Molecular Docking and Molecular Dynamics Studies of Antidiabetic Phenolic Compound Isolated from Leaf Extract of Englerophytum magalismontanum (Sond.) T.D.Penn.
Abstract
:1. Introduction
2. Results and Discussion
2.1. Plant Extraction and Biological Activities of Crude Methanol Extract and Its Fractions
2.1.1. Plant Extraction and Solvent-Solvent Fractionation of Crude Extract
2.1.2. Antioxidant Activity and TPC of Crude Extract and Its Fractions
2.1.3. α-Amylase and α-Glucosidase Inhibitory Activity of Crude Extract and Its Fraction
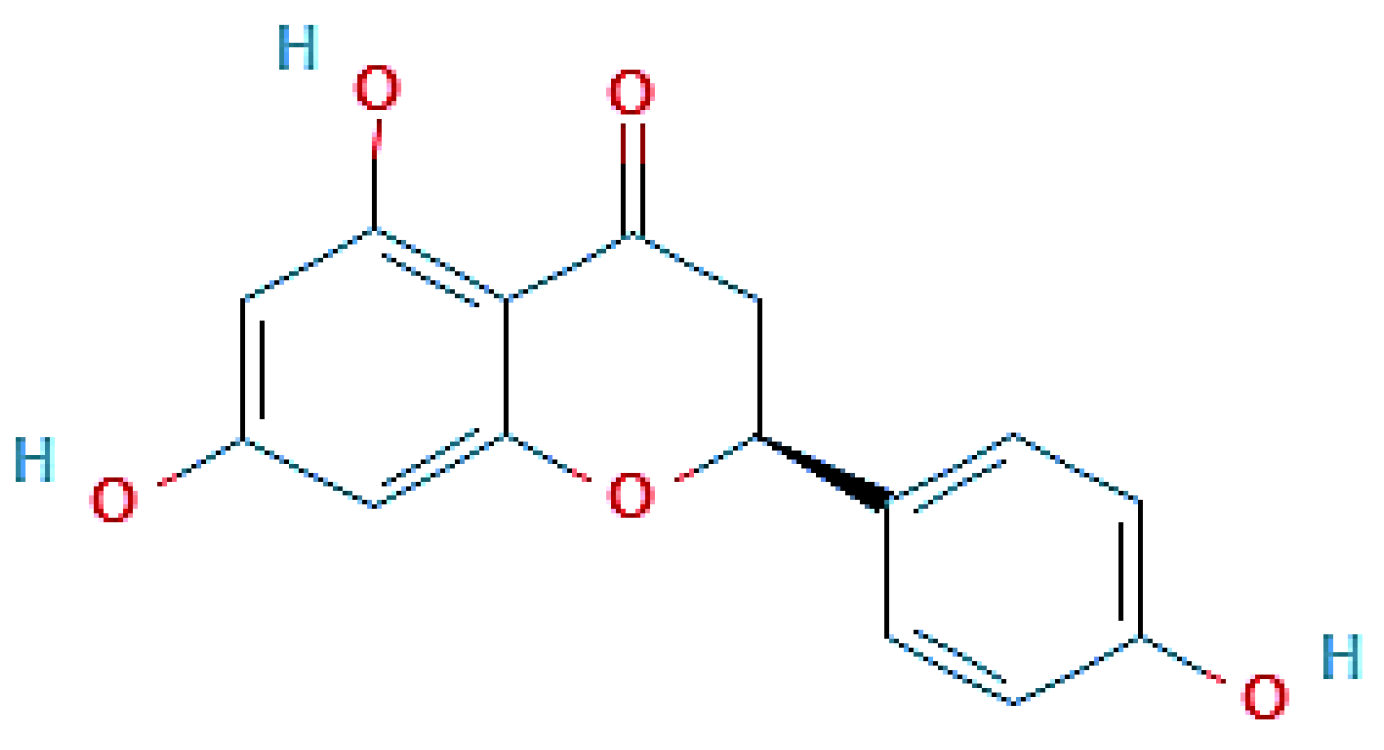
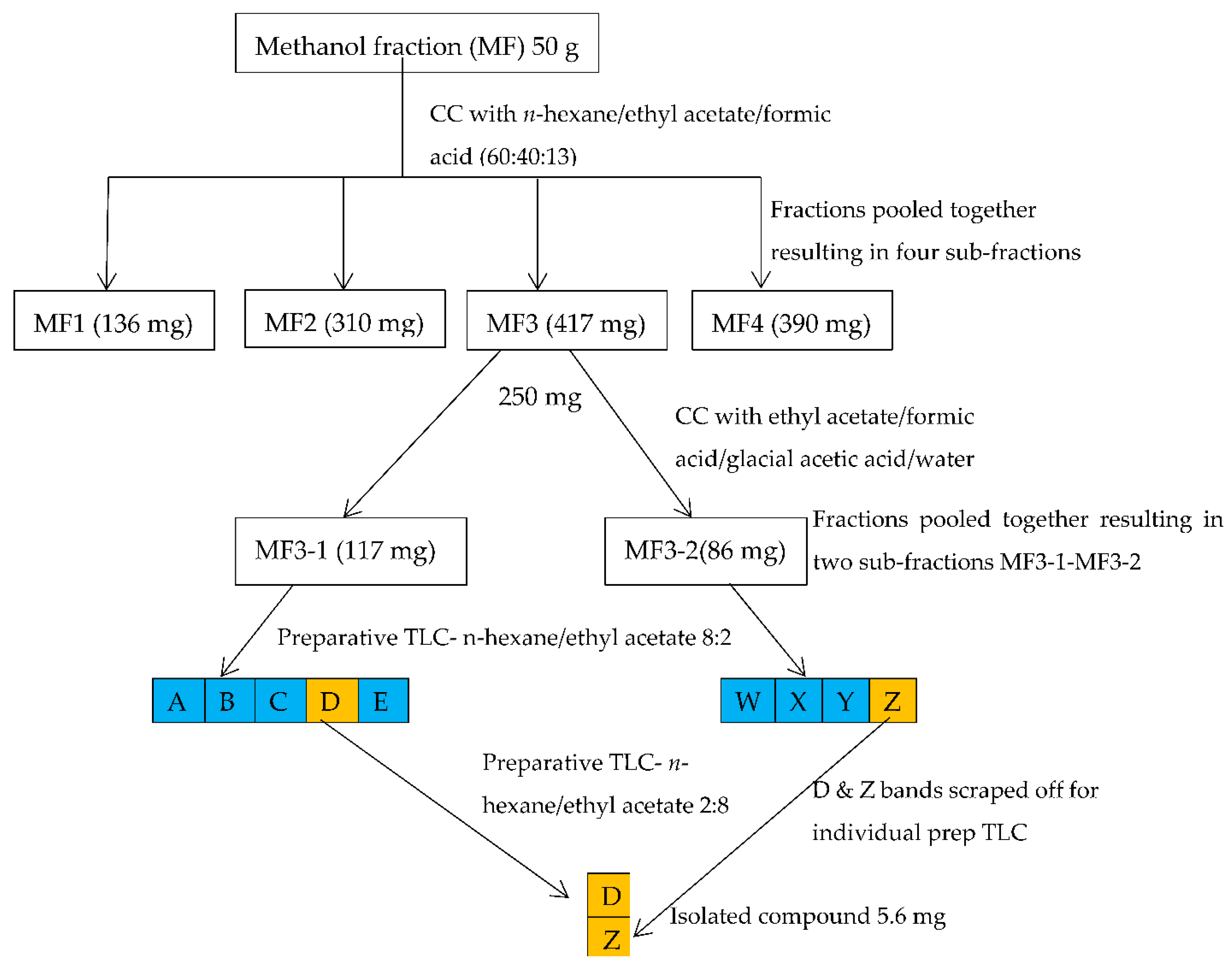
