Role of AI and Histopathological Images in Detecting Prostate Cancer: A Survey
Abstract
1. Introduction
2. Histopathology Images Background
2.1. Diagnostic Challenges Using Histopathological Images
2.1.1. Extremely Large Image Size
2.1.2. Insufficient Labeled Images
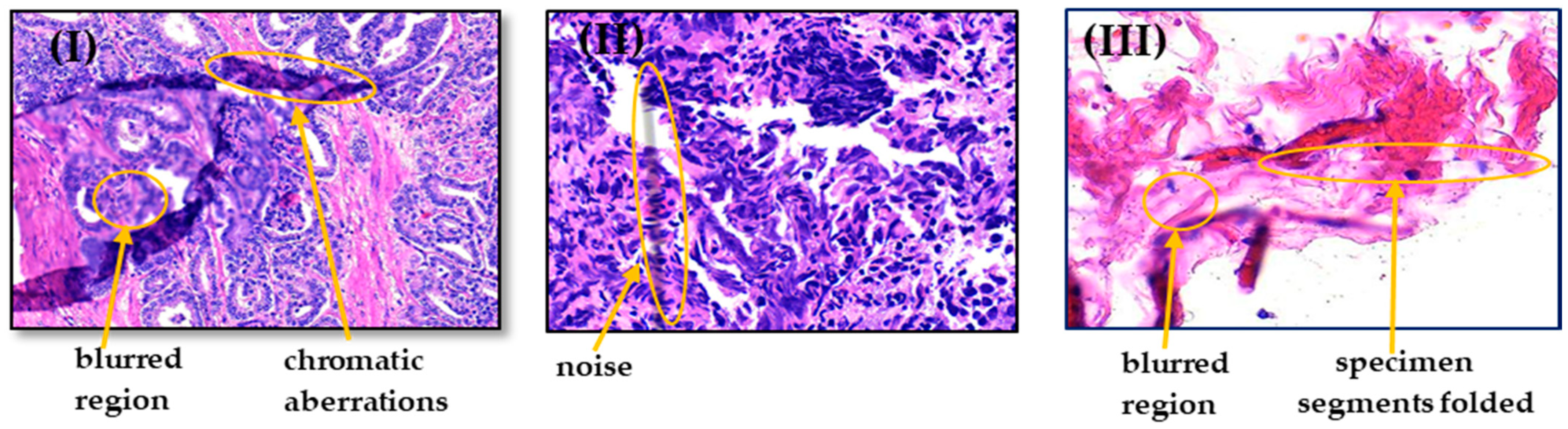
2.1.3. Artifacts and Color Variation
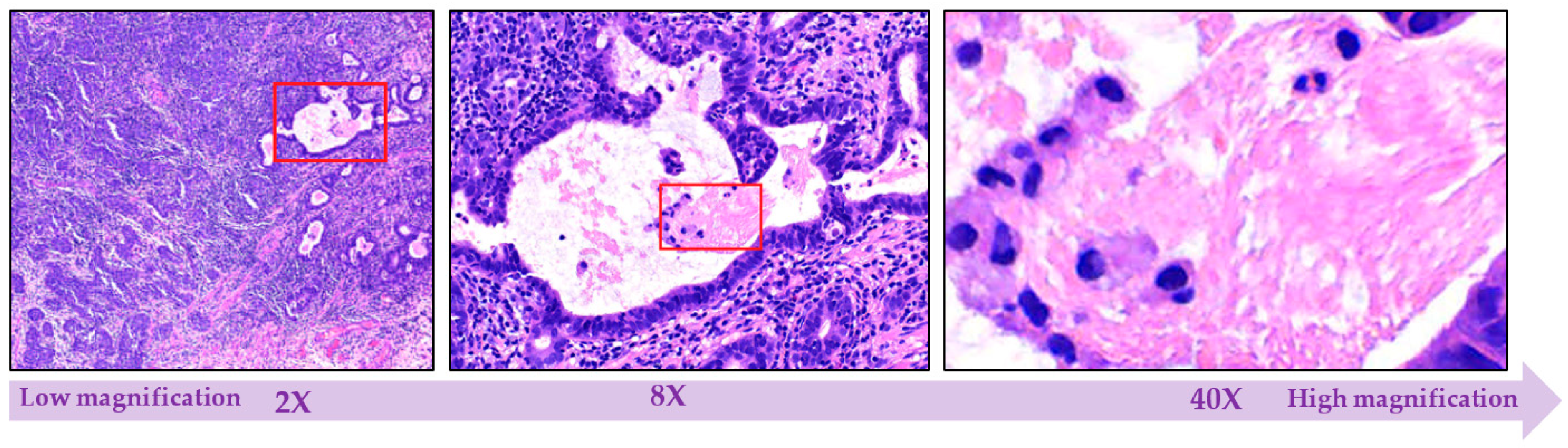
2.1.4. Multi-Level Magnification Led to Multi-Level Information
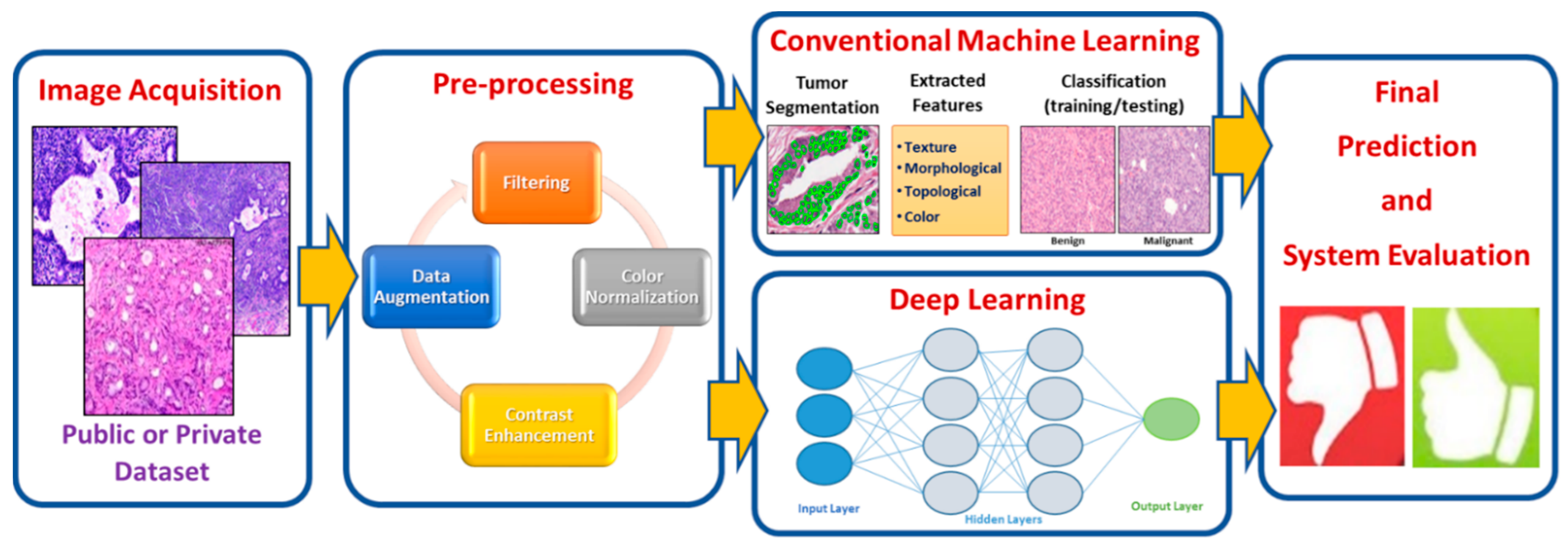
3. Histopathology Image Analysis Methodology
3.1. Image Acquisition
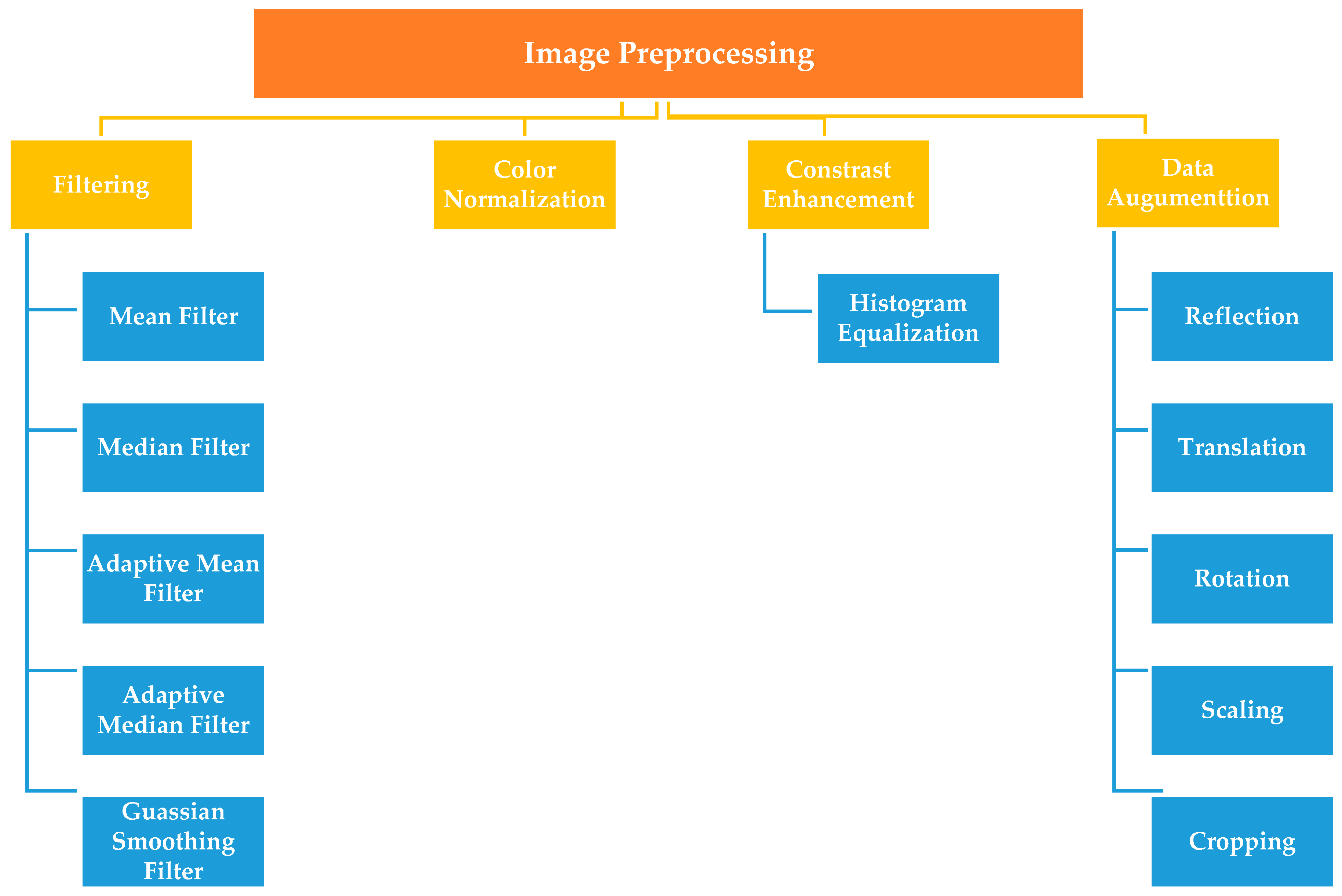
3.2. Image Preprocessing
3.2.1. Filtering
3.2.2. Color Normalization Techniques
3.2.3. Histogram Equalization
3.2.4. Data Augmentation
3.3. Traditional Machine Learning Techniques
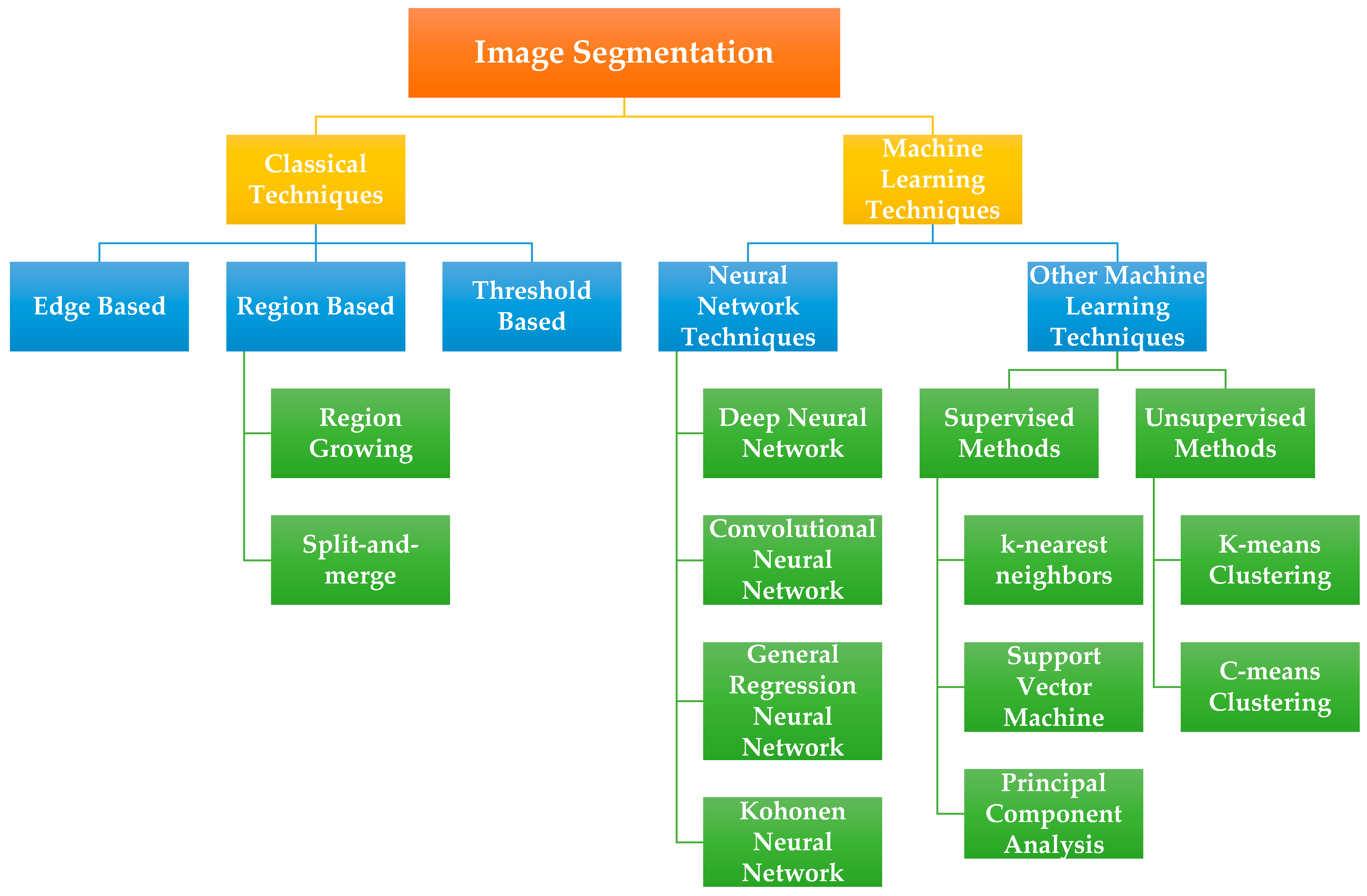
3.3.1. Image Segmentation
3.3.2. Feature Selection
3.3.3. Classification
3.4. Deep Learning-Based Techniques
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harmon, S.A.; Tuncer, S.; Sanford, T.; Choyke, P.L.; Turkbey, B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn. Interv. Radiol. 2019, 25, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Kalaw, E.M. Automated classification for pathological prostate images using AdaBoost-based Ensemble Learning. In Proceedings of the 2016 IEEE Symposium Series on Computational Intelligence (SSCI), Athens, Greece, 6–9 December 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Reda, I.; Ayinde, B.O.; Elmogy, M.; Shalaby, A.; El-Melegy, M.; El-Ghar, M.A.; El-Fetouh, A.A.; Ghazal, M.; El-Baz, A. A new CNN-based system for early diagnosis of prostate cancer. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 207–210. [Google Scholar]
- Ried, K.; Tamanna, T.; Matthews, S.; Eng, P.; Sali, A. New Screening Test Improves Detection of Prostate Cancer Using Circulating Tumor Cells and Prostate-Specific Markers. Front. Oncol. 2020, 10, 582. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Key Statistics for Prostate Cancer. Available online: http://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html (accessed on 29 January 2021).
