-Field Parameters for Sulfates and Sulfamates Based on Ab-Initio Calculations—Extensions of Amber and Charmm Fields. J. Comput. Chem. 1995, 16, 56–79. [Google Scholar] [CrossRef]
- Clerc, O.; Mariethoz, J.; Rivet, A.; Lisacek, F.; Perez, S.; Ricard-Blum, S. A pipeline to translate glycosaminoglycan sequences into 3D models. Application to the exploration of glycosaminoglycan conformational space. Glycobiology 2019, 29, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Mobius, K.; Nordsieck, K.; Pichert, A.; Samsonov, S.A.; Thomas, L.; Schiller, J.; Kalkhof, S.; Teresa Pisabarro, M.; Beck-Sickinger, A.G.; Huster, D. Investigation of lysine side chain interactions of interleukin-8 with heparin and other glycosaminoglycans studied by a methylation-NMR approach. Glycobiology 2013, 23, 1260–1269. [Google Scholar] [CrossRef] [PubMed]
- Blaum, B.S.; Deakin, J.A.; Johansson, C.M.; Herbert, A.P.; Barlow, P.N.; Lyon, M.; Uhrin, D. Lysine and arginine side chains in glycosaminoglycan-protein complexes investigated by NMR, cross-linking, and mass spectrometry: A case study of the factor H-heparin interaction. J. Am. Chem. Soc. 2010, 132, 6374–6381. [Google Scholar] [CrossRef]
- Khan, S.; Gor, J.; Mulloy, B.; Perkins, S.J. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: New insight into heparin-protein complexes. J. Mol. Biol. 2010, 395, 504–521. [Google Scholar] [CrossRef]
- Mikhailov, D.; Linhardt, R.J.; Mayo, K.H. NMR solution conformation of heparin-derived hexasaccharide. Biochem. J. 1997, 328, 51–61. [Google Scholar] [CrossRef]
- Mulloy, B.; Forster, M.J.; Jones, C.; Davies, D.B. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochem. J. 1993, 293, 849–858. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C. Interpreting protein/DNA interactions: Distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 2011, 39, 2483–2491. [Google Scholar] [CrossRef]
- Dragan, A.I.; Read, C.M.; Crane-Robinson, C. Enthalpy-entropy compensation: The role of solvation. Eur. Biophys. J. EBJ 2017, 46, 301–308. [Google Scholar] [CrossRef]
- Fukada, M.; Fujikawa, A.; Chow, J.P.; Ikematsu, S.; Sakuma, S.; Noda, M. Protein tyrosine phosphatase receptor type Z is inactivated by ligand-induced oligomerization. FEBS Lett. 2006, 580, 4051–4056. [Google Scholar] [CrossRef] [PubMed]
- Afratis, N.A.; Nikitovic, D.; Multhaupt, H.A.; Theocharis, A.D.; Couchman, J.R.; Karamanos, N.K. Syndecans—Key regulators of cell signaling and biological functions. FEBS J. 2017, 284, 27–41. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryan, E.O.; Jiang, Z.; Nguyen, H.; Wang, X. Interactions of Pleiotrophin with a Structurally Defined Heparin Hexasaccharide. Biomolecules 2022, 12, 50. https://doi.org/10.3390/biom12010050
Ryan EO, Jiang Z, Nguyen H, Wang X. Interactions of Pleiotrophin with a Structurally Defined Heparin Hexasaccharide. Biomolecules. 2022; 12(1):50. https://doi.org/10.3390/biom12010050
Chicago/Turabian StyleRyan, Eathen O., Zhoumai Jiang, Hoa Nguyen, and Xu Wang. 2022. "Interactions of Pleiotrophin with a Structurally Defined Heparin Hexasaccharide" Biomolecules 12, no. 1: 50. https://doi.org/10.3390/biom12010050
APA StyleRyan, E. O., Jiang, Z., Nguyen, H., & Wang, X. (2022). Interactions of Pleiotrophin with a Structurally Defined Heparin Hexasaccharide. Biomolecules, 12(1), 50. https://doi.org/10.3390/biom12010050