2.2. Isolation and Identification of the Structure of Active Compound
2.3. In Silico Activity of Acarbose and Naringenin against α-Glucosidase Receptor
2.3.1. Molecular Docking of Acarbose and Naringenin
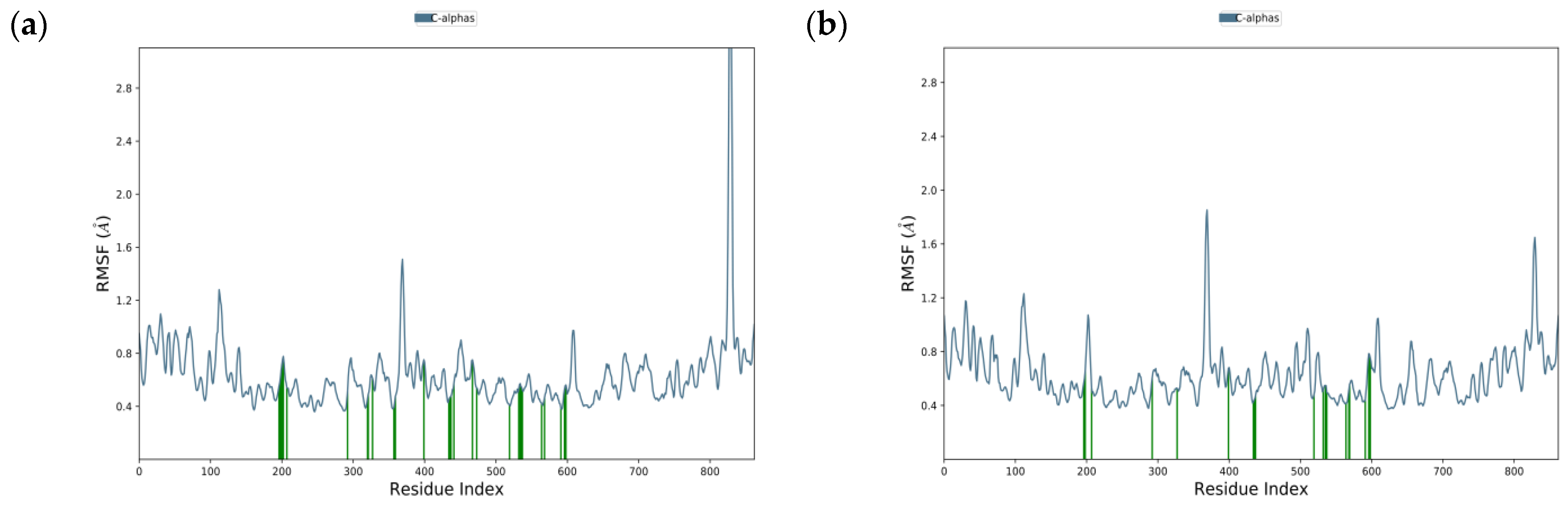
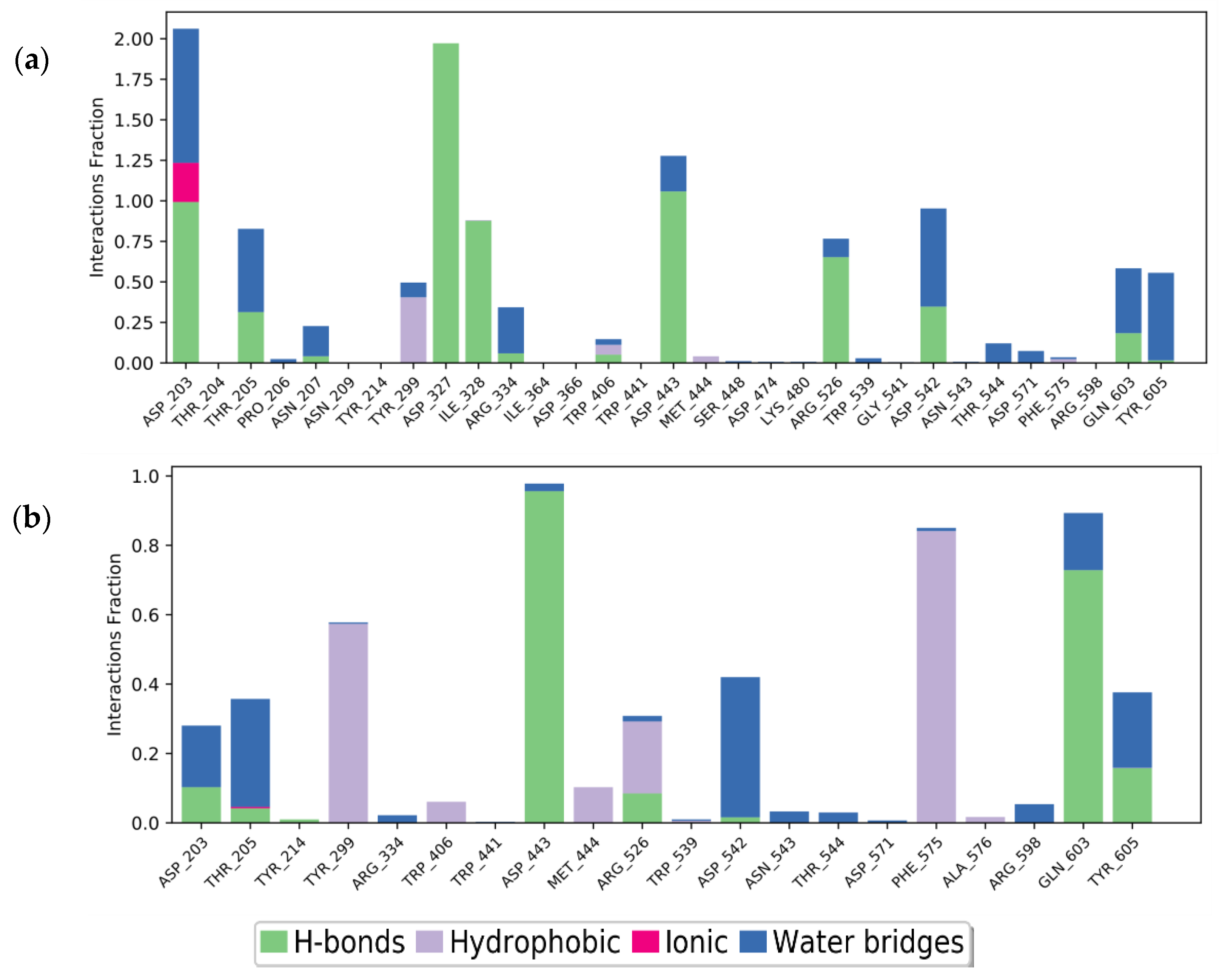
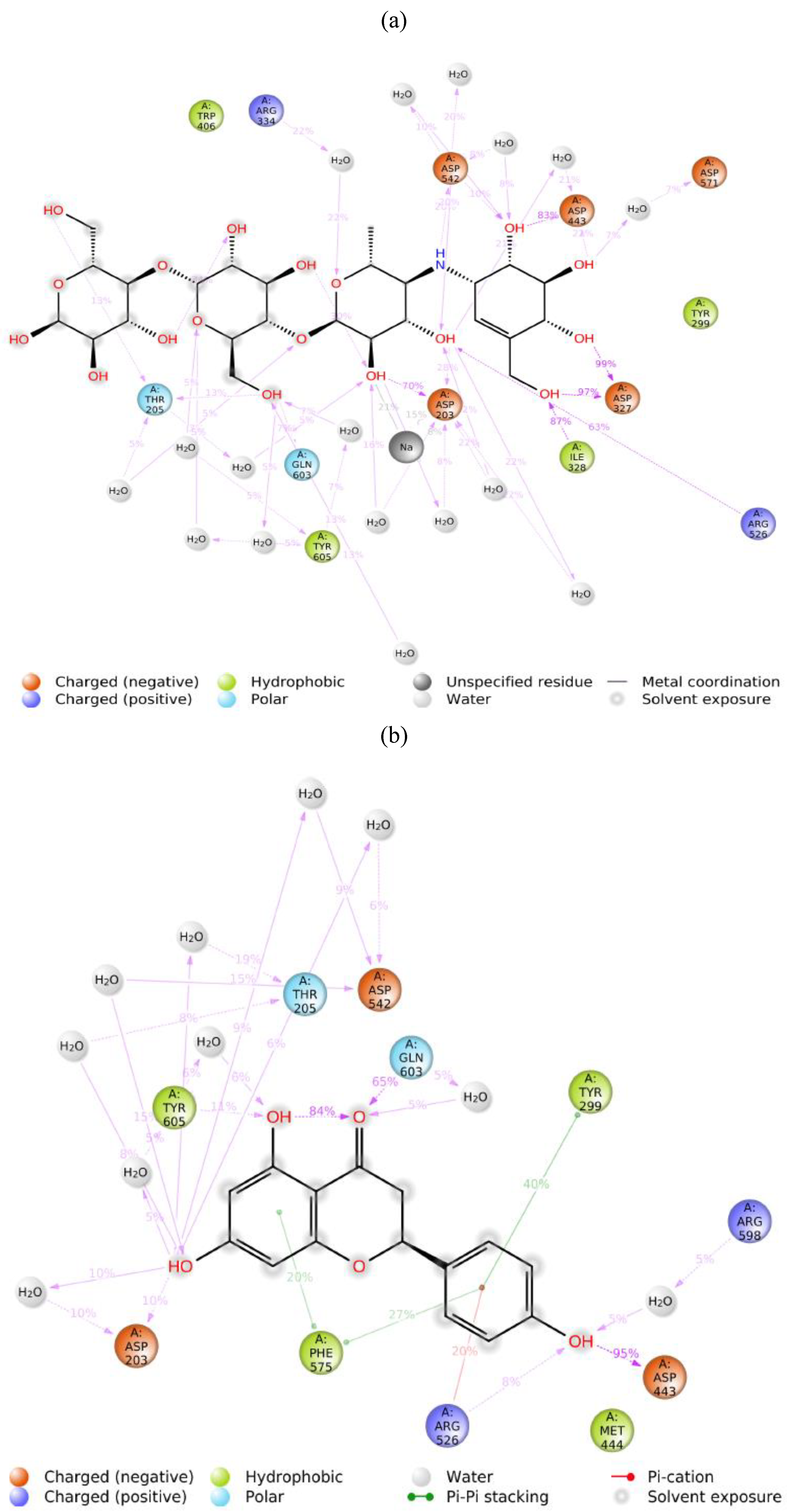
2.3.2. Molecular Dynamics of Acarbose and Naringenin
3. Materials and Methods
3.1. Chemicals and Apparatus
3.2. Plant Materials
3.3. Plant Extraction and Bio-Assay Guided Fractionation
3.4. Characterization of Chemical Structure of Compound
3.5. Total Polyphenolic Content (TPC)
