A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease
Abstract
1. Introduction
2. Signaling Pathways and Cellular Physiology Regulated by PTPRG
2.1. PTPRG Expression and Biochemical Features
2.2. PTPRG Affects Cell Differentiation
2.3. Cell Adhesion
2.4. PTPRG as HCO3− Sensor
3. Participation of PTPRG in Different Pathological Processes
3.1. Neuropsychiatric and Behavioral Disorders
3.2. Inflammation
3.3. Other Diseases
4. PTPRG Network’s Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| PTPRG | Protein tyrosine phosphatase receptor gamma |
| PTPRZ | Protein tyrosine phosphatase receptor zeta |
| LMWPTP | Low-molecular-weight tyrosine phosphatase |
| SNP | Single nucleotide polymorphism |
| TS | Tumor suppressor |
| CAH | Carbonic anhydrase like domain |
| CNTNs | Contactin protein family |
| PTKs | Protein tyrosine kinases |
| PTPs | Protein tyrosine phosphatases |
| PSTPs | Protein serine/threonine phosphatases |
| NGF | Neuronal growth factor |
| MNs | Motor neurons |
| ECD | Extracellular domain |
| ICD | Intracellular domain |
| WD | Wedge domain |
| TCF/LEF | T cell factor/lymphoid enhancer factor family |
| CML | Chronic myeloid leukemia |
| CPP | Cell penetrating peptide |
| LFA1 | Lymphocyte function-associated antigen 1 |
| VSMCs | Vascular smooth muscle cells |
| VCAM-1/ICAM-1 | Vascular cell adhesion protein 1/intracellular adhesion molecule 1 |
| OOE | Out-of-equilibrium tests |
| ASD | Autism spectrum disorder |
| AD | Alzheimer’s disease |
| LOAD | Alzheimer’s diagnosed is late onset |
| CNS | Central nervous system |
| LPS | Bacterial lipopolysaccharides |
| IR | Insulin receptor |
| T2DM | Type 2 diabetes mellitus |
| GDM | Gestational diabetes mellitus |
| NFLD | Nonalcoholic fatty liver disease |
| NLRP3 | NOD-type receptors 3 |
| SOD | Super-oxide dismutase |
| AKI | Acute kidney injury |
| CAT | Catalase |
| ROS | Reactive oxygen species |
| EGFR | Epidermal growth factor-receptor |
| FED | Fuchs’ endothelial dystrophy |
| KEGG | Kyoto Encyclopedia of Genes and Genome |
References
- Liberti, S.; Sacco, F.; Calderone, A.; Perfetto, L.; Iannuccelli, M.; Panni, S.; Santonico, E.; Palma, A.; Nardozza, A.P.; Castagnoli, L.; et al. HuPho: The human phosphatase portal. FEBS J. 2013, 280, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef]
- Krueger, N.X.; Saito, H. A human transmembrane protein-tyrosine-phosphatase, PTP zeta, is expressed in brain and has an N-terminal receptor domain homologous to carbonic anhydrases. Proc. Natl. Acad. Sci. USA 1992, 89, 7417–7421. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Barnea, G.; Silvennoinen, O.; Shaanan, B.; Honegger, A.M.; Canoll, P.D.; D’Eustachio, P.; Morse, B.; Levy, J.B.; Laforgia, S.; Huebner, K.; et al. Identification of a carbonic anhydrase-like domain in the extracellular region of RPTP gamma defines a new subfamily of receptor tyrosine phosphatases. Mol. Cell Biol. 1993, 13, 1497–1506. [Google Scholar] [CrossRef]
- Tonks, N.K. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013, 280, 346–378. [Google Scholar] [CrossRef]
- Sorio, C.; Melotti, P.; D’Arcangelo, D.; Mendrola, J.; Calabretta, B.; Croce, C.M.; Huebner, K. Receptor protein tyrosine phosphatase gamma, Ptp gamma, regulates hematopoietic differentiation. Blood 1997, 90, 49–57. [Google Scholar] [CrossRef]
- Jiang, G.; den Hertog, J.; Su, J.; Noel, J.; Sap, J.; Hunter, T. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-alpha. Nature 1999, 401, 606–610. [Google Scholar] [CrossRef]
