1. Introduction
Several deadly diseases endanger honeybees. Possibly one of the best known is Nosema. Nosema, which is also called Nosemiasis or Nosemosi [
1], is caused by two species of microsporidia,
Nosema apis (
N. apis) and Nosema ceraena (
N. ceraena) [
2]. Several works were published regarding the impact of Nosema disease on commerce, society and food, as shown in [
3,
4], and the disease is currently of one the major economic importance worldwide [
5]. The health of the two species of bees is a particular interest of biologists, not only because of their significant role in the economy and food production but also because of the vital role they give in the pollination of agricultural and horticultural crops. Many biological descriptions of its DNA and its behavior can be found in literature, for example in [
6,
7]. Furthermore, several recent works try to treat this disease using a chemical simulation, as presented in [
8,
9].
Furthermore, from a computer science point of view, honeybees are of significant interest. Several works were, for example, involved in bees and controlling their behavior [
10]. The study presented monitoring the behavior of bees to help people associated with beekeeping to manage their honey colonies and discover the bee disturbance caused by a pathogen, Colony Collapse Disorder (CCD) or colony health assessment. In [
11], many tools of image analysis were explored to study the honeybee auto grooming behavior. Chemical and gas sensors were used for measurement. Destructor infestations are applied inside the honeybee colony to detect disease. The study was based on measurements of the atmosphere of six beehives using six types of solid-state gas sensors during a 12-h experiment [
12]. Regarding the image processing of Nosema disease part, there are currently two major works. In [
13], the authors used the Scale Invariant Feature Transform to extract features from cell images. It is a technique that transforms image data into scale-invariant coordinates relative to local features. A segmentation technique and a support vector machine algorithm were then applied to microscopic processed images to automatically classify
N. apis and
N. ceranae microsporidia. In [
14], the authors used the image processing techniques to extract the most valuable features from Nosema microscopic images and apply an Artificial Neural Network (ANN) for the recognition, which was statistically evaluated using the cross-validation technique. The last two works used image processing tools for feature extraction and Support Vector Machine (SVM) and ANN for classification. Today the traditional tools of machine learning like ANN, Convolutional Neural Network (CNN), and SVM are frequently used in human disease detection [
15], especially in medical image classification of Heart diseases [
16], Alzheimer disease [
17] and Thorax diseases [
18]. Deep learning approaches were used in [
19] for semantic images segmentation. This work used the Atrous convolutional Neural Network for segmentation and some pre-trained NN for validation like PASCAL-Context, PASCAL-Person-Part and CityscapesDeep. In [
20], a method using a 2D overlapping ellipse was implemented using the tools of image processing and applied to the problem of segmenting potentially overlapping cells in fluorescence microscopy images. Deep learning is an end-to-end machine learning process that trains feature extraction together with the classification itself. Instead of organizing statistics to run through predefined equations, deep learning uses multiple layers of processing data and setting fundamental parameters on knowledge records, and it trains the computer to analyze and recognize data. Deep learning approaches are widely applied in the analysis of microscopic images in many fields: human microbiota [
21], material sciences [
22], microorganism detection [
23], cellular image processing [
24] and many other important works in this field. Deep learning techniques have accelerated with transfer learning the ability to recognize and classify several diseases. The objective of this paper is to validate this hypothesis.
All the methods of Nosema detection and recognition presented by the biologists in the literature were either molecular detections or genetic descriptions. This paper evaluates two different strategies for automatic identification of the Nosema cell disease based on the microscopic images. First, images of Nosema cells and the existing objects have been cropped from the principal microscopic images. Using these images, the first dataset has been built. Then, the obtained images were processed again and several different features have been extracted. These features were used to create a second dataset. The obtained databases were used for the evaluation recognition of the Nosema cells. The first approach uses a model, which uses the extracted features by an ANN and an SVM. The second approach uses the deep learning and transfer learning methods: first, CNN, and then pre-trained networks AlexNet, VGG-16 and VGG-19. The tools of transfer learning used by authors reached notable results as this is the first time they have been used for the purpose of Nosema cell recognition.
The main innovation of this paper is the evaluation of two different strategies of automatic detection and recognition Nosema cells from microscopic images and identification of the robust and successful approach as a robust methodology for automated identifying and recognizing Nosema cells versus the other existing objects in the same microscopic images.
The rest of the paper is organized as follow:
Section 2 describes the dataset preparation. In
Section 3 is described dataset, segmentation, features extraction, ANN training, the use of SVM, CNN, the use of Alex Net, VGG-16 and VGG-19. The experiments are described in
Section 4.
Section 5 discusses the obtained results. Finally, the paper is concluded.
2. Materials: Preparation of The Dataset
For the experiment, Nosema microscopic images were used. So far, it is not known whether these images contain a sufficient amount of information for accurate detection and recognition of the disease cells. It was only known that the important information was diffused all over the image and behind the majority of unimportant data. The used images in this work are 400 RGB images, encoded with JPEG and with a resolution of 2272 × 1704 pixels. Each sample was labelled by one of the 7 classes, according to the severity of the disease or the number of disease cells present in the microscopic image. From these 400 RGB images, a set of sub-images have been extracted. To do that, each microscopic image was divided into many smaller images forming subdivisions of the existing and clear objects. This first phase was done manually due to the low quality of input images by cropping the object of interest (i.e., cells). All the existing objects in the microscopic images were extracted as sub-images and labelled whether they stand for: Nosema(N) and not Nosema cells (n-N), see
Figure 1. The area chosen was as small as possible, where an isolated and clear microscopic cell is located. Then, in the second automatic phase, the selected objects are processed to prepare them for the segmentation process (see
Figure 1).
Based on the steps described above, a dataset containing 2000 sample images in total was created. It consists of 1000 Nosema cells samples and 1000 images, which are not Nosema cells, i.e., any other existing objects in the microscopic images.
Table 1 below shows information about the extracted sub-images for dataset construction.
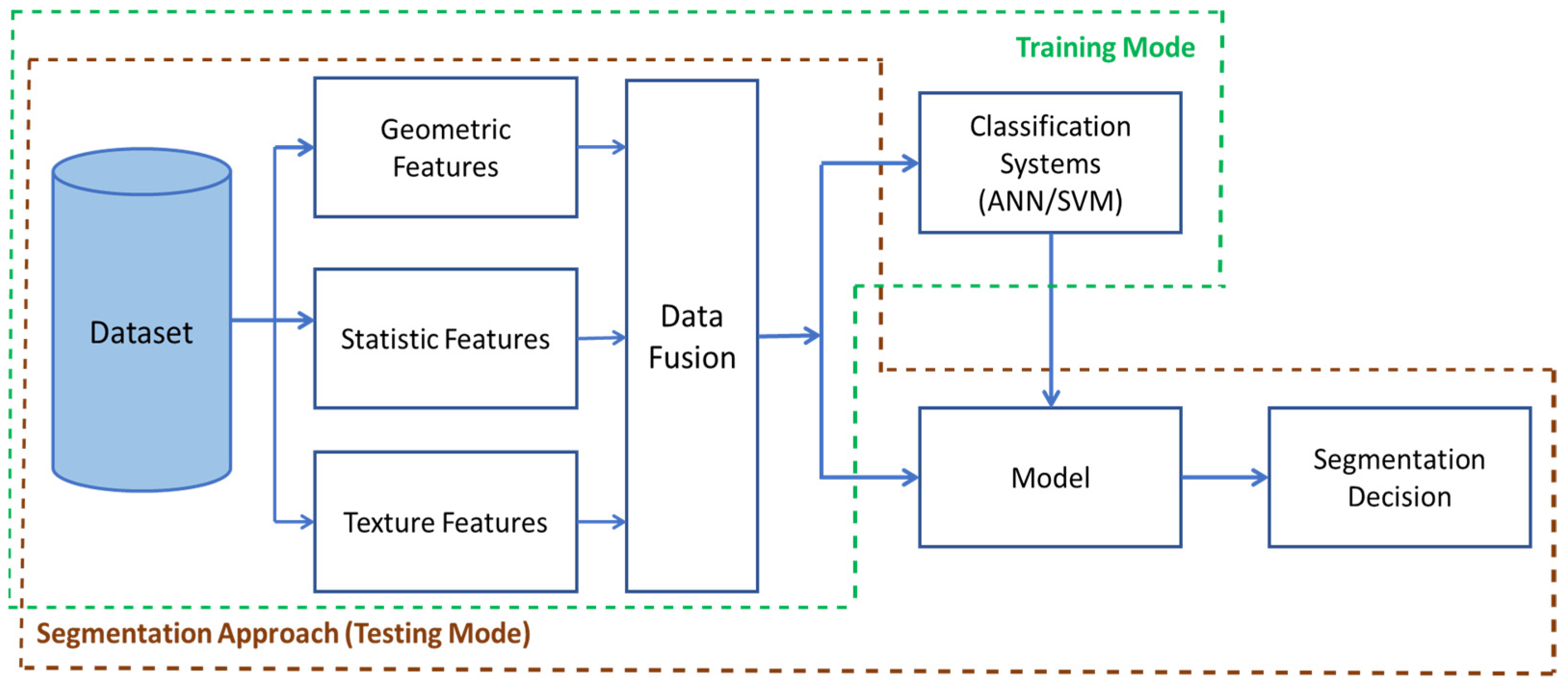
The microscopic sub-images were examined using two strategies:
The first strategy is based on an image processing approach, where features were extracted manually.
The second set of strategies is based on the use of the whole sub-image and the deep learning.
Figure 2 shows strategies covered in the paper.
4. Experimental Methodology and Results
For the statistical evaluation, the 10-fold cross-validation strategy was followed between 10% and 90%. Accuracy is used as a quality measure here. The experiments have been designed for machine learning approaches (SVM and ANN), transfer learning approaches (AlexNet, VGG-16 and VGG-19), and deep learning method with CNN.
The first experiment was done for ANN and SVM. For ANN, just a single hidden layer was used and only the number of neurons in the hidden layer was adjusted, using 15 or 19 neurons for the input layer and 1 neuron for the output layer (see
Table 2).
The next experiment used the deep learning method, in particular deep CNN classifier. The architecture of CNN had 3 convolutional blocks, which have been stacked with 3 × 3 filters followed by a 2 × 2 subsampling layer (max_pooling). In this way, increasing the number of filters increases the depth of the network, and a kind of cone is formed with increasingly reduced but more relevant characteristics. It should be noted that in convolutional layers, padding is used to ensure that the height and width of the output feature maps match the inputs. Finally, each layer will use the ReLU activation function. Additionally, dropout layers have been added that implement regularization. The dropout technique is a simple technique that will randomly remove nodes from the network and has the effect of regularization as the remaining nodes must adapt to compensate for the slack of the removed nodes and a layer of batch normalization. Batch normalization (batch_normalization) is a technique designed to automatically standardize inputs to a layer in a deep learning neural network and has the effect of speeding up the process of training a neural network and, in some cases, improving the performance of the model. Once the above has been commented on, in
Table 3, the architecture used for an 80 × 80 input image with three RGB channels is shown. The accuracy reached 92.50%.
Finally, the last experiment was for transfer learning approaches. AlexNet is known for its simplicity, but in the case of this experiment, it does not give an encouraging result. SGDM was the default and chosen optimizer for AlexNet. AlexNet does not require many options to work well, and the default training options were reserved. Sixty-four is the size of mini-bach and the initial learning rate was chosen as 0.001. The maximum number of epochs is fixed to 20; this chosen training options made the experiment faster (see
Table 4).
Table 5 describes the four cross-validation folders experiments and given accuracy by each one. As is shown in
Table 5, the third experiments in which the data were split between 70% for training and 30% for test and validation, give the best accuracy (87.48%) by 6 epochs number.
Only the last three layers of VGG-16 and VGG19 were modified to make them fit the target domain. The fully connected layer (FC) in both models has been changed to a new FC layer with an output size of 2 according to the 2 classes, which were needed to classify. Adam was the chosen optimizer, given his good learning rate and the specific adaptive nature of the learning rate parameters. For Adam, the initial learning rate was chosen as 0.0004; a small valor is a good option to increase the training time. The size of the mini-batch was fixed at 10. The validation information of the model is that given in the test. Thus, a learning factor of 10 is defined. The maximum number of epochs was fixed to 25 but during the simulation process, the number was variable according to the experiments carried out, but it was initialized in the first experiment to 6. Finally, a validation frequency set to 3. The trained options of the experiment are listed in
Table 4.
Detailed results for VGG-16 and VGG-19 neural networks are shown in
Table 4, and while the best simulation accuracy is given by VGG-16,
Figure 7 describes the followed steps using VGG16 to identify the Nosema and
Figure 8 shows the best accuracy. Three experiments have been implemented, but only those that gave good results with a similar number of epochs for the two pre-trained networks have been described in
Table 6. The data was split between training and validation, the experiments were conducted 30 times, following a 10-fold cross-validation process. The three last experiments gave the best accuracy; the first one took 70% of data for training and the 30% were for validation and the best accuracy was given by 6 epochs number. In the second experiment, 80% were placed for training, and the rest were for validation, the experiment was repeated several times with increasing the number of epochs and as
Table 6 shows, the best accuracy given by VGG-16 is 96.25% with 20 epochs, and for VGG-19, the highest accuracy is 93.50% with 25 epochs, and in the third experiment presented in the result section, the data were divided between 90% for training and 10% for testing, and the results made an accuracy fall.
Table 7 summarizes the main results of the different experiments. The best result is reached using VGG-16 with accuracy of 96.25%, and the lowest accuracy is given by ANN (83.20%). Those results will be discussed in the next section.
5. Discussion
This section discusses in detail the behavior and features of each experiment and it discusses compromise between accuracy and the robustness of the proposed methods was included. Besides, a comparison vs. the most representative publication on this topic (see
Table 8), with comparison vs. a previous work [
14], authors increased the dataset from 185 to 2000 images and the extracted features number from 9 to 19, and those features for the Nosema cell are related to several aspects of the image cell: geometric shape, statistical characteristics, texture and color features given by GLCM. Two strategies were followed to recognize Nosema; while only one was followed (ANN) in [
14]; the first strategy consists of the use of calculated characteristics by an ANN and an SVM and the second is based on sub-images extracted from treated microscopic images using an implemented CNN and the tools of transfer Learning. ANN used in [
14] gave a success rate of 91.1% in Nosema recognition. SVM also was used in [
13] to classify the two types of Nosema and other objects. The experiments reached relative and accurate values.
From
Table 2,
Table 3,
Table 4,
Table 5 and
Table 6, it can be concluded that whether it is the largest dataset or the smallest dataset, the level of learning of the network with transfer learning models is obviously better than the traditional models, especially ANNs are examined in this study and SVM which brought near results. Furthermore, one notes a clear rate of convergence of the transfer model VGG-16 and VGG-19 at the level of the provided results. In addition, these transfer models are a bit faster than ANN and SVM, at least in this case. CNN has demonstrated its effectiveness in this problem of recognizing or classifying Nosema cells as a deep learning model. CNN was almost comparable to VGG-19. On the other hand, it should be said that the training options for the ANNs, as well as the transfer learning algorithms, make a difference in the results.
In front of AlexNet, the VGG-16, VGG-19 and CNN have proven their strong effectiveness in this work in the classification of patterns, cells and objects.
For the features extraction part, several different features from the sub-images were evaluated: geometric, statistic, texture and GLCM features extracted from the yellow channel. This experiment used a large database, the results given by the ANN as well as by the SVM good since it is the first time. The quality of the microscopic images used in this work did not always help to extract clear and sharp objects. By calculating the results with a different number of features (15 and 19), the importance of the data extracted by the GLCM in the resulting amelioration was approved.
6. Conclusions
In order to identify Nosema cells, this experiment examined two strategies of classification: the traditional ones and the deep learning classifiers. Different experiments were implemented for both strategies, despite the noisy quality of the microscopic images used. The best accuracy for the recognition or classification of Nosema is reached by VGG-16, 96.25%, which is compared to state of the art is the most accurate methodology in this area so far.
The innovation of this proposal is to analyze and find the better option for this identification, checking different strategies to implement an automatic identification of Nosema cell, as was shown after experiments, and with good and robust accuracy. It was reached with VGG-16 architecture.
After reviewing the state-of-the-art material, it can be concluded that only a few automatic approaches have been introduced so far. Because of this, we contribute with a variety of explored classification methods and their accuracies. In particular, we would emphasize the difference between shallow ANNs with handcrafted features and end-to-end learning using the deep learning approach using CNN together with several transfer learning architectures.