3.6. DPPH (2,2-Diphenyl-1-picrylhydrazyl) Antioxidant Assay
3.7. Enzyme Inhibitory Activity
3.7.1. α-Amylase Inhibitory Assay
3.7.2. α-Glucosidase Inhibitory Assay
3.7.3. Calculation of IC50
3.8. In Silico Assay
3.8.1. Ligand and Receptor Preparations
3.8.2. Molecular Docking
3.8.3. Molecular Dynamics
3.9. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| MMFF94 | molecular modeling force field developed in 1994 |
| PDB ID | protein data bank identifier |
| NPT | isobaric-isothermal ensemble |
| PDBQT | protein data bank, partial charge (Q) and atom type (T) format |
| OPLS | optimized potentials for liquid simulations |
References
- Hawłas, H.J.; Lewenstein, K. Predictive algorithm for the insulin dose selection with continuous glucose monitoring system. In Mechatronics 2013—Recent Technological and Scientific Advances; Březina, T., Jabloński, R., Eds.; Springer: Cham, Switzerland, 2014; pp. 771–777. [Google Scholar]
- Yimam Ahmed, M.; Ejigu, S.H.; Zeleke, A.Z.; Hassen, M.Y. Glycemic control, diabetes complications and their determinants among ambulatory diabetes mellitus patients in Southwest Ethiopia: A prospective cross-sectional study. Diabetes Metab. Syndr. Obes. 2020, 13, 1089–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmet, P.; Alberti, K.G.M.M.; Shaw, J. Global and societal implications of the diabetes epidemic. Nature 2001, 414, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Salpea, P.; Karuranga, S.; Petersohn, I.; Malanda, B.; Gregg, E.W.; Unwin, N.; Wild, S.H.; Williams, R. Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2020, 162, 108086. [Google Scholar] [CrossRef] [Green Version]
- Davidson, M.B. Diabetes Mellitus: Diagnosis and Treatment, 3rd ed.; Churchill Livingstone: New York, NY, USA, 1991. [Google Scholar]
- World Health Organization. Diabetes Education and Prevention. 2010. Available online: www.who.org (accessed on 24 November 2013).