- Hoogland, A.M.; Kweldam, C.F.; Van Leenders, G.J.L.H. Prognostic Histopathological and Molecular Markers on Prostate Cancer Needle-Biopsies: A Review. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- de Matos, J.; Britto, A.D.S., Jr.; Oliveira, L.E.; Koerich, A.L. Histopathologic image processing: A review. arXiv 2019, arXiv:1904.07900. Available online: https://arxiv.org/abs/1904.07900 (accessed on 5 January 2021).
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Aswathy, M.; Jagannath, M. Detection of breast cancer on digital histopathology images: Present status and future possibilities. Inform. Med. Unlocked 2017, 8, 74–79. [Google Scholar] [CrossRef]
- Wang, S.; Burtt, K.; Turkbey, B.; Choyke, P.; Summers, R.M. Computer Aided-Diagnosis of Prostate Cancer on Multiparametric MRI: A Technical Review of Current Research. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Anuranjeeta; Shukla, K.K.; Tiwari, A.; Sharma, S. Classification of Histopathological Images of Breast Cancerous and Non Cancerous Cells based on Morphological Features. Biomed. Pharmacol. J. 2017, 10, 353–366. [Google Scholar] [CrossRef]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef]
- Madabhushi, A.; Agner, S.; Basavanhally, A.; Doyle, S.; Lee, G. Computer-aided prognosis: Predicting patient and disease outcome via quantitative fusion of multi-scale, multi-modal data. Comput. Med. Imaging Graph. 2011, 35, 506–514. [Google Scholar] [CrossRef]
- Rashid, S.; Fazli, L.; Boag, A.; Siemens, R.; Abolmaesumi, P.; Salcudean, S.E. Separation of Benign and Malignant Glands in Prostatic Adenocarcinoma. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; Springer International Publishing: Berlin/Heidelberg, Germany, 2013; pp. 461–468. [Google Scholar]
- Naik, S.; Doyle, S.; Agner, S.; Madabhushi, A.; Feldman, M.; Tomaszewski, J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Paris, France, 14–17 May 2008; pp. 284–287. [Google Scholar]
- Singh, M.; Kalaw, E.M.; Giron, D.M.; Chong, K.-T.; Tan, C.L.; Lee, H.K. Gland segmentation in prostate histopathological images. J. Med. Imaging 2017, 4, 027501. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Masood, K.; Irfan, M.; Draz, U.; Nagra, A.; Asif, M.; Alshehri, B.; Glowacz, A.; Tadeusiewicz, R.; Mahnashi, M.; et al. Multistage Segmentation of Prostate Cancer Tissues Using Sample Entropy Texture Analysis. Entropy 2020, 22, 1370. [Google Scholar] [CrossRef]
- Salman, S.; Ma, Z.; Mohanty, S.; Bhele, S.; Chu, Y.-T.; Knudsen, B.; Gertych, A. A Machine Learning Approach to Identify Prostate Cancer Areas in Complex Histological Images. In Intelligent and Fuzzy Techniques in Big Data Analytics and Decision Making; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2014; Volume 283, pp. 295–306. [Google Scholar]
- DiFranco, M.D.; O’Hurley, G.; Kay, E.W.; Watson, R.W.G.; Cunningham, P. Ensemble based system for whole-slide prostate cancer probability mapping using color texture features. Comput. Med. Imaging Graph. 2011, 35, 629–645. [Google Scholar] [CrossRef]
- Doyle, S.; Madabhushi, A.; Feldman, M.; Tomaszeweski, J. A Boosting Cascade for Automated Detection of Prostate Cancer from Digitized Histology. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Copenhagen, Denmark, 1–6 October 2006; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2006; Volume 9, pp. 504–511. [Google Scholar]
- Albashish, D.; Sahran, S.; Abdullah, A.; Adam, A.; Alweshah, M. A hierarchical classifier for multiclass prostate histopathology image gleason grading. J. Inf. Commun. Technol. 2018, 17, 323–346. [Google Scholar] [CrossRef]
- Sanghavi, F.M. Automated classification of histopathology images of prostate cancer using a Bag-of-Words approach. In Mobile Multimedia/Image Processing, Security, and Applications 2016; SPIE: Bellingham, WA, USA, 2016; Volume 9869, p. 98690. [Google Scholar] [CrossRef]
- Gurav, S.B.; Kulhalli, K.V.; Desai, V.V. Prostate cancer detection using histopathology images and classification using improved RideNN. Biomed. Eng. Appl. Basis Commun. 2019, 31. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Sarma, K.V.; Ho, K.C.; Shen, S.; Knudsen, B.S.; Gertych, A.; Arnold, C.W. Path R-CNN for Prostate Cancer Diagnosis and Gleason Grading of Histological Images. IEEE Trans. Med. Imaging 2019, 38, 945–954. [Google Scholar] [CrossRef]
- Mahmood, F.; Borders, D.; Chen, R.J.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep Adversarial Training for Multi-Organ Nuclei Segmentation in Histopathology Images. IEEE Trans. Med. Imaging 2020, 39, 3257–3267. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, G.; Liu, Z.; Xiong, Z.; Hu, J. A deep learning algorithm for one-step contour aware nuclei segmentation of histopathology images. Med. Biol. Eng. Comput. 2019, 57, 2027–2043. [Google Scholar] [CrossRef]
- García, G.; Colomer, A.; Naranjo, V. First-Stage Prostate Cancer Identification on Histopathological Images: Hand-Driven versus Automatic Learning. Entropy 2019, 21, 356. [Google Scholar] [CrossRef] [PubMed]
- Arvaniti, E.; Fricker, K.S.; Moret, M.; Rupp, N.; Hermanns, T.; Fankhauser, C.; Wey, N.; Wild, P.J.; Rüschoff, J.H.; Claassen, M. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bulten, W.; Pinckaers, H.; van Boven, H.; Vink, R.; de Bel, T.; van Ginneken, B.; van der Laak, J.; de Kaa, C.H.; Litjens, G. Automated gleason grading of prostate biopsies using deep learning. arXiv 2019, arXiv:1907.07980. Available online: https://arxiv.org/abs/1907.07980 (accessed on 10 January 2021).
- Kallen, H.; Molin, J.; Heyden, A.; Lundstrom, C.; Astrom, K. Towards grading gleason score using generically trained deep convolutional neural networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1163–1167. [Google Scholar]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Van De Kaa, C.H.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Dohmgörgen, T.; Toma, M.; Kristiansen, G. High-accuracy prostate cancer pathology using deep learning. Nat. Mach. Intell. 2020, 2, 1–8. [Google Scholar] [CrossRef]
- Duran-Lopez, L.; Dominguez-Morales, J.; Rios-Navarro, A.; Gutierrez-Galan, D.; Jimenez-Fernandez, A.; Vicente-Diaz, S.; Linares-Barranco, A. Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed. Sensors 2021, 21, 1122. [Google Scholar] [CrossRef]
- Nir, G.; Hor, S.; Karimi, D.; Fazli, L.; Skinnider, B.F.; Tavassoli, P.; Turbin, D.; Villamil, C.F.; Wang, G.; Wilson, R.S.; et al. Automatic grading of prostate cancer in digitized histopathology images: Learning from multiple experts. Med. Image Anal. 2018, 50, 167–180. [Google Scholar] [CrossRef]
- Tariq, M.; Iqbal, S.; Ayesha, H.; Abbas, I.; Ahmad, K.T.; Niazi, M.F.K. Medical image based breast cancer diagnosis: State of the art and future directions. Expert Syst. Appl. 2021, 167, 114095. [Google Scholar] [CrossRef]
- Jimenez-del-Toro, O.; Otálora, S.; Andersson, M.; Eurén, K.; Hedlund, M.; Rousson, M.; Atzori, M. Analysis of histopathology images: From traditional machine learning to deep learning. In Biomedical Texture Analysis; Academic Press: Cambridge, MA, USA, 2017; pp. 281–314. [Google Scholar]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Belsare, A. Histopathological Image Analysis Using Image Processing Techniques: An Overview. Signal Image Process. Int. J. 2012, 3, 23–36. [Google Scholar] [CrossRef]
- Arevalo, J.; Cruz-Roa, A.; González, F.A. Histopathology image representation for automatic analysis: A state-of-the-art review. Rev. Med. 2014, 22, 79–91. [Google Scholar] [CrossRef]
- Jothi, J.A.A.; Rajam, V.M.A. A survey on automated cancer diagnosis from histopathology images. Artif. Intell. Rev. 2017, 48, 31–81. [Google Scholar] [CrossRef]
- Das, A.; Nair, M.S.; Peter, S.D. Computer-Aided Histopathological Image Analysis Techniques for Automated Nuclear Atypia Scoring of Breast Cancer: A Review. J. Digit. Imaging 2020, 33, 1–31. [Google Scholar] [CrossRef]
- Madabhushi, A. Digital pathology image analysis: Opportunities and challenges. Imaging Med. 2009, 1, 7–10. [Google Scholar] [CrossRef]
- Humphrey, P.A. Histopathology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030411. [Google Scholar] [CrossRef]
- Mosquera-Lopez, C.; Agaian, S.; Velez-Hoyos, A.; Thompson, I. Computer-Aided Prostate Cancer Diagnosis from Digitized Histopathology: A Review on Texture-Based Systems. IEEE Rev. Biomed. Eng. 2015, 8, 98–113. [Google Scholar] [CrossRef]
- Li, C.; Chen, H.; Li, X.; Xu, N.; Hu, Z.; Xue, D.; Qi, S.; Ma, H.; Zhang, L.; Sun, H. A review for cervical histopathology image analysis using machine vision approaches. Artif. Intell. Rev. 2020, 53, 4821–4862. [Google Scholar] [CrossRef]
- Krithiga, R.; Geetha, P. Breast Cancer Detection, Segmentation and Classification on Histopathology Images Analysis: A Systematic Review. Arch. Comput. Methods Eng. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Van Booven, D.J.; Kuchakulla, M.; Pai, R.; Frech, F.S.; Ramasahayam, R.; Reddy, P.; Parmar, M.; Ramasamy, R.; Arora, H. A Systematic Review of Artificial Intelligence in Prostate Cancer. Res. Rep. Urol. 2021, 13, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.; Hwang, M.; Shah, K.; Madabhushi, A.; Feldman, M.; Tomaszeweski, J. Automated grading of prostate cancer using architectural and textural image features. In Proceedings of the 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Arlington, VA, USA, 12–15 April 2007; pp. 1284–1287. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Yao, K.; Zynger, D.L.; Clinton, S.K.; Chen, J.; Koyuturk, M.; LaFramboise, T.; Gurcan, M. Visually Meaningful Histopathological Features for Automatic Grading of Prostate Cancer. IEEE J. Biomed. Health Inform. 2017, 21, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Khurd, P.; Bahlmann, C.; Maday, P.; Kamen, A.; Gibbs-Strauss, S.; Genega, E.M.; Frangioni, J.V. Computer-aided Gleason grading of prostate cancer histopathological images using texton forests. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 636–639. [Google Scholar] [CrossRef]
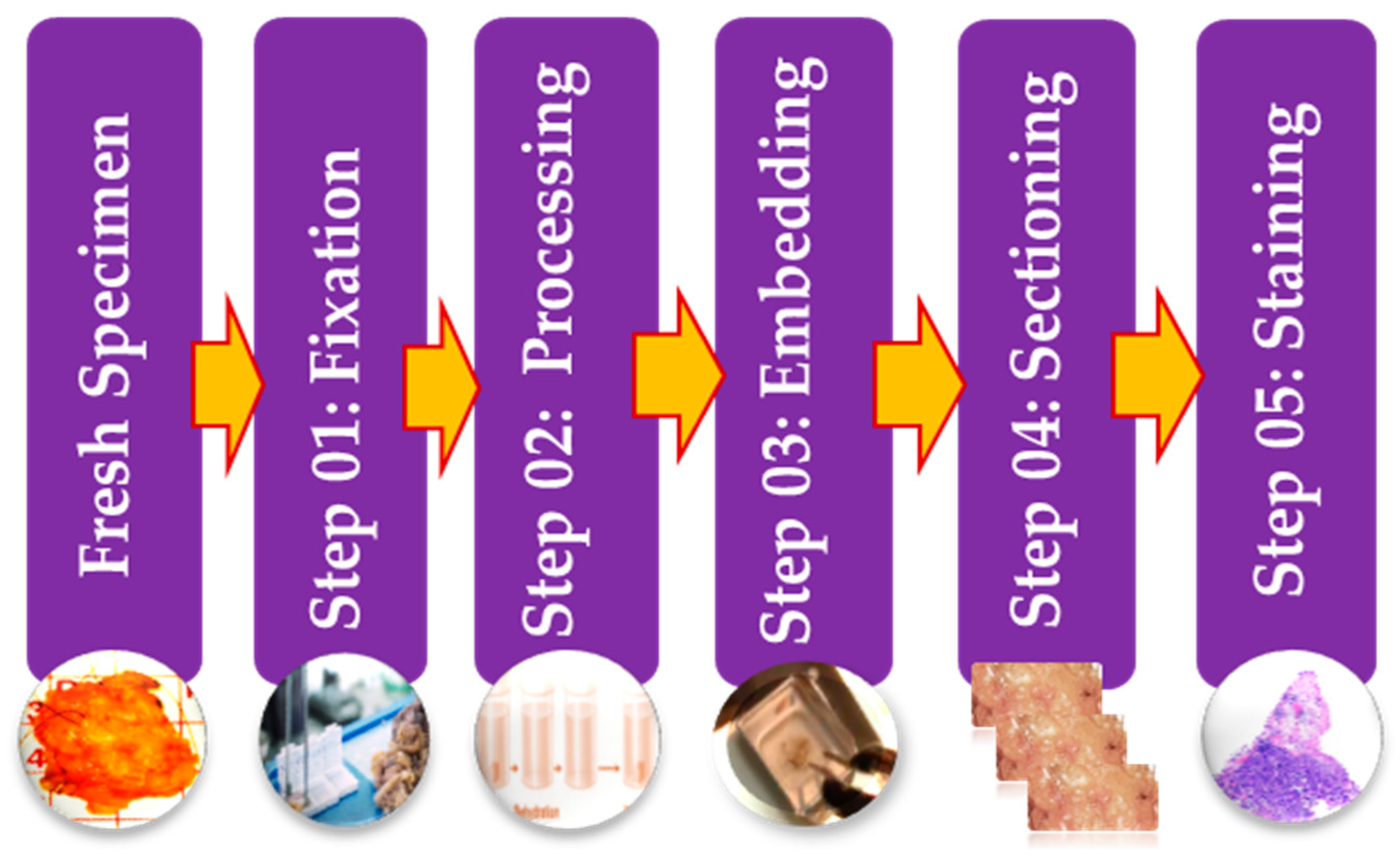
- Slaoui, M.; Fiette, L. Histopathology Procedures: From Tissue Sampling to Histopathological Evaluation. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2010; Volume 691, pp. 69–82. [Google Scholar]
- Cahill, L.C.; Fujimoto, J.G.; Giacomelli, M.G.; Yoshitake, T.; Wu, Y.; Lin, D.I.; Ye, H.; Carrasco-Zevallos, O.M.; Wagner, A.A.; Rosen, S. Comparing histologic evaluation of prostate tissue using nonlinear microscopy and paraffin H&E: A pilot study. Mod. Pathol. 2019, 32, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jia, Z.; Wang, L.-B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I.-C. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 1–17. [Google Scholar] [CrossRef]
- Zangeneh, E.; Rahmati, M.; Mohsenzadeh, Y. Low resolution face recognition using a two-branch deep convolutional neural network architecture. Expert Syst. Appl. 2020, 139, 112854. [Google Scholar] [CrossRef]
- Kramberger, T.; Potočnik, B. LSUN-Stanford Car Dataset: Enhancing Large-Scale Car Image Datasets Using Deep Learning for Usage in GAN Training. Appl. Sci. 2020, 10, 4913. [Google Scholar] [CrossRef]
- Peng, Y.; Jiang, Y.; Eisengart, L.; Healy, M.A.; Straus, F.H.; Yang, X.J. Computer-aided identification of prostatic adenocarcinoma: Segmentation of glandular structures. J. Pathol. Inform. 2011, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Song, F.; Wang, Y.; Dong, H.; Guo, Y.; Liu, J. Breast cancer histopathology image classification through assembling multiple compact CNNs. BMC Med. Inform. Decis. Mak. 2019, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Park, H.-G.; Kim, C.-H.; Prakash, D.; Madusanka, N.; So, J.-H.; Cho, N.-H.; Choi, H.-K. Quantitative Analysis of Benign and Malignant Tumors in Histopathology: Predicting Prostate Cancer Grading Using SVM. Appl. Sci. 2019, 9, 2969. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef]
- Veta, M.M.; Pluim, J.P.W.; Van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Şerbănescu, M.-S.; Manea, N.C.; Streba, L.; Belciug, S.; Pleşea, I.E.; Pirici, I.; Bungărdean, R.M.; Pleşea, R.M. Automated Gleason grading of prostate cancer using transfer learning from general-purpose deep-learning networks. Rom. J. Morphol. Embryol. Rev. Roum. Morphol. Embryol. 2020, 61, 149–155. [Google Scholar] [CrossRef]
- Arvaniti, E.; Claassen, M. Coupling weak and strong supervision for classification of prostate cancer histopathology images. arXiv 2018, arXiv:1811.07013. Available online: https://arxiv.org/abs/1811.07013 (accessed on 2 January 2021).
- A Sharif, S.M.; Naqvi, R.A.; Biswas, M. Learning Medical Image Denoising with Deep Dynamic Residual Attention Network. Mathematics 2020, 8, 2192. [Google Scholar] [CrossRef]
- Çelik, G.; Talu, M.F. Resizing and cleaning of histopathological images using generative adversarial networks. Phys. A Stat. Mech. Its Appl. 2020, 554, 122652. [Google Scholar] [CrossRef]
- Arif, M.; Rajpoot, N. Classification of potential nuclei in prostate histology images using shape manifold learning. In Proceedings of the 2007 International Conference on Machine Vision, Isalambad, Pakistan, 28–29 December 2007; pp. 113–118. [Google Scholar]
- Jafari-Khouzani, K.; Soltanian-Zadeh, H. Multiwavelet grading of pathological images of prostate. IEEE Trans. Biomed. Eng. 2003, 50, 697–704. [Google Scholar] [CrossRef]
- Li, X.; Plataniotis, K.N. A Complete Color Normalization Approach to Histopathology Images Using Color Cues Computed From Saturation-Weighted Statistics. IEEE Trans. Biomed. Eng. 2015, 62, 1862–1873. [Google Scholar] [CrossRef] [PubMed]
- Piórkowski, A. Color Normalization-Based Nuclei Detection in Images of Hematoxylin and Eosin-Stained Multi Organ Tissues. In Proceedings of the International Conference on Image Processing and Communications, Bydgoszcz, Poland, 11–13 September 2019; pp. 57–64. [Google Scholar]
- Xiao, Y.; Decenciere, E.; Velasco-Forero, S.; Burdin, H.; Bornschlogl, T.; Bernerd, F.; Warrick, E.; Baldeweck, T. A New Color Augmentation Method for Deep Learning Segmentation of Histological Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 886–890. [Google Scholar] [CrossRef]
- Vicory, J.; Couture, H.D.; Thomas, N.E.; Borland, D.; Marron, J.; Woosley, J.; Niethammer, M. Appearance normalization of histology slides. Comput. Med. Imaging Graph. 2015, 43, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yang, J. Multi-level magnification correlation hashing for scalable histopathological image retrieval. Neurocomputing 2019, 351, 134–145. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- McClure, P.; Elnakib, A.; El-Ghar, M.A.; Khalifa, F.; Soliman, A.; El-Diasty, T.; Suri, J.S.; Elmaghraby, A.; El-Baz, A. In-Vitro and In-Vivo Diagnostic Techniques for Prostate Cancer: A Review. J. Biomed. Nanotechnol. 2014, 10, 2747–2777. [Google Scholar] [CrossRef] [PubMed]
- Reda, I.; Khalil, A.; Elmogy, M.; El-Fetouh, A.A.; Shalaby, A.; El-Ghar, M.A.; Elmaghraby, A.; Ghazal, M.; El-Baz, A. Deep Learning Role in Early Diagnosis of Prostate Cancer. Technol. Cancer Res. Treat. 2018, 17, 1533034618775530. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Prostate Fused-MRI-Pathology. Available online: https://wiki.cancerimagingarchive.net/display/Public/Prostate+Fused-MRI-Pathology (accessed on 27 March 2021).
- TCGA-PRAD. Available online: https://wiki.cancerimagingarchive.net/display/Public/TCGA-PRAD (accessed on 27 March 2021).
- Prostate cANcer graDe Assessment (PANDA) Challenge. Available online: https://www.kaggle.com/c/prostate-cancer-grade-assessment/data (accessed on 27 March 2021).
- PESO: Prostate Epithelium Segmentation on H&E-Stained Prostatectomy Whole Slide Images. Available online: https://zenodo.org/record/1485967#.YF945q8zbIU (accessed on 27 March 2021).
- Jain, R.; Kasturi, R.; Schunck, B.G. Machine Vision; McGraw-Hill International Edition: New York, NY, USA, 1995. [Google Scholar]
- Hoshyar, A.N.; Al-Jumaily, A.; Hoshyar, A.N. The Beneficial Techniques in Preprocessing Step of Skin Cancer Detection System Comparing. Procedia Comput. Sci. 2014, 42, 25–31. [Google Scholar] [CrossRef]
- Patidar, P.; Gupta, M.; Srivastava, S.; Nagawat, A.K. Image De-noising by Various Filters for Different Noise. Int. J. Comput. Appl. 2010, 9, 45–50. [Google Scholar] [CrossRef]
- Lee, G.; Singanamalli, A.; Wang, H.; Feldman, M.D.; Master, S.R.; Shih, N.N.C.; Spangler, E.; Rebbeck, T.; Tomaszewski, J.E.; Madabhushi, A. Supervised multi-view canonical correlation analysis (sMVCCA): Integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans. Med Imaging 2014, 34, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Gurcan, M.N.; Boucheron, L.E.; Can, A.; Madabhushi, A.; Rajpoot, N.M.; Yener, B. Histopathological Image Analysis: A Review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Meer, P.; Foran, D.J. Unsupervised segmentation based on robust estimation and color active contour models. IEEE Trans. Inf. Technol. Biomed. 2005, 9, 475–486. [Google Scholar] [CrossRef]
- Bautista, P.A.; Hashimoto, N.; Yagi, Y. Color standardization in whole slide imaging using a color calibration slide. J. Pathol. Inform. 2014, 5, 4. [Google Scholar] [CrossRef]
- Zuo, C.; Chen, Q.; Sui, X. Range Limited Bi-Histogram Equalization for image contrast enhancement. Optik 2013, 124, 425–431. [Google Scholar] [CrossRef]
- Tam, A.; Barker, J.; Rubin, D.L. A method for normalizing pathology images to improve feature extraction for quantitative pathology. Med. Phys. 2016, 43, 528–537. [Google Scholar] [CrossRef]
- Shanmugavadivu, P.; Balasubramanian, K. Particle swarm optimized multi-objective histogram equalization for image enhancement. Opt. Laser Technol. 2014, 57, 243–251. [Google Scholar] [CrossRef]
- Nanni, L.; Brahnam, S.; Ghidoni, S.; Maguolo, G. General purpose (GenP) bioimage ensemble of handcrafted and learned features with data augmentation. arXiv 2019, arXiv:1904.08084. Available online: https://arxiv.org/abs/1904.08084 (accessed on 30 January 2021).