- Majeti, R.; Bilwes, A.M.; Noel, J.P.; Hunter, T.; Weiss, A. Dimerization-induced inhibition of receptor protein tyrosine phosphatase function through an inhibitory wedge. Science 1998, 279, 88–91. [Google Scholar] [CrossRef]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.A.; Muller, S.; Knapp, S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.B.; Canoll, P.D.; Silvennoinen, O.; Barnea, G.; Morse, B.; Honegger, A.M.; Huang, J.T.; Cannizzaro, L.A.; Park, S.H.; Druck, T.; et al. The cloning of a receptor-type protein tyrosine phosphatase expressed in the central nervous system. J. Biol. Chem. 1993, 268, 10573–10581. [Google Scholar] [CrossRef]
- Tamura, H.; Fukada, M.; Fujikawa, A.; Noda, M. Protein tyrosine phosphatase receptor type Z is involved in hippocampus-dependent memory formation through dephosphorylation at Y1105 on p190 RhoGAP. Neurosci. Lett. 2006, 399, 33–38. [Google Scholar] [CrossRef]
- Lamprianou, S.; Vacaresse, N.; Suzuki, Y.; Meziane, H.; Buxbaum, J.D.; Schlessinger, J.; Harroch, S. Receptor protein tyrosine phosphatase gamma is a marker for pyramidal cells and sensory neurons in the nervous system and is not necessary for normal development. Mol. Cell Biol. 2006, 26, 5106–5119. [Google Scholar] [CrossRef]
- Lorenzetto, E.; Moratti, E.; Vezzalini, M.; Harroch, S.; Sorio, C.; Buffelli, M. Distribution of different isoforms of receptor protein tyrosine phosphatase gamma (Ptprg-RPTP gamma) in adult mouse brain: Upregulation during neuroinflammation. Brain Struct. Funct. 2014, 219, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Vezzalini, M.; Mombello, A.; Menestrina, F.; Mafficini, A.; Della Peruta, M.; van Niekerk, C.; Barbareschi, M.; Scarpa, A.; Sorio, C. Expression of transmembrane protein tyrosine phosphatase gamma (PTPgamma) in normal and neoplastic human tissues. Histopathology 2007, 50, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Lissandrini, D.; Vermi, W.; Vezzalini, M.; Sozzani, S.; Facchetti, F.; Bellone, G.; Mafficini, A.; Gentili, F.; Ennas, M.G.; Tecchio, C.; et al. Receptor-type protein tyrosine phosphatase gamma (PTPgamma), a new identifier for myeloid dendritic cells and specialized macrophages. Blood 2006, 108, 4223–4231. [Google Scholar] [CrossRef]
- Mafficini, A.; Vezzalini, M.; Zamai, L.; Galeotti, L.; Bergamini, G.; Della Peruta, M.; Melotti, P.; Sorio, C. Protein Tyrosine Phosphatase Gamma (PTPgamma) is a Novel Leukocyte Marker Highly Expressed by CD34 Precursors. Biomark. Insights 2007, 2, 218–225. [Google Scholar] [CrossRef]
- Vezzalini, M.; Mafficini, A.; Tomasello, L.; Lorenzetto, E.; Moratti, E.; Fiorini, Z.; Holyoake, T.L.; Pellicano, F.; Krampera, M.; Tecchio, C.; et al. A new monoclonal antibody detects downregulation of protein tyrosine phosphatase receptor type gamma in chronic myeloid leukemia patients. J. Hematol. Oncol. 2017, 10, 129. [Google Scholar] [CrossRef]
- Sorio, C.; Mendrola, J.; Lou, Z.; LaForgia, S.; Croce, C.M.; Huebner, K. Characterization of the receptor protein tyrosine phosphatase gene product PTP gamma: Binding and activation by triphosphorylated nucleosides. Cancer Res. 1995, 55, 4855–4864. [Google Scholar]
- Yang, X.; Zhao, Y.; Sun, Q.; Yang, Y.; Gao, Y.; Ge, W.; Liu, J.; Xu, X.; Weng, D.; Wang, S.; et al. Adenine nucleotide-mediated regulation of hepatic PTP1B activity in mouse models of type 2 diabetes. Diabetologia 2019, 62, 2106–2117. [Google Scholar] [CrossRef]
- Zhang, W.; Savelieva, K.V.; Tran, D.T.; Pogorelov, V.M.; Cullinan, E.B.; Baker, K.B.; Platt, K.A.; Hu, S.; Rajan, I.; Xu, N.; et al. Characterization of PTPRG in knockdown and phosphatase-inactive mutant mice and substrate trapping analysis of PTPRG in mammalian cells. PLoS ONE 2012, 7, e45500. [Google Scholar] [CrossRef]
- Shintani, T.; Maeda, N.; Noda, M. Receptor-like protein tyrosine phosphatase gamma (RPTPgamma), but not PTPzeta/RPTPbeta, inhibits nerve-growth-factor-induced neurite outgrowth in PC12D cells. Dev. Neurosci. 2001, 23, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, H.; Hurley, M.; Gibson, A.; Panova, V.; Tchetchelnitski, V.; Barr, A.; Stoker, A.W. Receptor tyrosine phosphatase PTPgamma is a regulator of spinal cord neurogenesis. Mol. Cell Neurosci. 2011, 46, 469–482. [Google Scholar] [CrossRef]
- Tomasello, L.; Vezzalini, M.; Boni, C.; Bonifacio, M.; Scaffidi, L.; Yassin, M.; Al-Dewik, N.; Takam Kamga, P.; Krampera, M.; Sorio, C. Regulative Loop between beta-catenin and Protein Tyrosine Receptor Type gamma in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2020, 21, 2298. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.G.; Van Vactor, D. Receptor protein tyrosine phosphatases in nervous system development. Physiol. Rev. 2003, 83, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Fukada, M.; Fujikawa, A.; Chow, J.P.; Ikematsu, S.; Sakuma, S.; Noda, M. Protein tyrosine phosphatase receptor type Z is inactivated by ligand-induced oligomerization. FEBS Lett. 2006, 580, 4051–4056. [Google Scholar] [CrossRef]
- Peles, E.; Nativ, M.; Campbell, P.L.; Sakurai, T.; Martinez, R.; Lev, S.; Clary, D.O.; Schilling, J.; Barnea, G.; Plowman, G.D.; et al. The carbonic anhydrase domain of receptor tyrosine phosphatase beta is a functional ligand for the axonal cell recognition molecule contactin. Cell 1995, 82, 251–260. [Google Scholar] [CrossRef]
- Chatterjee, M.; Schild, D.; Teunissen, C.E. Contactins in the central nervous system: Role in health and disease. Neural Regen. Res. 2019, 14, 206–216. [Google Scholar] [CrossRef]
- Bouyain, S.; Watkins, D.J. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 2443–2448. [Google Scholar] [CrossRef]
- Nikolaienko, R.M.; Hammel, M.; Dubreuil, V.; Zalmai, R.; Hall, D.R.; Mehzabeen, N.; Karuppan, S.J.; Harroch, S.; Stella, S.L.; Bouyain, S. Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues. J. Biol. Chem. 2016, 291, 21335–21349. [Google Scholar] [CrossRef] [PubMed]
- Mercati, O.; Danckaert, A.; Andre-Leroux, G.; Bellinzoni, M.; Gouder, L.; Watanabe, K.; Shimoda, Y.; Grailhe, R.; De Chaumont, F.; Bourgeron, T.; et al. Contactin 4, -5 and -6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biol. Open 2013, 2, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Akasaka, K.; Lee, S.; Kobayashi, S.; Kawano, H.; Murayama, S.; Takahashi, N.; Hashimoto, K.; Kano, M.; Asano, M.; et al. Impaired motor coordination in mice lacking neural recognition molecule NB-3 of the contactin/F3 subgroup. J. Neurobiol. 2003, 56, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Groen, A.; Overvoorde, J.; van der Wijk, T.; den Hertog, J. Redox regulation of dimerization of the receptor protein-tyrosine phosphatases RPTPalpha, LAR, RPTPmu and CD45. FEBS J. 2008, 275, 2597–2604. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Bilwes, A.; Hunter, T.; Noel, J.P. The structure of the membrane distal phosphatase domain of RPTPalpha reveals interdomain flexibility and an SH2 domain interaction region. Biochemistry 2003, 42, 7904–7914. [Google Scholar] [CrossRef]
- Bouyain, S.; Watkins, D.J. Identification of tyrosine phosphatase ligands for contactin cell adhesion molecules. Commun. Integr. Biol. 2010, 3, 284–286. [Google Scholar] [CrossRef]
- Nolte, M.A.; Margadant, C. Activation and suppression of hematopoietic integrins in hemostasis and immunity. Blood 2020, 135, 7–16. [Google Scholar] [CrossRef]
- Mirenda, M.; Toffali, L.; Montresor, A.; Scardoni, G.; Sorio, C.; Laudanna, C. Protein tyrosine phosphatase receptor type gamma is a JAK phosphatase and negatively regulates leukocyte integrin activation. J. Immunol. 2015, 194, 2168–2179. [Google Scholar] [CrossRef]
- Montresor, A.; Toffali, L.; Fumagalli, L.; Constantin, G.; Rigo, A.; Ferrarini, I.; Vinante, F.; Laudanna, C. Activation of Protein Tyrosine Phosphatase Receptor Type gamma Suppresses Mechanisms of Adhesion and Survival in Chronic Lymphocytic Leukemia Cells. J. Immunol. 2021, 207, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Montresor, A.; Toffali, L.; Rigo, A.; Ferrarini, I.; Vinante, F.; Laudanna, C. CXCR4- and BCR-triggered integrin activation in B-cell chronic lymphocytic leukemia cells depends on JAK2-activated Bruton’s tyrosine kinase. Oncotarget 2018, 9, 35123–35140. [Google Scholar] [CrossRef]
- Aspatwar, A.; Tolvanen, M.E.; Ortutay, C.; Parkkila, S. Carbonic anhydrase related proteins: Molecular biology and evolution. Subcell. Biochem. 2014, 75, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Skelton, L.A.; Xu, L.; Chandler, M.P.; Berthiaume, J.M.; Boron, W.F. Role of Receptor Protein Tyrosine Phosphatase gamma in Sensing Extracellular CO2 and HCO3. J. Am. Soc. Nephrol. 2016, 27, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Boedtkjer, E.; Hansen, K.B.; Boedtkjer, D.M.B.; Aalkjaer, C.; Boron, W.F. Extracellular HCO3− is sensed by mouse cerebral arteries: Regulation of tone by receptor protein tyrosine phosphatase gamma. J. Cereb. Blood Flow Metab. 2016, 36, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.B.; Staehr, C.; Rohde, P.D.; Homilius, C.; Kim, S.; Nyegaard, M.; Matchkov, V.V.; Boedtkjer, E. PTPRG is an ischemia risk locus essential for HCO3−-dependent regulation of endothelial function and tissue perfusion. Elife 2020, 9, e57553. [Google Scholar] [CrossRef]
- Druck, T.; Kastury, K.; Hadaczek, P.; Podolski, J.; Toloczko, A.; Sikorski, A.; Ohta, M.; LaForgia, S.; Lasota, J.; McCue, P.; et al. Loss of heterozygosity at the familial RCC t(3;8) locus in most clear cell renal carcinomas. Cancer Res. 1995, 55, 5348–5353. [Google Scholar]
- LaForgia, S.; Morse, B.; Levy, J.; Barnea, G.; Cannizzaro, L.A.; Li, F.; Nowell, P.C.; Boghosian-Sell, L.; Glick, J.; Weston, A.; et al. Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc. Natl. Acad. Sci. USA 1991, 88, 5036–5040. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, D.; Parsons, D.W.; Bardelli, A.; Sager, J.; Szabo, S.; Ptak, J.; Silliman, N.; Peters, B.A.; van der Heijden, M.S.; et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004, 304, 1164–1166. [Google Scholar] [CrossRef]
- Della Peruta, M.; Martinelli, G.; Moratti, E.; Pintani, D.; Vezzalini, M.; Mafficini, A.; Grafone, T.; Iacobucci, I.; Soverini, S.; Murineddu, M.; et al. Protein tyrosine phosphatase receptor type {gamma} is a functional tumor suppressor gene specifically downregulated in chronic myeloid leukemia. Cancer Res. 2010, 70, 8896–8906. [Google Scholar] [CrossRef]
- Drube, J.; Ernst, T.; Pfirrmann, M.; Albert, B.V.; Drube, S.; Reich, D.; Kresinsky, A.; Halfter, K.; Sorio, C.; Fabisch, C.; et al. PTPRG and PTPRC modulate nilotinib response in chronic myeloid leukemia cells. Oncotarget 2018, 9, 9442–9455. [Google Scholar] [CrossRef]
- Shu, S.T.; Sugimoto, Y.; Liu, S.; Chang, H.L.; Ye, W.; Wang, L.S.; Huang, Y.W.; Yan, P.; Lin, Y.C. Function and regulatory mechanisms of the candidate tumor suppressor receptor protein tyrosine phosphatase gamma (PTPRG) in breast cancer cells. Anticancer Res. 2010, 30, 1937–1946. [Google Scholar]
- Boni, C.; Sorio, C. The role of the tumor suppressor gene Protein Tyrosine Phosphatase Gamma (PTPRG) in cancer. Front. Cell Dev. Biol. 2021. [Google Scholar] [CrossRef]
- Le, H.T.; Maksumova, L.; Wang, J.; Pallen, C.J. Reduced NMDA receptor tyrosine phosphorylation in PTPalpha-deficient mouse synaptosomes is accompanied by inhibition of four src family kinases and Pyk2: An upstream role for PTPalpha in NMDA receptor regulation. J. Neurochem. 2006, 98, 1798–1809. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Craig, A.M. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: Presynaptic hubs for synapse organization. Trends Neurosci. 2013, 36, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J.; Weinberger, D.R. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol. Psychiatry 2005, 10, 40–68. [Google Scholar] [CrossRef]
- Takahashi, N.; Nielsen, K.S.; Aleksic, B.; Petersen, S.; Ikeda, M.; Kushima, I.; Vacaresse, N.; Ujike, H.; Iwata, N.; Dubreuil, V.; et al. Loss of function studies in mice and genetic association link receptor protein tyrosine phosphatase alpha to schizophrenia. Biol. Psychiatry 2011, 70, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Cressant, A.; Dubreuil, V.; Kong, J.; Kranz, T.M.; Lazarini, F.; Launay, J.M.; Callebert, J.; Sap, J.; Malaspina, D.; Granon, S.; et al. Loss-of-function of PTPR gamma and zeta, observed in sporadic schizophrenia, causes brain region-specific deregulation of monoamine levels and altered behavior in mice. Psychopharmacology 2017, 234, 575–587. [Google Scholar] [CrossRef]
- Gandawijaya, J.; Bamford, R.A.; Burbach, J.P.H.; Oguro-Ando, A. Cell Adhesion Molecules Involved in Neurodevelopmental Pathways Implicated in 3p-Deletion Syndrome and Autism Spectrum Disorder. Front. Cell Neurosci. 2020, 14, 611379. [Google Scholar] [CrossRef]
- Zuko, A.; Kleijer, K.T.E.; Oguro-Ando, A.; Kas, M.J.H.; van Daalen, E.; van der Zwaag, B.; Burbach, J.P.H. Contactins in the neurobiology of autism. Eur. J. Pharmacol. 2013, 719, 63–74. [Google Scholar] [CrossRef]
- Kranz, T.M.; Berns, A.; Shields, J.; Rothman, K.; Walsh-Messinger, J.; Goetz, R.R.; Chao, M.V.; Malaspina, D. Phenotypically distinct subtypes of psychosis accompany novel or rare variants in four different signaling genes. EBioMedicine 2016, 6, 206–214. [Google Scholar] [CrossRef]
- Kranz, T.M.; Harroch, S.; Manor, O.; Lichtenberg, P.; Friedlander, Y.; Seandel, M.; Harkavy-Friedman, J.; Walsh-Messinger, J.; Dolgalev, I.; Heguy, A.; et al. De novo mutations from sporadic schizophrenia cases highlight important signaling genes in an independent sample. Schizophr. Res. 2015, 166, 119–124. [Google Scholar] [CrossRef]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar] [CrossRef]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef]
- Hamshere, M.L.; Green, E.K.; Jones, I.R.; Jones, L.; Moskvina, V.; Kirov, G.; Grozeva, D.; Nikolov, I.; Vukcevic, D.; Caesar, S.; et al. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br. J. Psychiatry 2009, 195, 23–29. [Google Scholar] [CrossRef]
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Herold, C.; Hooli, B.V.; Mullin, K.; Liu, T.; Roehr, J.T.; Mattheisen, M.; Parrado, A.R.; Bertram, L.; Lange, C.; Tanzi, R.E. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol. Psychiatry 2016, 21, 1608–1612. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef]
- Suzuki, K. Chronic Inflammation as an Immunological Abnormality and Effectiveness of Exercise. Biomolecules 2019, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Maeda, N.; Nishiwaki, T.; Noda, M. Characterization of rat receptor-like protein tyrosine phosphatase gamma isoforms. Biochem. Biophys. Res. Commun. 1997, 230, 419–425. [Google Scholar] [CrossRef]
- Campan, M.; Yoshizumi, M.; Seidah, N.G.; Lee, M.E.; Bianchi, C.; Haber, E. Increased proteolytic processing of protein tyrosine phosphatase mu in confluent vascular endothelial cells: The role of PC5, a member of the subtilisin family. Biochemistry 1996, 35, 3797–3802. [Google Scholar] [CrossRef]
- Kapp, K.; Siemens, J.; Haring, H.U.; Lammers, R. Proteolytic processing of the protein tyrosine phosphatase alpha extracellular domain is mediated by ADAM17/TACE. Eur J. Cell Biol. 2012, 91, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Streuli, M.; Krueger, N.X.; Ariniello, P.D.; Tang, M.; Munro, J.M.; Blattler, W.A.; Adler, D.A.; Disteche, C.M.; Saito, H. Expression of the receptor-linked protein tyrosine phosphatase LAR: Proteolytic cleavage and shedding of the CAM-like extracellular region. EMBO J. 1992, 11, 897–907. [Google Scholar] [CrossRef]
- Eguchi, A.; Wree, A.; Feldstein, A.E. Biomarkers of liver cell death. J. Hepatol. 2014, 60, 1063–1074. [Google Scholar] [CrossRef]
- Moratti, E.; Vezzalini, M.; Tomasello, L.; Giavarina, D.; Sorio, C. Identification of protein tyrosine phosphatase receptor gamma extracellular domain (sPTPRG) as a natural soluble protein in plasma. PLoS ONE 2015, 10, e0119110. [Google Scholar] [CrossRef]
- Chow, J.P.; Fujikawa, A.; Shimizu, H.; Suzuki, R.; Noda, M. Metalloproteinase- and gamma-secretase-mediated cleavage of protein-tyrosine phosphatase receptor type Z. J. Biol. Chem. 2008, 283, 30879–30889. [Google Scholar] [CrossRef] [PubMed]
- Yamanoi, Y.; Fujii, M.; Murakami, Y.; Nagai, K.; Hoshi, K.; Hashimoto, Y.; Honda, T.; Saito, K.; Kitazume, S. Soluble protein tyrosine phosphatase receptor type Z (PTPRZ) in cerebrospinal fluid is a potential diagnostic marker for glioma. Neurooncol. Adv. 2020, 2, vdaa055. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef]
- Wu, H.; Ballantyne, C.M. Skeletal muscle inflammation and insulin resistance in obesity. J. Clin. Investig. 2017, 127, 43–54. [Google Scholar] [CrossRef]
- Crunkhorn, S. Metabolic Disease: Protein tyrosine phosphatase inhibitor reverses diabetes. Nat. Rev. Drug Discov. 2017, 16, 312–313. [Google Scholar] [CrossRef]
- Stanford, S.M.; Aleshin, A.E.; Zhang, V.; Ardecky, R.J.; Hedrick, M.P.; Zou, J.; Ganji, S.R.; Bliss, M.R.; Yamamoto, F.; Bobkov, A.A.; et al. Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase. Nat. Chem. Biol. 2017, 13, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Brenachot, X.; Ramadori, G.; Ioris, R.M.; Veyrat-Durebex, C.; Altirriba, J.; Aras, E.; Ljubicic, S.; Kohno, D.; Fabbiano, S.; Clement, S.; et al. Hepatic protein tyrosine phosphatase receptor gamma links obesity-induced inflammation to insulin resistance. Nat. Commun. 2017, 8, 1820. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, E.S.; Yoo, J.; Kim, Y. Predicting risk of type 2 diabetes mellitus in Korean adults aged 40-69 by integrating clinical and genetic factors. Prim. Care Diabetes 2019, 13, 3–10. [Google Scholar] [CrossRef]
- Pineda-Cortel, M.R.B.; Bunag, J.A.A.; Mamerto, T.P.; Abulencia, M.F.B. Differential gene expression and network-based analyses of the placental transcriptome reveal distinct potential biomarkers for gestationaldiabetes mellitus. Diabetes Res. Clin. Pract. 2021, 180, 109046. [Google Scholar] [CrossRef]
- Arrese, M.; Cabrera, D.; Kalergis, A.M.; Feldstein, A.E. Innate Immunity and Inflammation in NAFLD/NASH. Dig. Dis. Sci. 2016, 61, 1294–1303. [Google Scholar] [CrossRef]
- Gallego, P.; Castejon-Vega, B.; Del Campo, J.A.; Cordero, M.D. The Absence of NLRP3-inflammasome Modulates Hepatic Fibrosis Progression, Lipid Metabolism, and Inflammation in KO NLRP3 Mice during Aging. Cells 2020, 9, 2148. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; James, M.T. Acute Kidney Injury. Ann. Intern Med. 2018, 168, 837. [Google Scholar] [CrossRef] [PubMed]