- Shai, L.J.; Masoko, P.; Mokgotho, M.P.; Magano, S.R.; Mogale, A.M.; Boaduo, N.; Eloff, J.N. Yeast alpha glucosidase inhibitory and antioxidant activities of six medicinal plants collected in Phalaborwa, South Africa. S. Afr. J. Bot. 2010, 76, 465–470. [Google Scholar] [CrossRef] [Green Version]
- Ojewole, J.A. Antinociceptive, anti-inflammatory and antidiabetic properties of Hypoxis hemerocallidea Fisch. & C.A. Mey. (Hypoxidaceae) corm (‘African Potato’) aqueous extract in mice and rats. J. Ethnopharmacol. 2006, 103, 126–134. [Google Scholar] [PubMed]
- Sabiu, S.; Balogun, F.O.; Amoo, S.O. Phenolics profiling of Carpobrotus edulis (L.) N.E.Br. and insights into molecular dynamics of their significance in type 2 diabetes therapy and its retinopathy complication. Molecules 2021, 26, 4867. [Google Scholar] [CrossRef]
- Laporta, O.; Pérez-Fons, L.; Mallvia, R.; Caturla, N.; Micol, V. Isolation, characterization and antioxidant capacity assessment of the bioactive compounds derived from Hypoxis rooperi corm extract (African potato). Food Chem. 2007, 101, 1425–1437. [Google Scholar] [CrossRef]
- Olaokun, O.O.; Mkolo, N.M.; Mogale, M.A.; King, P.H. Phytochemical screening, antioxidant, anti-inflammatory and glucose utilization activities of three South African plants used traditionally to treat diseases. Biol. Med. 2017, 9, 412. [Google Scholar] [CrossRef]
- Zadeh, M.A.M.; Kargarfard, M.; Marandi, S.M.; Habibi, A. Diets along with interval training regimes improves inflammatory & anti-inflammatory condition in obesity with type 2 diabetes subjects. J. Diabetes Metab. Disord. 2018, 17, 253–267. [Google Scholar] [CrossRef]
- Maurya, R.; Sebastian, P.; Namdeo, M.; Devender, M.; Gertler, A. COVID-19 severity in obesity: Leptin and inflammatory cytokine interplay in the link between high morbidity and mortality. Front. Immunol. 2021, 12, 649359. [Google Scholar] [CrossRef]
- Hutchings, A.; Scott, A.H.; Lewis, G.; Cunningham, A.B. Zulu Medicinal Plants: An Inventory; University of Natal Press: Pietermaritzburg, South Africa, 1996. [Google Scholar]
- Van Wyk, B.E.; Van Oudshoorn, B.; Gericke, N. Medicinal Plants of South Africa, 1st ed.; Briza Publications: Pretoria, South Africa, 1997. [Google Scholar]
- Watt, J.M.; Breyer-Brandwijk, M.G. The Medicinal and Poisonous Plants of Southern and Eastern Africa; Eing an Account of Their Medicinal and Other Uses, Chemical Composition, Pharmacological Effects and Toxicology in Man and Animal, 2nd ed.; E & S Livingstone: London, UK, 1962. [Google Scholar]
- Van der Merwe, D.; Swan, G.E.; Botha, C.J. Use of ethnoveterinary medicinal plants in cattle by Setswana-speaking people in the Madikwe area of the North West province of South Africa. J. S. Afr. Vet. Assoc. 2001, 72, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adebayo, S.A.; Amoo, S.O. South African botanical resources: A gold mine of natural pro-inflammatory enzyme inhibitors? S. Afr. J. Bot. 2019, 123, 214–227. [Google Scholar] [CrossRef]
- Phan, L.T.M.; Nguyen, K.T.P.; Vuong, H.T.; Tran, D.D.; Nguyen, T.X.P.; Hoang, M.N.; Mai, T.P.; Nguyen, H.H. Supercritical fluid extraction of polyphenols from Vietnamese Callisia fragrans leaves and antioxidant activity of the extract. J. Chem. 2020, 17, 2020. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Lin, L.G.; Ye, W.C. Techniques for extraction and isolation of natural products: A comprehensive review. Chin. Med. 2018, 13, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Truong, D.H.; Ta, N.T.A.; Pham, T.V.; Huynh, T.D.; Do, Q.T.G.; Dinh, N.C.G.; Dang, C.D.; Nguyen, T.K.C.; Bui, A.V. Effects of solvent—Solvent fractionation on the total terpenoid content and in vitro anti-inflammatory activity of Serevenia buxifolia bark extract. Food Sci. Nutr. 2021, 9, 1720–1735. [Google Scholar] [CrossRef]