- Shin, H.-C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical Image Synthesis for Data Augmentation and Anonymization Using Generative Adversarial Networks. In Tools and Algorithms for the Construction and Analysis of Systems; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018; pp. 1–11. [Google Scholar]
- Sandfort, V.; Yan, K.; Pickhardt, P.J.; Summers, R.M. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci. Rep. 2019, 9, 16884. [Google Scholar] [CrossRef]
- Liu, S.; Shah, Z.; Sav, A.; Russo, C.; Berkovsky, S.; Qian, Y.; Coiera, E.; Di Ieva, A. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci. Rep. 2020, 10, 7733. [Google Scholar] [CrossRef]
- Ataky, S.T.M.; De Matos, J.; Britto, A.D.S.; Oliveira, L.E.S.; Koerich, A.L. Data Augmentation for Histopathological Images Based on Gaussian-Laplacian Pyramid Blending. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [Google Scholar] [CrossRef]
- Chauhan, N.K.; Singh, K. A Review on Conventional Machine Learning vs Deep Learning. In Proceedings of the 2018 International Conference on Computing, Power and Communication Technologies (GUCON), Greater Noida, India, 28–29 September 2018; pp. 347–352. [Google Scholar]
- Nielsen, B.; Albregtsen, F.; Danielsen, H.E. Automatic segmentation of cell nuclei in Feulgen-stained histological sections of prostate cancer and quantitative evaluation of segmentation results. Cytom. Part A 2012, 81, 588–601. [Google Scholar] [CrossRef]
- Simon, I.; Pound, C.R.; Partin, A.W.; Clemens, J.Q.; Christens-Barry, W.A. Automated image analysis system for detecting boundaries of live prostate cancer cells. Cytometry 1998, 31, 287–294. [Google Scholar] [CrossRef]
- Nguyen, K.; Sabata, B.; Jain, A.K. Prostate cancer grading: Gland segmentation and structural features. Pattern Recognit. Lett. 2012, 33, 951–961. [Google Scholar] [CrossRef]
- Nguyen, K.; Jain, A.K.; Allen, R.L. Automated Gland Segmentation and Classification for Gleason Grading of Prostate Tissue Images. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 1497–1500. [Google Scholar]
- Ayyad, S.M.; Saleh, A.I.; Labib, L.M.; Aiyad, S.M. A new distributed feature selection technique for classifying gene expression data. Int. J. Biomath. 2019, 12. [Google Scholar] [CrossRef]
- Ayyad, S.M.; Saleh, A.I.; Labib, L.M. Gene expression cancer classification using modified K-Nearest Neighbors technique. Biosystems 2019, 176, 41–51. [Google Scholar] [CrossRef]
- Albashish, D.; Sahran, S.; Abdullah, A.; Adam, A.; Shukor, N.A.; Pauzi, S.H.M. Multi-scoring feature selection method based on SVM-RFE for prostate cancer diagnosis. In Proceedings of the 2015 International Conference on Electrical Engineering and Informatics (ICEEI), Denpasar, Indonesia, 10–11 August 2015; pp. 682–686. [Google Scholar]
- Peyret, R.; Bouridane, A.; Khelifi, F.; Tahir, M.A.; Al-Maadeed, S. Automatic classification of colorectal and prostatic histologic tumor images using multiscale multispectral local binary pattern texture features and stacked generalization. Neurocomputing 2018, 275, 83–93. [Google Scholar] [CrossRef]
- Sparks, R.; Madabhushi, A. Content-based image retrieval utilizing explicit shape descriptors: Applications to breast MRI and prostate histopathology. SPIE Med. Imaging 2011, 7962, 79621. [Google Scholar] [CrossRef]
- Tabesh, A.; Teverovskiy, M. Tumor Classification in Histological Images of Prostate Using Color Texture. In Proceedings of the 2006 Fortieth Asilomar Conference on Signals, Systems and Computers, Pacific Grove, CA, USA, 29 October–1 November 2006; pp. 841–845. [Google Scholar]
- Akakin, H.C.; Gurcan, M.N. Content-Based Microscopic Image Retrieval System for Multi-Image Queries. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 758–769. [Google Scholar] [CrossRef]
- Lopez, C.M.; Agaian, S.; Sanchez, I.; Almuntashri, A.; Zinalabdin, O.; Al Rikabi, A.; Thompson, I. Exploration of efficacy of gland morphology and architectural features in prostate cancer gleason grading. In Proceedings of the 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Seoul, Korea, 14–17 October 2012; pp. 2849–2854. [Google Scholar]
- Shaban, W.M.; Rabie, A.H.; Saleh, A.I.; Abo-Elsoud, M. A new COVID-19 Patients Detection Strategy (CPDS) based on hybrid feature selection and enhanced KNN classifier. Knowl. -Based Syst. 2020, 205, 106270. [Google Scholar] [CrossRef] [PubMed]
- Ayyad, S.M.; Saleh, A.I.; Labib, L.M. Classification techniques in gene expression microarray data. Int. J. Comput. Sci. Mob. Comput. 2018, 7, 52–56. [Google Scholar]
- Castanho, M.; Hernandes, F.; De Ré, A.; Rautenberg, S.; Billis, A. Fuzzy expert system for predicting pathological stage of prostate cancer. Expert Syst. Appl. 2013, 40, 466–470. [Google Scholar] [CrossRef]
- Shaban, W.M.; Rabie, A.H.; Saleh, A.I.; Abo-Elsoud, M. Detecting COVID-19 patients based on fuzzy inference engine and Deep Neural Network. Appl. Soft Comput. 2020, 99, 106906. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Yong, S.-P. A comparison of deep learning and hand crafted features in medical image modality classification. In Proceedings of the 2016 3rd International Conference on Computer and Information Sciences (ICCOINS), Kuala Lumpur, Malaysia, 15–17 August 2016; pp. 633–638. [Google Scholar]
- Swiderska-Chadaj, Z.; De Bel, T.; Blanchet, L.; Baidoshvili, A.; Vossen, D.; Van Der Laak, J.; Litjens, G. Impact of rescanning and normalization on convolutional neural network performance in multi-center, whole-slide classification of prostate cancer. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
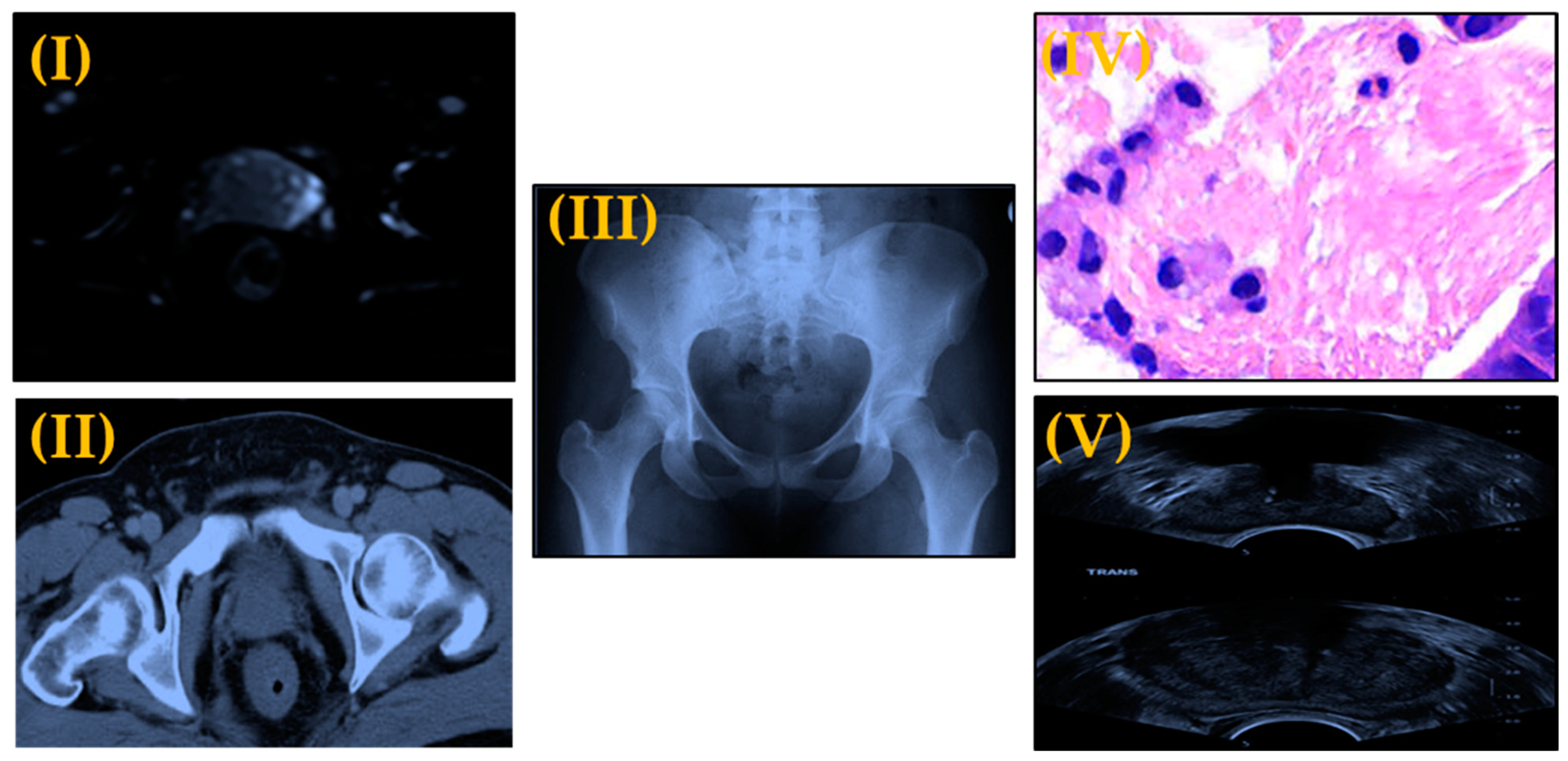
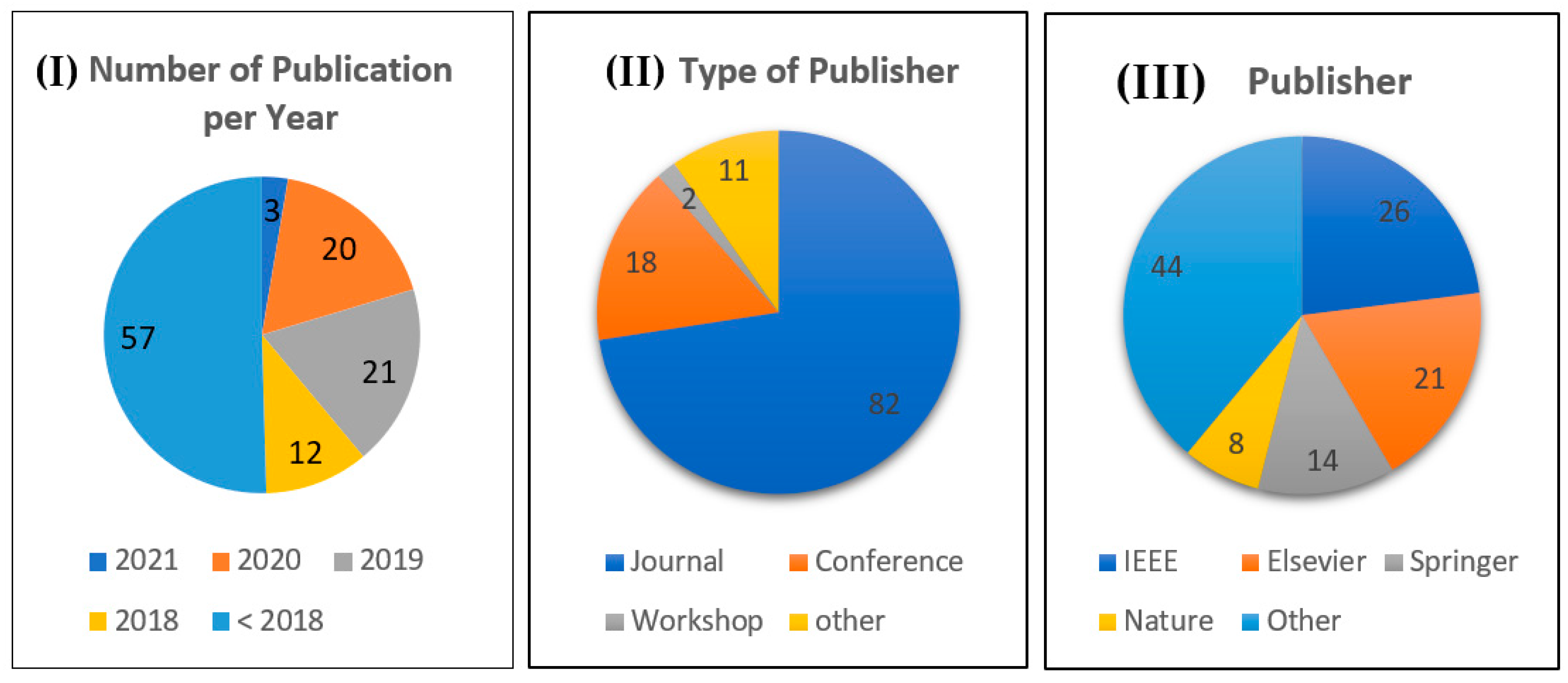
| Reference | Study Aim | Year | Strength | Weakness | Number of Patients |
|---|---|---|---|---|---|
| [2] | Automated classification using AdaBoost-based Ensemble Learning | 2016 | They integrated various feature descriptors, different color channels, and classifiers. | The algorithm able to discover only the critical regions on the digital slides | 50 |
| [14] | A novel technique of labeling individual glands as malignant or benign was proposed. | 2013 | The technique can detect individual malignant gland units without relying on the neighboring histology and/or the spatial extent of the cancer. | It applied on a small number of radical prostatectomy patients | 8 |
| [15] | Methodology for automated gland and nuclei segmentation | 2008 | They incorporate low-, high-level knowledge, and structural constraints imposed via domain knowledge. | They focused on a smaller cohort of cancer images and the dataset is private | 44 |
| [16] | A new automated method for gland segmentation | 2017 | This method texture- and gland structure-based methods | The method failed in the images with the cribriform pattern. They validated data using 2-fold cross validation | 10 |
| [17] | Multistage Segmentation Using Sample Entropy Texture Analysis | 2020 | An added advantage of performing multistage segmentation using sample entropy values is that one could easily separate epithelial nuclei from the stroma nuclei in standard H&E stained images without using any additional immunohistochemical (IHC) markers. | It requires identifying sample entropy features | 25 |
| [18] | A new approach to identify prostate cancer areas in complex | 2014 | It utilizes the differential information embedded in the intensity characteristics of H&E images to quickly classify areas of the prostate tissue | Classification performance is tested using only KNN algorithm | 20 |
| [19] | Ensemble based system for feature selection and classification | 2011 | They addressed the possibility of missing tumor regions through the use of tile-based probabilities and heat maps. | They focused only on texture feature selection and not used a voting schema for the ensemble classifier to enhance the probability scores | 14 |
| [20] | A novel fully automated CAD system | 2006 | The proposed system represents the first attempt to automatically analyse histopathology across multiple scales | Their system trained using only 3 images | 6 |
| [21] | A new multiclass approach | 2018 | It obtained improved grading results | It was evaluated based on its impact on the performance of the ensemble framework only | 213 |
| [22] | A bag-of-words approach to classify images using SpeededUp Robust Features (SURF) | 2016 | The drawbacks of scale-invariant feature transform descriptor is overcome by the SURF descriptors causing an enhanced output accuracy | More features needed to be integrated with their feature extraction process to enhance accuracy of the classification | 75 |
| [23] | An automatic method for segmentation and classification (Integration of Salp Swarm Optimization Algorithm and Rider Optimization Algorithm) | 2019 | Less time complexity | The maximal accuracy, sensitivity, and specificity does not exceed 90% | 20 |
| [24] | A new region-based convolutional neural network framework for multi-task prediction | 2018 | The model achieved a detection accuracy 99.07% with an average area under the curve of 0.998 | They didn’t have patient-level information with which to perform a more rigorous patient-level stratification. | 40 |
| [25] | An approach to nuclei segmentation using a conditional generative adversarial network | 2019 | It enforces higher-order consistency and captures better results when compared to conventional CNN models. | The model trained on small annotated patches | 34 |
| [26] | Deep neural network algorithm for segmentation of individual nuclei | 2019 | A simple, fast, and parameter-free postprocessing procedure is done to get the final segmented nuclei as one 1000 × 1000 image can be segmented in less than 5 s. | The model is trained on a small number of images and has been tested on the images that may have different appearances | 30 |
| [27] | Two novel approaches (combination of 4 types of feature descriptors, advanced machine-learning classifiers) to automatically identify prostate cancer | 2019 | They apply for the first time on prostate segmented glands, deep-learning algorithms modifying the popular VGG19 neural network. | The hand-driven learning approach employs SVM, where selecting the suitable kernel function could be tricky | 35 |
| [28] | Automated Gleason grading via deep learning | 2018 | The study showed promising results especially for cases with heterogeneous Gleason patterns | The model trained on small mini patches at each iteration | 886 |
| [29] | A deep learning system using the U-Net | 2019 | The system outperformed 10 out of 15 pathologists | The system was built upon three pretrained preprocessing modules, each of which still required pixel-wise annotations. | 1243 |
| [30] | Predicting Gleason Score Using OverFeat Trained Deep CNN as feature extractor | 2016 | It is quite effective, even without from-scratch training on WSI tiles. Processing time is low | Small size of patches | 213 |
| [31] | CNN to idiomatically identify the features | 2016 | The system is not constrained to H&E stained images and could easily be applied to immunohistochemistry | Some detection errors happen at the boundaries of the tissue | 254 |
| [32] | DL model to detect cancer based on NASNetLarge architecture and high-quality annotated training dataset | 2020 | The model demonstrated its strong ability in prediction as accuracy attained 98% | The availability of fully digitalized cohorts represents a bottleneck | 400 |
| [33] | A novel benchmark was designed for measuring and comparing the performances of different CNN models with the proposed PROMETEO | 2021 | Average processing time is less compared to other architectures | The network validated on 3-fold cross-validation method | 470 |
| [34] | Novel features that include spatial inter-nuclei statistics and intra-nuclei properties for discriminating high-grade prostate cancer patterns | 2018 | The system tackled the inter-observer variability in prostate grading and can lead to a consensus-based training that improves both classification | lack examples of the highest grades of disease | 56 |
| Dataset | URL | Magnification | Year | Dataset Size | Number of Patients |
|---|---|---|---|---|---|
| Annotated dataset | [75] | 40× | 2017 | 4 images for training and 2 for validation | 6 |
| Prostate Fused-MRI-Pathology | [76] | 20× | Last modified 2021 | comprises a total of 28 3 Tesla T1-weighted, T2-weighted, Diffusion weighted and Dynamic Contrast Enhanced prostate MRI along with accompanying digitized histopathology images | 28 |
| TCGA-PRAD project | [77] | 40× | Last modified 2020 | It includes includes 368 digitized prostate pathology slides | 14 |
| Prostate cANcer graDe Assessment (PANDA) Challenge | [78] | 20× | 2020 | It consists of 11.000 cases for training, 400 cases for public test set, and 400 cases for private test set | NA |
| PESO dataset | [79] | 10× | 2019 | It consists of 62 case for the training set and 40 case for the testing set | 102 |
| Features Type | Reference | Year | Accuracy Result |
|---|---|---|---|
| Texture | [56] | 2011 | The AUC value is 0.91 for the first database and 0.96 for the second database. |
| [102] | 2015 | The proposed method outperforms the classic SVM-RFE in accuracy and reducing redundancy. | |
| [103] | 2018 | The proposed method attained a classification accuracy around 99%. | |
| Topological | [13] | 2011 | The model attainted an average accuracy 90%. |
| [50] | 2011 | The test classification results have an average of 96.76% | |
| [49] | 2017 | The developed way achieved 93.0% training accuracy and 97.6% testing accuracy, for the tested cases. | |
| Morphological | [15] | 2007 | Average accuracy for prostate cancer classification was 92.48% |
| [104] | 2011 | The system achieved 0.55 under the precision recall curve measure | |
| [58] | 2019 | The prediction model resulted an average accuracy of 90.2% | |
| Color | [98] | 2012 | The proposed method attained an average of 86% accuracy in classifying a tissue pattern into different classes. |
| [105] | 2006 | They achieved accuracy of 91.3% | |
| Color & Texture | [106] | 2012 | The algorithm achieved an average of 86% and 93% of classification accuracy. |
| [107] | 2012 | Classification accuracies are 97.6%, 96.6% and 87.3% when differentiating Gleason 4 versus Gleason 3, Gleason 5 versus Gleason 3, and Gleason 5 versus Gleason 4. | |
| Topological & Morphological & Texture | [48] | 2007 | SVM classifier applied to test the accuracy of the extracted features and achieved about 93% when differentiating among Gleason grade 3 and stroma, 92.4% among epithelium and stroma, and 76.9% among Gleason 4 and 3. |
| [27] | 2019 | The proposed model using hand-crafted features achieved an average accuracy of 94.6%. |
| Classifier | Reference | Year | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|---|
| KNN | [66] | 2003 | - | 0.917 | - | - |
| [18] | 2014 | - | 0.76 | - | - | |
| SVM | [48] | 2007 | - | 0.876 | - | - |
| [14] | 2013 | 0.75 | - | 0.83 | 0.81 | |
| [13] | 2019 | 0.98 ± 0.011 for artefacts versus glands 0.92 ± 0.04 for benign versus pathological | 0.95 ± 0.02 for artefacts versus glands 0.88 ± 0.07 for benign versus pathological | 0.95 ± 0.03 for artefacts versus glands 0.87 ± 0.07 for benign versus pathological | 0.94 ± 0.01 for artefacts versus glands 0.80 ± 0.06 for benign versus pathological | |
| [58] | 2019 | - | 0.655 (one-shot classification) 0.92 (Binary classification) | - | - | |
| Bag-of-Words | [22] | 2016 | - | 0.901 | 0.905 | 0.79 |
| MLA | [21] | 2018 | - | 0.883 | 0.94 | 0.876 |
| Boosting Cascade | [20] | 2006 | - | 0.88 | - | - |
| SVM and Random Forest | [19] | 2011 | 0.95 | - | 0.91 | 0.89 |
| Fuzzy Set Theory + Genetic Algorithm | [110] | 2013 | 0.824 | - | 0.95714 | 0.7097 |
| Adaboost | [2] | 2016 | - | 0.978 | - | - |
| Method | Reference | Year | Accuracy Result | Software | ||
|---|---|---|---|---|---|---|
| CNN | [31] | 2016 | AUC ranges from 0.88 to 0.99. | N/A | ||
| CNN built upon VGG19 | [27] | 2019 | Average accuracy of classifying Artefacts vs. Glands is 95.4%, average accuracy of classifying Benign vs. Pathological is 88.3%, Average accuracy of Multi-class classification is 87.6% | Matlab 2018b + Python 3.5 with Keras library and Tensorflow as backend. | ||
| Pretrained CNN | [30] | 2016 | The classification accuracy per image patch is 81%, while for the whole images, the classification accuracy is 89%. | N/A | ||
| Different CNN Architectures | ResNet-50 | [28] | 2018 | They evaluated their results using test cohort and they observed that MobileNet attained the best performance on the validation set | Python 3 with Keras library and tensorflow as backend. Some analysis was done in R by the help of using survminer and survival packages. | |
| MobileNet | ||||||
| Inception-V3 | ||||||
| DenseNet-121 | ||||||
| VGG-16 | ||||||
| U-Net | [29] | 2020 | The developed model achieved accuracy of 99% for biopsies containing tumor and a specificity of 82%. | Tensorflow and Keras | ||
| SSA-RideNN | [23] | 2019 | The technique achieved maximal accuracy of 89.6% and sensitivity of 89.1%, and specificity of 85.9% | Matlab | ||
| SVM | [34] | 2018 | They used Cohen’s kappa coefficient to evaluate the performance. The highest value attained is 0.52 by logistic regression, while 0.37 is attained by using CNN. | Matlab | ||
| Random forest | ||||||
| linear discriminant analysis | ||||||
| logistic regression | ||||||
| CNN | ||||||
| Different CNN Architectures | EfficientNet | [113] | 2020 | UNet attained the best result of AUC about 0.98 | N/A | |
| DenseNet | ||||||
| U-Net | ||||||
| cGAN | [25] | 2018 | The proposed technique achieved F1-score 85.7% for prostate dataset | Pytorch 0.4 | ||
| NB that utilizes CNN | [26] | 2019 | Their proposed model achieves 81.3% precision, 91.4% in recall, and 85.4% in F1. | Python 2.7 with Keras library and Tensorflow | ||
| Path RCNN | [24] | 2019 | Path RCNN attained accuracy of 99% and a mean of area under the curve of 0.99. | Python and Tensorflow backend | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayyad, S.M.; Shehata, M.; Shalaby, A.; Abou El-Ghar, M.; Ghazal, M.; El-Melegy, M.; Abdel-Hamid, N.B.; Labib, L.M.; Ali, H.A.; El-Baz, A. Role of AI and Histopathological Images in Detecting Prostate Cancer: A Survey. Sensors 2021, 21, 2586. https://doi.org/10.3390/s21082586
Ayyad SM, Shehata M, Shalaby A, Abou El-Ghar M, Ghazal M, El-Melegy M, Abdel-Hamid NB, Labib LM, Ali HA, El-Baz A. Role of AI and Histopathological Images in Detecting Prostate Cancer: A Survey. Sensors. 2021; 21(8):2586. https://doi.org/10.3390/s21082586
Chicago/Turabian StyleAyyad, Sarah M., Mohamed Shehata, Ahmed Shalaby, Mohamed Abou El-Ghar, Mohammed Ghazal, Moumen El-Melegy, Nahla B. Abdel-Hamid, Labib M. Labib, H. Arafat Ali, and Ayman El-Baz. 2021. "Role of AI and Histopathological Images in Detecting Prostate Cancer: A Survey" Sensors 21, no. 8: 2586. https://doi.org/10.3390/s21082586
APA StyleAyyad, S. M., Shehata, M., Shalaby, A., Abou El-Ghar, M., Ghazal, M., El-Melegy, M., Abdel-Hamid, N. B., Labib, L. M., Ali, H. A., & El-Baz, A. (2021). Role of AI and Histopathological Images in Detecting Prostate Cancer: A Survey. Sensors, 21(8), 2586. https://doi.org/10.3390/s21082586