- Gabarre, P.; Dumas, G.; Dupont, T.; Darmon, M.; Azoulay, E.; Zafrani, L. Acute kidney injury in critically ill patients with COVID-19. Intensiv. Care Med. 2020, 46, 1339–1348. [Google Scholar] [CrossRef]
- Newbury, L.J.; Simpson, K.; Khalid, U.; John, I.; de Rivera, L.B.; Lu, Y.A.; Lopez-Anton, M.; Watkins, W.J.; Jenkins, R.H.; Fraser, D.J.; et al. miR-141 mediates recovery from acute kidney injury. Sci. Rep. 2021, 11, 16499. [Google Scholar] [CrossRef]
- Liu, M.; Yang, R.; Urrehman, U.; Ye, C.; Yan, X.; Cui, S.; Hong, Y.; Gu, Y.; Liu, Y.; Zhao, C.; et al. MiR-19b suppresses PTPRG to promote breast tumorigenesis. Oncotarget 2016, 7, 64100–64108. [Google Scholar] [CrossRef]
- Yu, C.; Tian, F.; Liu, J.; Su, M.; Wu, M.; Zhu, X.; Qian, W. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Prolif. 2019, 52, e12610. [Google Scholar] [CrossRef]
- Liu, A.; Sun, Y.; Yu, B. MicroRNA-208a Correlates Apoptosis and Oxidative Stress Induced by H2O2 through Protein Tyrosine Kinase/Phosphatase Balance in Cardiomyocytes. Int. Heart J. 2018, 59, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Matthaei, M.; Hribek, A.; Clahsen, T.; Bachmann, B.; Cursiefen, C.; Jun, A.S. Fuchs Endothelial Corneal Dystrophy: Clinical, Genetic, Pathophysiologic, and Therapeutic Aspects. Annu Rev. Vis. Sci. 2019, 5, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Baratz, K.H.; Tosakulwong, N.; Ryu, E.; Brown, W.L.; Branham, K.; Chen, W.; Tran, K.D.; Schmid-Kubista, K.E.; Heckenlively, J.R.; Swaroop, A.; et al. E2-2 protein and Fuchs’s corneal dystrophy. N. Engl. J. Med. 2010, 363, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Kuot, A.; Hewitt, A.W.; Griggs, K.; Klebe, S.; Mills, R.; Jhanji, V.; Craig, J.E.; Sharma, S.; Burdon, K.P. Association of TCF4 and CLU polymorphisms with Fuchs’ endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur. J. Hum. Genet. 2012, 20, 632–638. [Google Scholar] [CrossRef]
- Lau, L.C.; Ma, L.; Young, A.L.; Rong, S.S.; Jhanji, V.; Brelen, M.E.; Pang, C.P.; Chen, L.J. Association of common variants in TCF4 and PTPRG with Fuchs’ corneal dystrophy: A systematic review and meta-analysis. PLoS ONE 2014, 9, e109142. [Google Scholar] [CrossRef]
- Wang, K.J.; Jhanji, V.; Chen, J.; Law, R.W.; Leung, A.T.; Zhang, M.; Wang, N.; Pang, C.P.; Yam, G.H. Association of transcription factor 4 (TCF4) and protein tyrosine phosphatase, receptor type G (PTPRG) with corneal dystrophies in southern Chinese. Ophthalmic Genet. 2014, 35, 138–141. [Google Scholar] [CrossRef]
- Merico, D.; Zarrei, M.; Costain, G.; Ogura, L.; Alipanahi, B.; Gazzellone, M.J.; Butcher, N.J.; Thiruvahindrapuram, B.; Nalpathamkalam, T.; Chow, E.W.; et al. Whole-Genome Sequencing Suggests Schizophrenia Risk Mechanisms in Humans with 22q11.2 Deletion Syndrome. G3 Genes Genomes Genet. 2015, 5, 2453–2461. [Google Scholar] [CrossRef] [PubMed]
- Malaspina, D.; Kranz, T.M.; Heguy, A.; Harroch, S.; Mazgaj, R.; Rothman, K.; Berns, A.; Hasan, S.; Antonius, D.; Goetz, R.; et al. Prefrontal neuronal integrity predicts symptoms and cognition in schizophrenia and is sensitive to genetic heterogeneity. Schizophr. Res. 2016, 172, 94–100. [Google Scholar] [CrossRef][Green Version]

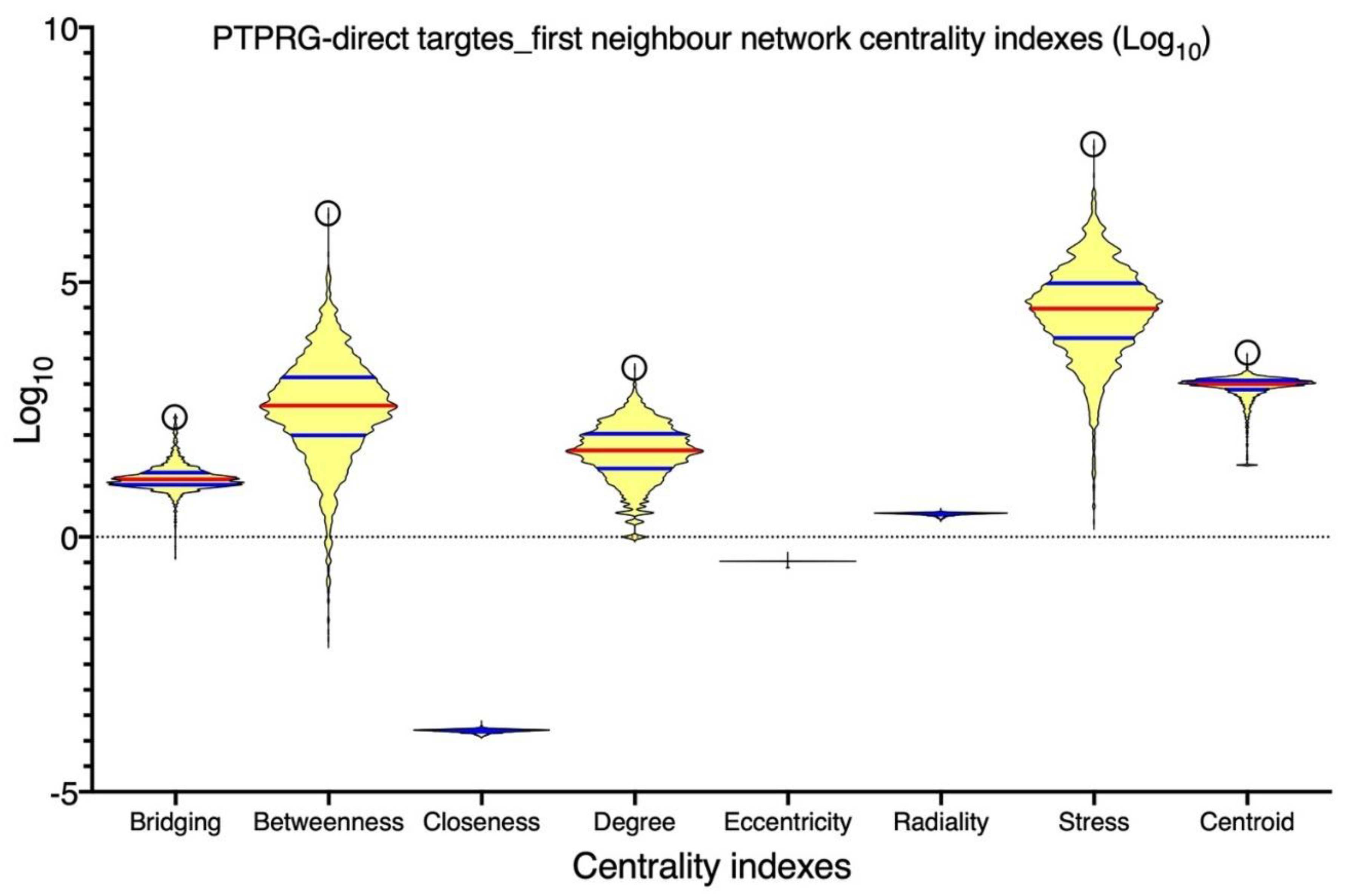
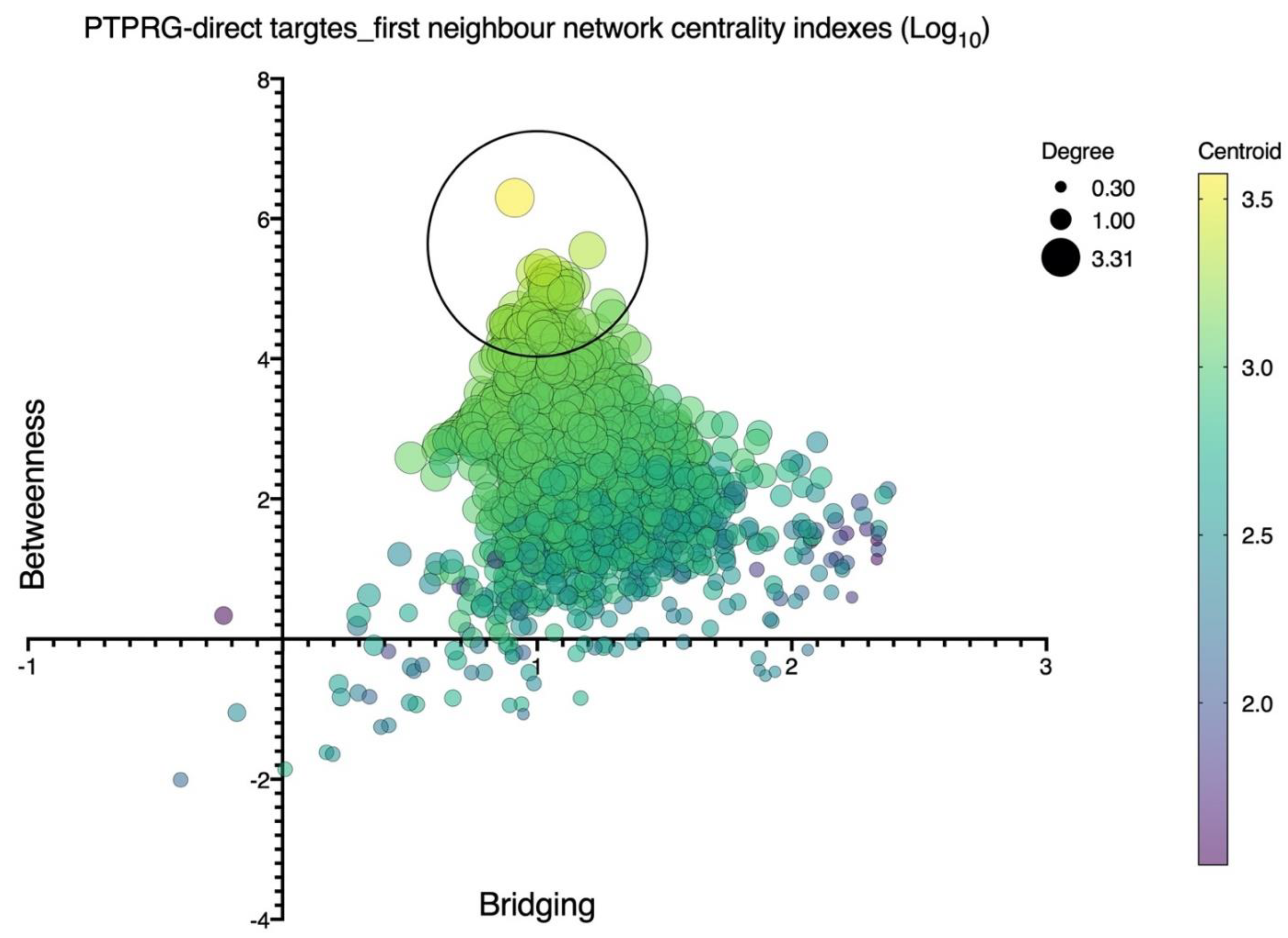
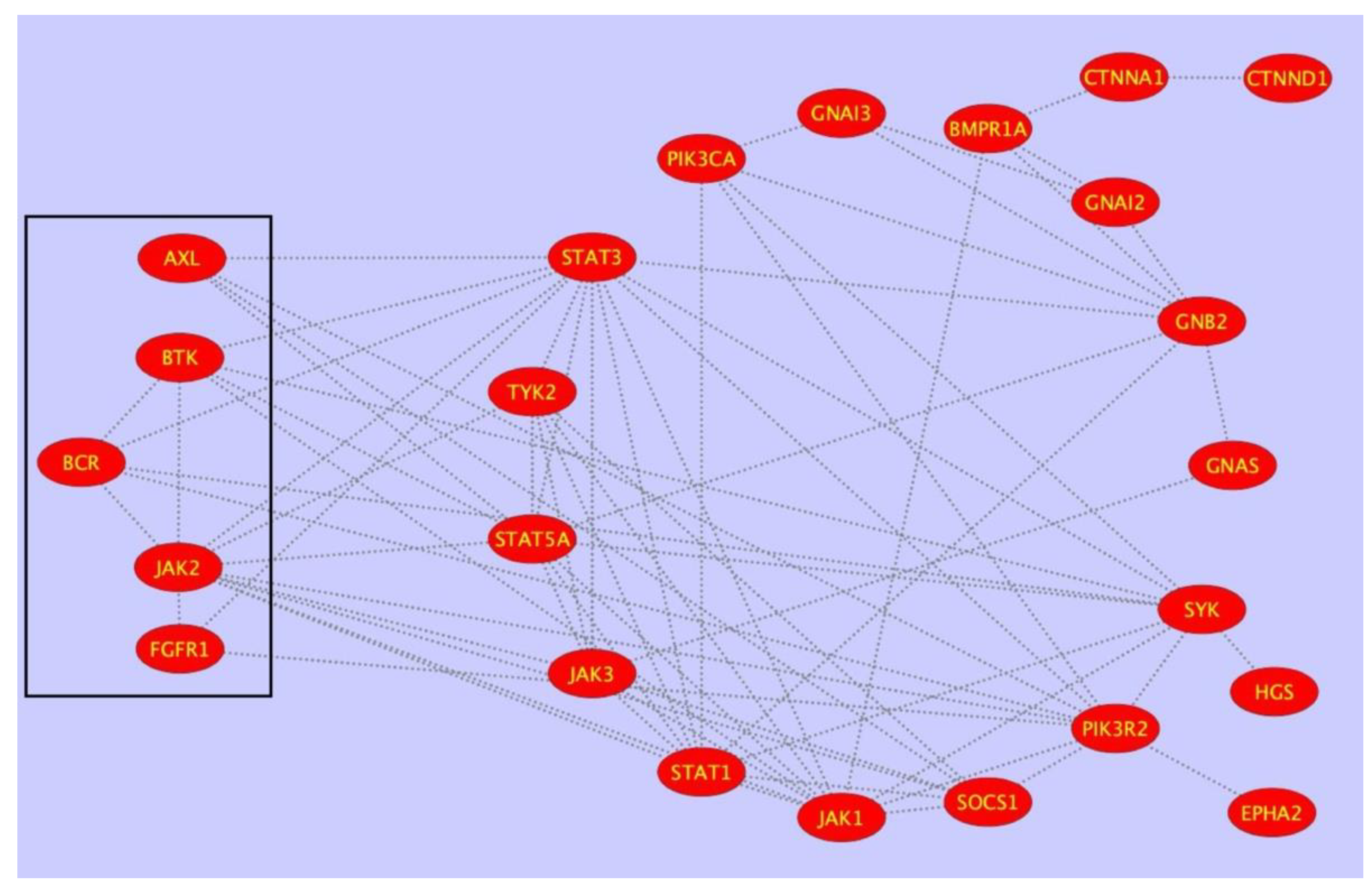
| Disease | Role of PTPRG | References |
|---|---|---|
| Cancers | Tumor suppressor role in many malignancies with a reported oncogenic role in glioblastoma | [51] |
| Neuropsychiatric and behavioral disorders | PTPRG variants have been associated to neuropsychiatric pathologies such as autism and schizophrenia | [56,59,60,96,97] |
| Inflammation | At the hepatic level, the overexpression of PTPRG correlates with inflammatory processes especially in obese subjects. Increased levels of the ECD portion in serum constitutes a potential biomarker of liver damage. | [73,80,81,82,84] |
| Acute kidney injury | PTPRG shutdown by mir-141 is associated to accelerated fibrotic process in AKI-prone kidney. | [87] |
| Fuchs’ endothelial dystrophy | Possible correlation between FED and PTPRG | [93] |
| Pathway or Process (KEGG) | XD-Score | q-Value | Overlap/Size |
|---|---|---|---|
| Acute myeloid leukemia | 0.24613 | 0.00665 | 4/52 |
| Jak-STAT signaling pathway | 0.23695 | 0.00000 | 10/134 |
| Pancreatic cancer | 0.22415 | 0.00177 | 5/70 |
| Type II diabetes mellitus | 0.21751 | 0.03097 | 3/43 |
| Fc epsilon RI signaling pathway | 0.18459 | 0.01242 | 4/65 |
| Leukocyte transendothelial migration | 0.18334 | 0.00111 | 6/98 |
| Primary immunodeficiency | 0.18086 | 0.11451 | 2/33 |
| Endometrial cancer | 0.17844 | 0.04415 | 3/50 |
| B cell receptor signaling pathway | 0.17032 | 0.01248 | 4/69 |
| Chronic myeloid leukemia | 0.17032 | 0.01248 | 4/69 |
| Chemokine signaling pathway | 0.15020 | 0.00003 | 9/170 |
| Long term depression | 0.14897 | 0.05952 | 3/57 |
| Aldosterone-regulated sodium reabsorption | 0.14897 | 0.13898 | 2/38 |
| Progesterone-mediated oocyte maturation | 0.14097 | 0.01893 | 4/79 |
| Chagas disease | 0.14046 | 0.00665 | 5/99 |
| Pathway or Process (REACTOME) | XD-Score | q-Value | Overlap/Size |
| G ALPHA Z SIGNALLING EVENTS | 0.63202 | 0.05139 | 2/12 |
| ADP SIGNALLING THROUGH P2Y PURINOCEPTOR 12 | 0.59693 | 0.01136 | 3/19 |
| DOWN STREAM SIGNAL TRANSDUCTION | 0.55359 | 0.00022 | 5/34 |
| G PROTEIN ACTIVATION | 0.44535 | 0.02116 | 3/25 |
| TIE2 SIGNALING | 0.43594 | 0.09034 | 2/17 |
| SIGNAL AMPLIFACTION | 0.37915 | 0.02764 | 3/29 |
| COLLAGEN MEDIATED ACTIVATION CASCADE | 0.34631 | 0.12357 | 2/21 |
| PI3K CASCADE | 0.34035 | 0.03184 | 3/32 |
| GAB1 SIGNALOSOME | 0.32899 | 0.64986 | 1/11 |
| GS APLHA MEDIATED EVENTS IN GLUCAGONE SIGNALLING | 0.32899 | 0.12357 | 2/22 |
| PLATELED ACTIVATION TRIGGERS | 0.32899 | 0.00131 | 5/55 |
| Pathway or Process (Go Data base) | XD-Score | q-Value | Overlap/Size |
| 1-phosphatidylinositol-3-kinase activity | 0.77762 | 0.01530 | 2/10 |
| Insulin receptor substrate binding | 0.59301 | 0.02376 | 2/13 |
| Non-membrane spanning protein tyrosine kinase activity | 0.57762 | 0.00000 | 6/40 |
| G-protein beta/gamma-subunit complex binding | 0.57762 | 0.00119 | 3/20 |
| Phosphatidylserine binding | 0.37762 | 0.54997 | 1/10 |
| Cadherin binding | 0.37762 | 0.04768 | 2/20 |
| Peptide hormone receptor binding | 0.37762 | 0.54997 | 1/10 |
| Guanyl nucleotide binding | 0.35262 | 0.00443 | 3/32 |
| RNA polymerase II core promoter sequence-specific DNA binding | 0.34126 | 0.05334 | 2/22 |
| Insulin-like growth factor receptor binding | 0.31096 | 0.55963 | 1/12 |
| Gamma-catenin binding | 0.31096 | 0.55963 | 1/12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boni, C.; Laudanna, C.; Sorio, C. A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease. Biomolecules 2022, 12, 84. https://doi.org/10.3390/biom12010084
Boni C, Laudanna C, Sorio C. A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease. Biomolecules. 2022; 12(1):84. https://doi.org/10.3390/biom12010084
Chicago/Turabian StyleBoni, Christian, Carlo Laudanna, and Claudio Sorio. 2022. "A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease" Biomolecules 12, no. 1: 84. https://doi.org/10.3390/biom12010084
APA StyleBoni, C., Laudanna, C., & Sorio, C. (2022). A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease. Biomolecules, 12(1), 84. https://doi.org/10.3390/biom12010084