- Aksoy, L.; Kolay, E.; Ağılönü, Y.; Aslan, Z.; Kargıoğlu, M. Free radical scavenging activity, total phenolic content, total antioxidant status, and total oxidant status of endemic Thermopsis turcica. Saudi J. Biol. Sci. 2013, 20, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Mc Cune, L.M.; Johns, T. Antioxidant activity in medical plants associated with the symptoms of diabetes mellitus used by the indigenous people of the North American boreal forest. J. Ethnopharmacol. 2002, 82, 197–205. [Google Scholar] [CrossRef]
- Olszowy, M.; Dawidowicz, A.L. Is it possible to use the DPPH and ABTS methods for reliable estimation of antioxidant power of colored compounds? Chem. Pap. 2018, 72, 393–400. [Google Scholar] [CrossRef]
- Khoddami, A.; Wilkes, M.A.; Roberts, T.H. Techniques for analysis of plant phenolic compounds. Molecules 2013, 18, 2328–2375. [Google Scholar] [CrossRef]
- Praveena, B.; Pradeep, S.N. Antioxidant and antibacterial activities in the leaf extracts of Indian borage (Plectranthus amboinicus). Food Nutr. Sci. 2012, 2012, 17508. [Google Scholar] [CrossRef] [Green Version]
- Huda-Faujan, N.; Noriham, A.; Norrakiah, A.S.; Babji, A.S. Antioxidant activity of plants methanolic extracts containing phenolic compounds. Afr. J. Biotechnol. 2009, 8, 484–489. Available online: http://www.academicjournals.org/AJB (accessed on 20 February 2020).
- Kwon, Y.I.; Apostolidis, E.; Shetty, K. In vitro studies of eggplant (Solanum melongena) phenolics as inhibitors of key enzymes relevant for type 2 diabetes and hypertension. Bioresour. Technol. 2008, 99, 2981–2988. [Google Scholar] [CrossRef] [PubMed]
- Lordan, S.; Smyth, T.J.; Soler-Vila, A.; Stanton, C.; Ross, R.P. The α-amylase and α-glucosidase inhibitory effects of Irish seaweed extracts. Food Chem. 2013, 141, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.C.; Anguenot, R.; Fillion, C.; Beaulieu, M.; Bérubé, J.; Richard, D. Effect of a commercially-available algal phlorotannins extract on digestive enzymes and carbohydrate absorption in vivo. Food Res. Int. 2011, 44, 3026–3029. [Google Scholar] [CrossRef]
- Quan, N.V.; Xuan, T.D.; Tran, H.D.; Thuy, N.T.D.; Trang, L.T.; Huong, C.T.; Andriana, Y.; Tuyen, P.T. Antioxidant, α-amylase and α-glucosidase inhibitory activities and potential constituents of Canarium tramdenum bark. Molecules 2019, 24, 605. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Hung, T.M.; Phuong, P.T.; Ngoc, T.M.; Min, B.S.; Song, K.S.; Seong, Y.H.; Bae, K. Anti-inflammatory activity of flavonoids from Populus davidiana. Arch. Pharm. Res. 2006, 29, 1102–1108. [Google Scholar] [CrossRef]
- Malla, S.; Pandey, R.P.; Kim, B.G.; Sohng, J.K. Regiospecific modifications of naringenin for astragalin production in Escherichia coli. Biotechnol. Bioeng. 2013, 110, 2525–2535. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Fact. 2015, 14, 178. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Kim, B.G.; Lee, Y.; Ryu, J.Y.; Lim, Y.; Hur, H.G.; Ahn, J.H. Regiospecific methylation of naringenin to ponciretin by soybean O-methyltransferase expressed in Escherichia coli. J. Biotechnol. 2005, 119, 155–162. [Google Scholar] [CrossRef]
- Ibrahim, A.R.S. Sulfation of naringenin by Cunninghamella elegans. Phytochemistry 2000, 53, 209–212. [Google Scholar] [CrossRef]
- Pari, L.; Amudha, K. Hepatoprotective role of naringin on nickel-induced toxicity in male Wistar rats. