Soil Properties and Bacterial Communities Associated with the Rhizosphere of the Common Bean after Using Brachiaria brizantha as a Service Crop: A 10-Year Field Experiment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiment
2.2. Rhizosphere Soil Sampling and Sample Preparation
2.3. Rhizosphere Soil Chemical and Physical Analyses
2.4. Rhizosphere Soil Microbial Biomass and Respiration
2.5. Rhizosphere Soil Enzymatic Activities
2.6. Rhizosphere Soil DNA Extraction
2.7. Quantification of Bacterial and Fungal Genes
2.8. High Throughput Sequencing of 16S rRNA Gene Amplicons and Bioinformatic Analysis
2.9. Statistical and Bioinformatic Analyses
3. Results
3.1. Effect of the Inclusion of B. brizantha Service Crop on Soil Chemical and Physical Parameters
3.2. Rhizosphere Soil Microbial Biomass and Respiration
3.3. Rhizosphere Soil Enzyme Activities
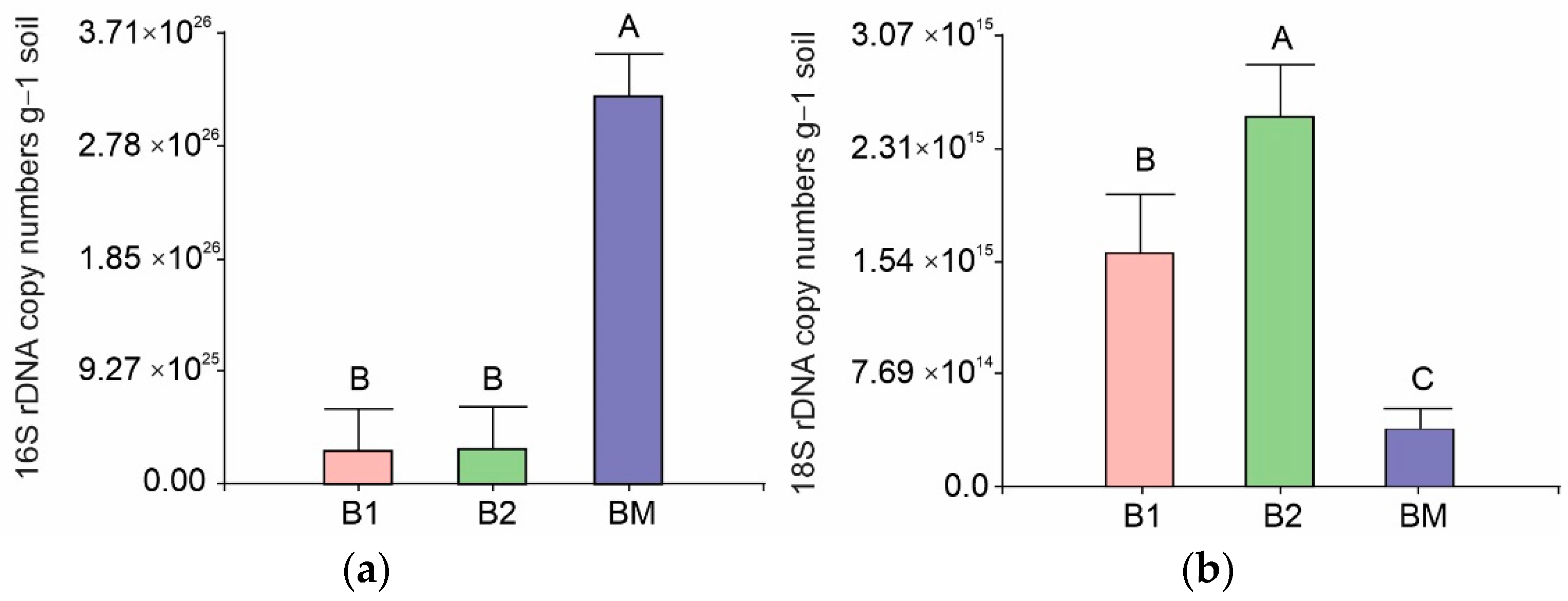
3.4. Bacterial and Fungal Abundance Via qPCR
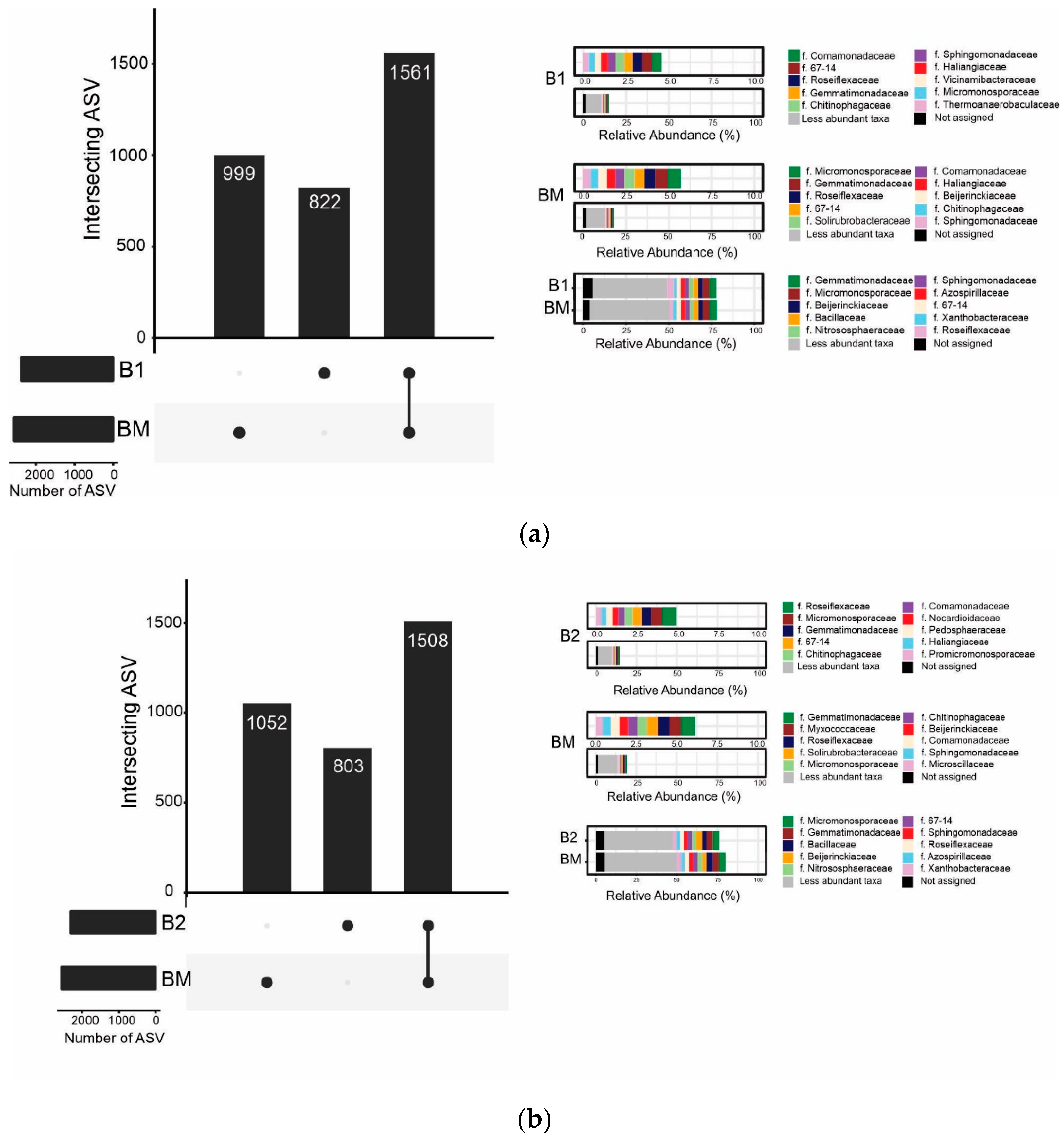
3.5. Bacterial Community Structure and Composition: Abundance and Diversity
3.6. Predicted Functions of the Bacterial Community
3.7. Variation in Rhizosphere Soil Properties under Different Service Crop Treatments
3.8. Relationships among Rhizosphere Soil Properties and Bacterial Communities
4. Discussion
4.1. Effects of B. brizantha on Rhizosphere Soil Chemical and Physical Properties
4.2. Effects of B. brizantha on Soil Microbial Activity
4.3. Effects of B. brizantha on Bacterial Community Structure, Abundance, and Diversity
4.4. Predicted Functions of the Bacterial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Broughton, W.J.; Hernández, G.; Blair, M.; Beebe, S.; Gepts, P.; Vanderleyden, J. Beans (Phaseolus spp.)—Model Food Legumes. Plant Soil 2003, 252, 55–128. [Google Scholar] [CrossRef] [Green Version]
- Vargas Gil, S.; Pastor, S.; March, G.J. Quantitative Isolation of Biocontrol Agents Trichoderma Spp., Gliocladium spp. and Actinomycetes from Soil with Culture Media. Microbiol. Res. 2009, 164, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Perez Brandan, C.; Chavarría, D.; Huidobro, J.; Meriles, J.M.; Perez Brandan, C.; Vargas Gil, S. Influence of a Tropical Grass (Brachiaria Brizantha Cv. Mulato) as Cover Crop on Soil Biochemical Properties in a Degraded Agricultural Soil. Eur. J. Soil Biol. 2017, 83, 84–90. [Google Scholar] [CrossRef]
- Pérez-Brandán, C.; Huidobro, J.; Grümberg, B.; Scandiani, M.M.; Luque, A.G.; Meriles, J.M.; Vargas-Gil, S. Soybean Fungal Soil-Borne Diseases: A Parameter for Measuring the Effect of Agricultural Intensification on Soil Health. Can. J. Microbiol. 2014, 60, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Malézieux, E. Designing Cropping Systems from Nature. Agron. Sustain. Dev. 2012, 32, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Pulleman, M.M.; de Boer, W.; Giller, K.E.; Kuyper, T.W. Soil Biodiversity and Nature-Mimicry in Agriculture; the Power of Metaphor? Outlook Agric. 2022, 51, 75–90. [Google Scholar] [CrossRef]
- Kaye, J.P.; Quemada, M. Using Cover Crops to Mitigate and Adapt to Climate Change. A Review. Agron. Sustain. Dev. 2017, 37, 4. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Singh, A.; Kahlon, C.S.; Brar, A.S.; Grover, K.K.; Dia, M.; Steiner, R.L. The Role of Cover Crops towards Sustainable Soil Health and Agriculture—A Review Paper. Am. J. Plant Sci. 2018, 9, 1935–1951. [Google Scholar] [CrossRef] [Green Version]
- Ogilvie, C.M.; Deen, W.; Martin, R.C. Service Crop Management to Maximize Crop Water Supply and Improve Agroecosystem Resilience: A Review. J. Soil Water Conserv. 2019, 74, 389–404. [Google Scholar] [CrossRef]
- Tiemann, L.K.; Grandy, A.S.; Atkinson, E.E.; Marin-Spiotta, E.; Mcdaniel, M.D. Crop Rotational Diversity Enhances Belowground Communities and Functions in an Agroecosystem. Ecol. Lett. 2015, 18, 761–771. [Google Scholar] [CrossRef]
- Steenwerth, K.; Belina, K.M. Cover Crops and Cultivation: Impacts on Soil N Dynamics and Microbiological Function in a Mediterranean Vineyard Agroecosystem. Appl. Soil Ecol. 2008, 40, 370–380. [Google Scholar] [CrossRef]
- Poeplau, C.; Don, A. Agriculture, Ecosystems and Environment Carbon Sequestration in Agricultural Soils via Cultivation of Cover Crops—A Meta-Analysis. Agric. Ecosyst. Environ. 2015, 200, 33–41. [Google Scholar] [CrossRef]
- Schipanski, M.E.; Barbercheck, M.; Douglas, M.R.; Finney, D.M.; Haider, K.; Kaye, J.P.; Kemanian, A.R.; Mortensen, D.A.; Ryan, M.R.; Tooker, J.; et al. A Framework for Evaluating Ecosystem Services Provided by Cover Crops in Agroecosystems. Agric. Syst. 2014, 125, 12–22. [Google Scholar] [CrossRef]
- Mazzoncini, M.; Sapkota, T.B.; Bàrberi, P.; Antichi, D.; Risaliti, R. Long-Term Effect of Tillage, Nitrogen Fertilization and Cover Crops on Soil Organic Carbon and Total Nitrogen Content. Soil Tillage Res. 2011, 114, 165–174. [Google Scholar] [CrossRef]
- Moukanni, N.; Brewer, K.M.; Gaudin, A.C.M.; O’Geen, A.T. Optimizing Carbon Sequestration Through Cover Cropping in Mediterranean Agroecosystems: Synthesis of Mechanisms and Implications for Management. Front. Agron. 2022, 4, 844166. [Google Scholar] [CrossRef]
- Chahal, I.; Vyn, R.J.; Mayers, D.; Van Eerd, L.L. Cumulative Impact of Cover Crops on Soil Carbon Sequestration and Profitability in a Temperate Humid Climate. Sci. Rep. 2020, 10, 13381. [Google Scholar] [CrossRef]
- Seitz, D.; Fischer, L.M.; Dechow, R.; Wiesmeier, M.; Don, A. The Potential of Cover Crops to Increase Soil Organic Carbon Storage in German Croplands. Plant Soil 2022. [Google Scholar] [CrossRef]
- Castiglioni, M.G.; Kraemer, F.B. Short-Term Effect of Cover Crops on Aggregate Stability Assessed by Two Techniques. Cienc. del Suelo 2019, 37, 298–314. [Google Scholar]
- Abán, C.L.; Verdenelli, R.; Gil, S.V.; Huidobro, D.J.; Meriles, J.M.; Brandan, C.P. Service Crops Improve a Degraded Monoculture System by Changing Common Bean Rhizospheric Soil Microbiota and Reducing Soil-Borne Fungal Diseases. FEMS Microbiol. Ecol. 2021, 97, fiaa258. [Google Scholar] [CrossRef]
- Chavarría, D.N.; Verdenelli, R.A.; Muñoz, E.J.; Conforto, C.; Restovich, S.B.; Andriulo, A.E.; Meriles, J.M.; Vargas-Gil, S. Soil Microbial Functionality in Response to the Inclusion of Cover Crop Mixtures in Agricultural Systems. Spanish J. Agric. Res. 2016, 14, e0304. [Google Scholar] [CrossRef] [Green Version]
- Arango, J.; Moreta, D.; Núñez, J.; Hartmann, K.; Domínguez, M.; Ishitani, M.; Miles, J.; Subbarao, G.; Peters, M.; Rao, I. Developing Methods to Evaluate Phenotypic Variability in Biological Nitrification Inhibition (BNI) Capacity of Brachiaria Grasses. Trop. Grassl.-Forrajes Trop. 2014, 2, 6–8. [Google Scholar] [CrossRef]
- Moreta, D.E.; Arango, J.; Sotelo, M.; Vergara, D.; Rincón, A.; Ishitani, M.; Castro, A.; Miles, J.; Peters, M.; Tohme, J.; et al. Biological Nitrification Inhibition (BNI) in Brachiaria Pastures: A Novel Strategy to Improve Eco-Efficiency of Crop-Livestock Systems and to Mitigate Climate Change. Trop. Grassl.-Forrajes Trop. 2014, 2, 88–91. [Google Scholar] [CrossRef]
- Berg, G.; Zachow, C.; Müller, H.; Philipps, J.; Tilcher, R. Next-Generation Bio-Products Sowing the Seeds of Success for Sustainable Agriculture. Agronomy 2013, 3, 648–656. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; Van Veen, J.A.; Tsai, S.M. Taxonomical and Functional Microbial Community Selection in Soybean Rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Hinsinger, P. The Rhizosphere: Complex by Design. Plant Soil 2008, 312, 1–6. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Babalola, O.O. The Rhizosphere Microbial Complex in Plant Health: A Review of Interaction Dynamics. J. Integr. Agric. 2022, 21, 2168–2182. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant Species and Soil Type Cooperatively Shape the Structure and Function of Microbial Communities in the Rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delitte, M.; Caulier, S.; Bragard, C.; Desoignies, N. Plant Microbiota Beyond Farming Practices: A Review. Front. Sustain. Food Syst. 2021, 5, 624203. [Google Scholar] [CrossRef]
- Vargas Gil, S.; Meriles, J.; Conforto, C.; Basanta, M.; Radl, V.; Hagn, A.; Schloter, M.; March, G.J. Response of Soil Microbial Communities to Different Management Practices in Surface Soils of a Soybean Agroecosystem in Argentina. Eur. J. Soil Biol. 2011, 47, 55–60. [Google Scholar] [CrossRef]
- Nurulita, Y.; Adetutu, E.M.; Gunawan, H.; Zul, D.; Ball, A.S. Restoration of Tropical Peat Soils: The Application of Soil Microbiology for Monitoring the Success of the Restoration Process. Agric. Ecosyst. Environ. 2016, 216, 293–303. [Google Scholar] [CrossRef]
- Volante, J.N.; Paruelo, J.M. Is Forest or Ecological Transition Taking Place? Evidence for the Semiarid Chaco in Argentina. J. Arid Environ. 2015, 123, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Walkley, A.; Black, I.A. An Examination of the Degtjareff Method for Determining Soil Organic Matter, and a Proposed Modification of the Chromic Acid Titration Method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Use of P-Nitrophenyl Phosphate for Assay of Soil Phosphatase Activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of Total, Organic, and Available Forms of Phosphorus in Soils. Soil Sci. 1945, 59, 39–45. [Google Scholar] [CrossRef]
- Blake, G.; Hartge, K. Bulk Density; Soil Science Society of America Press: Madison, WI, USA, 1986. [Google Scholar]
- Corvalan, E.; Franzoni, A.; Huidobro, J. Microsieves Method to Estimate Soil Aggregate Stability 1−2 mm. In Proceedings of the 14th Soil Science Congress, Mar del Plata, Argentina, 14 April 2000. [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An Extraction Method for Measuring Soil Microbial Biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Brookes, P.C.; Joergensen, R.G. Microbial Biomass Measurements by Fumigation-Extraction. In Microbiological Methods for Assessing Soil Quality; CABI: Wallingford, UK, 2005; pp. 77–83. [Google Scholar]
- Alef, K. Soil Respiration. In Methods in Applied Soil Microbiology and Biochemistry; Alef, K., Nannipieri, P., Eds.; Academic Press: London, UK, 1995; pp. 214–219. [Google Scholar]
- Adam, G.; Duncan, H. Development of a Sensitive and Rapid Method for the Measurement of Total Microbial Activity Using Fluorescein Diacetate (FDA) in a Range of Soils. Soil Biol. Biochem. 2001, 33, 943–951. [Google Scholar] [CrossRef]
- García, C.; Hernandez, T.; Costa, F. Potential Use of Dehydrogenase Activity as an Index of Microbial Activity in Degraded Soils. Commun. Soil Sci. Plant Anal. 1997, 28, 123–134. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, B.; Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of Soil Microbial Community Structure by Use of Taxon-Specific Quantitative PCR Assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May, L.A.; Smiley, B.; Schmidt, M.G. Comparative Denaturing Gradient Gel Electrophoresis Analysis of Fungal Communities Associated with Whole Plant Corn Silage. Can. J. Microbiol. 2001, 47, 829–841. [Google Scholar] [CrossRef]
- Liu, M.; Klemens, E.; Zhang, B.; Holzhauer, S.I.J.; Li, Z.P.; Zhang, T.L.; Rauch, S. Effect of Intensive Inorganic Fertilizer Application on Microbial Properties in a Paddy Soil of Subtropical China. Agric. Sci. China 2011, 10, 1758–1764. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-Specific Primer and Probe Sets to Detect Methanogenic Communities Using Quantitative Real-Time Polymerase Chain Reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S RRNA Gene Sequences from Microbial Communities. Curr. Protoc. Microbiol. 2012, 27, 1E–5. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat Versión 2017. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. 2017. Available online: http://www.infostat.com.ar (accessed on 13 October 2022).
- Paulson, J.N.; Colin Stine, O.; Bravo, H.C.; Pop, M. Differential Abundance Analysis for Microbial Marker-Gene Surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Package ‘Corrplot. ’ Statistician 2012, 56, 316–324. [Google Scholar]
- Frasier, I.; Noellemeyer, E.; Figuerola, E.; Erijman, L.; Permingeat, H.; Quiroga, A. High Quality Residues from Cover Crops Favor Changes in Microbial Community and Enhance C and N Sequestration. Glob. Ecol. Conserv. 2016, 6, 242–256. [Google Scholar] [CrossRef]
- Miltner, A.; Bombach, P.; Schmidt-Brücken, B.; Kästner, M. SOM Genesis: Microbial Biomass as a Significant Source. Biogeochemistry 2012, 111, 41–55. [Google Scholar] [CrossRef]
- Kallenbach, C.M.; Frey, S.D.; Grandy, A.S. Direct Evidence for Microbial-Derived Soil Organic Matter Formation and Its Ecophysiological Controls. Nat. Commun. 2016, 7, 13630. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct Soil Microbial Diversity under Long-Term Organic and Conventional Farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Bonfim, E.M.S.; Freire, F.J.; Santos, M.V.F.; Silva, T.J.A.; Freire, M.B.G.S. Soil and Plant Phosphorus Critical Levels for Brachiaria Brizantha Related to Physical and Chemical Characteristics of Soils in the State of Pernambuco, Brazil. Rev. Bras. Cienc. do Solo 2004, 28, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Martins, L.E.C.; Monteiro, F.A.; Pedreira, B.C. Metabolic Change, Tillering and Root System of Brachiaria Brizantha in Response To Phosphorus and Zinc Nutrition. J. Plant Nutr. 2014, 37, 509–519. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.; Sun, J.; Wang, X.; Zhou, W. Responses of Extracellular Enzyme Activities and Microbial Community in Both the Rhizosphere and Bulk Soil to Long-Term Fertilization Practices in a Fluvo-Aquic Soil. Geoderma 2012, 173–174, 330–338. [Google Scholar] [CrossRef]
- Garcia-Franco, N.; Martínez-Mena, M.; Goberna, M.; Albaladejo, J. Changes in Soil Aggregation and Microbial Community Structure Control Carbon Sequestration after Afforestation of Semiarid Shrublands. Soil Biol. Biochem. 2015, 87, 110–121. [Google Scholar] [CrossRef]
- Rillig, M.C.; Muller, L.A.H.; Lehmann, A. Soil Aggregates as Massively Concurrent Evolutionary Incubators. ISME J. 2017, 11, 1943–1948. [Google Scholar] [CrossRef]
- Blanco-Canqui, H.; Lal, R. Mechanisms of Carbon Sequestration in Soil Aggregates. CRC Crit. Rev. Plant Sci. 2004, 23, 481–504. [Google Scholar] [CrossRef]
- Villasica, L.J.; Lina, S.; Asio, V. Aggregate Stability Affects Carbon Sequestration Potential of Different Tropical Soils. Ann. Trop. Res. 2018, 40, 71–88. [Google Scholar] [CrossRef]
- Liu, M.; Han, G.; Zhang, Q. Effects of Soil Aggregate Stability on Soil Organic Carbon and Nitrogen under Land Use Change in an Erodible Region in Southwest China. Int. J. Environ. Res. Public Health 2019, 16, 3809. [Google Scholar] [CrossRef] [Green Version]
- Hashim, Z.E.; Alzubaidi, L.A.; Al-Madhhachi, A.T. The Influence of Microbiology on Soil Aggregation Stability. IOP Conf. Ser. Mater. Sci. Eng. 2020, 870, 012110. [Google Scholar] [CrossRef]
- Lehmann, A.; Zheng, W.; Rillig, M.C. Soil Biota Contributions to Soil Aggregation. Nat. Ecol. Evol. 2017, 1, 1828–1835. [Google Scholar] [CrossRef] [Green Version]
- García-González, I.; Hontoria, C.; Gabriel, J.L.; Alonso-Ayuso, M.; Quemada, M. Cover Crops to Mitigate Soil Degradation and Enhance Soil Functionality in Irrigated Land. Geoderma 2018, 322, 81–88. [Google Scholar] [CrossRef]
- Lehman, R.M.; Acosta-Martinez, V.; Buyer, J.S.; Cambardella, C.A.; Collins, H.P.; Ducey, T.F.; Halvorson, J.J.; Jin, V.L.; Johnson, J.M.F.; Kremer, R.J.; et al. Soil Biology for Resilient, Healthy Soil. J. Soil Water Conserv. 2015, 70, 12A–18A. [Google Scholar] [CrossRef]
- Schnurer, J.; Rosswall, T. Fluorescein Diacetate Hydrolysis as a Measure of Total Microbial Activity in Soil and Litter. Appl. Environ. Microbiol. 1982, 43, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Abán, C.L.; Pérez Brandan, C.; Verdenelli, R.; Huidobro, J.; Meriles, J.M.; Vargas Gil, S. Changes in Microbial and Physicochemical Properties under Cover Crop Inclusion in a Degraded Common Bean Monoculture System. Eur. J. Soil Biol. 2021, 107, 103365. [Google Scholar] [CrossRef]
- Gabbarini, L.A.; Figuerola, E.; Frene, J.P.; Robledo, N.B.; Ibarbalz, F.M.; Babin, D.; Smalla, K.; Erijman, L.; Wall, L.G. Impacts of Switching Tillage to No-Tillage and Vice Versa on Soil Structure, Enzyme Activities and Prokaryotic Community Profiles in Argentinean Semi-Arid Soils. FEMS Microbiol. Ecol. 2021, 97, fiab025. [Google Scholar] [CrossRef] [PubMed]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial Indicators for Soil Quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Martínez, V.; Acosta-Mercado, D.; Sotomayor-Ramírez, D.; Cruz-Rodríguez, L. Microbial Communities and Enzymatic Activities under Different Management in Semiarid Soils. Appl. Soil Ecol. 2008, 38, 249–260. [Google Scholar] [CrossRef]
- Mendes, L.W.; Raaijmakers, J.M.; De Hollander, M.; Mendes, R.; Tsai, S.M. Influence of Resistance Breeding in Common Bean on Rhizosphere Microbiome Composition and Function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef] [Green Version]
- Sousa, R.M.S.; Mendes, L.W.; Antunes, J.E.L.; Oliveira, L.M.D.S.; Sousa, A.M.D.C.B.; Gomes, R.L.F.; Lopes, A.C.D.A.; Araújo, F.F.; Melo, V.M.M.; Araujo, A.S.F. Diversity and Structure of Bacterial Community in Rhizosphere of Lima Bean. Appl. Soil Ecol. 2020, 150, 103490. [Google Scholar] [CrossRef]
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the Bacterial Community of Soybean Rhizospheres during Growth in the Field. PLoS ONE 2014, 9, e100709. [Google Scholar] [CrossRef] [Green Version]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.W. Effects of Actinobacteria on Plant Disease Suppression and Growth Promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A Global Atlas of the Dominant Bacteria Found in Soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, M.; Yang, Z.; Cong, M.; Zhu, X.; Jia, H. Soil Microbial Community Response to Nitrogen Application on a Swamp Meadow in the Arid Region of Central Asia. Front. Microbiol. 2022, 12, 4126. [Google Scholar] [CrossRef]
- Garland, G.; Edlinger, A.; Banerjee, S.; Degrune, F.; García-Palacios, P.; Pescador, D.S.; Herzog, C.; Romdhane, S.; Saghai, A.; Spor, A.; et al. Crop Cover Is More Important than Rotational Diversity for Soil Multifunctionality and Cereal Yields in European Cropping Systems. Nat. Food 2021, 2, 28–37. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; Heijden, M.G.A. Keystone Taxa as Drivers of Microbiome Structure and Functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Prakash, A.; Johri, B.N. Bacteria in Agrobiology: Crop Ecosystems. In Bacteria in Agrobiology: Crop Ecosystems; Maheshwari, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 37–59. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rengel, Z.; Zhang, K.; Jin, K.; Lyu, Y.; Zhang, L.; Cheng, L.; Zhang, F.; Shen, J. Ensuring Future Food Security and Resource Sustainability: Insights into the Rhizosphere. iScience 2022, 25, 104168. [Google Scholar] [CrossRef] [PubMed]
- Attia, S.; Russel, J.; Mortensen, M.S.; Madsen, J.S.; Sørensen, S.J. Unexpected Diversity among Small-Scale Sample Replicates of Defined Plant Root Compartments. ISME J. 2022, 16, 997–1003. [Google Scholar] [CrossRef]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and Exudates of the Root and Rhizosphere of Brachypodium Distachyon, a Model for Wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef] [Green Version]
- Donn, S.; Kirkegaard, J.A.; Perera, G.; Richardson, A.E.; Watt, M. Evolution of Bacterial Communities in the Wheat Crop Rhizosphere. Environ. Microbiol. 2015, 17, 610–621. [Google Scholar] [CrossRef]
- Chamberlain, L.A.; Bolton, M.L.; Cox, M.S.; Suen, G.; Conley, S.P.; Ané, J.M. Crop Rotation, but Not Cover Crops, Influenced Soil Bacterial Community Composition in a Corn-Soybean System in Southern Wisconsin. Appl. Soil Ecol. 2020, 154, 103603. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the Core Arabidopsis Thaliana Root Microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Azarbad, H.; Tremblay, J.; Bainard, L.D.; Yergeau, E. Relative and Quantitative Rhizosphere Microbiome Profiling Results in Distinct Abundance Patterns. Front. Microbiol. 2022, 12, 4054. [Google Scholar] [CrossRef] [PubMed]
- Jian, C.; Luukkonen, P.; Yki-Järvinen, H.; Salonen, A.; Korpela, K. Quantitative PCR Provides a Simple and Accessible Method for Quantitative Microbiota Profiling. PLoS ONE 2020, 15, e0227285. [Google Scholar] [CrossRef] [Green Version]
- Maul, J.E.; Buyer, J.S.; Lehman, R.M.; Culman, S.; Blackwood, C.B.; Roberts, D.P.; Zasada, I.A.; Teasdale, J.R. Microbial Community Structure and Abundance in the Rhizosphere and Bulk Soil of a Tomato Cropping System That Includes Cover Crops. Appl. Soil Ecol. 2014, 77, 42–50. [Google Scholar] [CrossRef]
- Strickland, M.S.; Rousk, J. Considering Fungal: Bacterial Dominance in Soils—Methods, Controls, and Ecosystem Implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- de Vries, F.T.; Hoffland, E.; van Eekeren, N.; Brussaard, L.; Bloem, J. Fungal/Bacterial Ratios in Grasslands with Contrasting Nitrogen Management. Soil Biol. Biochem. 2006, 38, 2092–2103. [Google Scholar] [CrossRef] [Green Version]
- Chavarría, D.N.; Verdenelli, R.A.; Serri, D.L.; Restovich, S.B.; Andriulo, A.E.; Meriles, J.M.; Vargas-Gil, S. Effect of Cover Crops on Microbial Community Structure and Related Enzyme Activities and Macronutrient Availability. Eur. J. Soil Biol. 2016, 76, 74–82. [Google Scholar] [CrossRef]
- Serri, D.L.; Pérez-Brandan, C.; Meriles, J.M.; Salvagiotti, F.; Bacigaluppo, S.; Malmantile, A.; Vargas-Gil, S. Development of a Soil Quality Index for Sequences with Different Levels of Land Occupation Using Soil Chemical, Physical and Microbiological Properties. Appl. Soil Ecol. 2022, 180, 104621. [Google Scholar] [CrossRef]
- Liang, S.; Deng, J.; Jiang, Y.; Wu, S.; Zhou, Y.; Zhu, W. Functional Distribution of Bacterial Community under Different Land Use Patterns Based on Faprotax Function Prediction. Polish J. Environ. Stud. 2020, 29, 1245–1261. [Google Scholar] [CrossRef]
- Liu, L.; Sun, F.; Zhao, H.; Mi, H.; He, S.; Chen, Y.; Liu, Y.; Lan, H.; Zhang, M.; Wang, Z. Compositional Changes of Sedimentary Microbes in the Yangtze River Estuary and Their Roles in the Biochemical Cycle. Sci. Total Environ. 2021, 760, 143383. [Google Scholar] [CrossRef]
- Mohapatra, B.; Saha, A.; Chowdhury, A.N.; Kar, A.; Kazy, S.K.; Sar, P. Geochemical, Metagenomic, and Physiological Characterization of the Multifaceted Interaction between Microbiome of an Arsenic Contaminated Groundwater and Aquifer Sediment. J. Hazard. Mater. 2021, 412, 125099. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Blagodatskaya, E. Microbial Hotspots and Hot Moments in Soil: Concept & Review. Soil Biol. Biochem. 2015, 83, 184–199. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the Nitrogen Cycle. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea Predominate among Ammonia-Oxidizing Prokaryotes in Soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef]
- Subbarao, G.V.; Nakahara, K.; Hurtado, M.P.; Ono, H.; Moreta, D.E.; Salcedo, A.F.; Yoshihashi, A.T.; Ishikawa, T.; Ishitani, M.; Ohnishi-Kameyama, M.; et al. Evidence for Biological Nitrification Inhibition in Brachiaria Pastures. Proc. Natl. Acad. Sci. USA 2009, 106, 17302–17307. [Google Scholar] [CrossRef]







| B1 | B2 | BM | |
|---|---|---|---|
| Chemical parameters | |||
| SOC (%) a | 1.24 ± 0.12 ab | 1.42 ± 0.04 a | 0.99 ± 0.18 b |
| OM (%) b | 2.13 ± 0.2 ab | 2.44 ± 0.07 a | 1.7 ± 0.32 b |
| Total N (%) c | 0.11 ± 0 ab | 0.12 ± 0.01 a | 0.09 ± 0.02 b |
| C/N d | 11 ± 1 ab | 12 ± 0 a | 10.67 ± 0.58 b |
| eP (p.p.m.) e | 22 ± 1.73 a | 20.33 ± 5.03 a | 23.33 ± 3.79 a |
| Physical parameters | |||
| Bulk density | 1.42 ± 0.04 b | 1.36 ± 0.07 b | 1.66 ± 0.11 a |
| Agregate stability | 36.99 ± 8.17 b | 50 ± 1.93 a | 14.75 ± 1.47 c |
| WHC (%) f | 32.67 ± 1.53 a | 31.33 ± 1.15 ab | 28 ± 2.65 b |
| pH | 7.07 ± 0.38 a | 6.83 ± 0.06 a | 7.07 ± 0.06 a |
| EC (mmhos/cm) g | 0.65 ± 0.23 a | 0.37 ± 0.02 ab | 0.27 ± 0.08 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abán, C.L.; Larama, G.; Ducci, A.; Huidobro, J.; Abanto, M.; Vargas-Gil, S.; Pérez-Brandan, C. Soil Properties and Bacterial Communities Associated with the Rhizosphere of the Common Bean after Using Brachiaria brizantha as a Service Crop: A 10-Year Field Experiment. Sustainability 2023, 15, 488. https://doi.org/10.3390/su15010488
Abán CL, Larama G, Ducci A, Huidobro J, Abanto M, Vargas-Gil S, Pérez-Brandan C. Soil Properties and Bacterial Communities Associated with the Rhizosphere of the Common Bean after Using Brachiaria brizantha as a Service Crop: A 10-Year Field Experiment. Sustainability. 2023; 15(1):488. https://doi.org/10.3390/su15010488
Chicago/Turabian StyleAbán, Carla L., Giovanni Larama, Antonella Ducci, Jorgelina Huidobro, Michel Abanto, Silvina Vargas-Gil, and Carolina Pérez-Brandan. 2023. "Soil Properties and Bacterial Communities Associated with the Rhizosphere of the Common Bean after Using Brachiaria brizantha as a Service Crop: A 10-Year Field Experiment" Sustainability 15, no. 1: 488. https://doi.org/10.3390/su15010488





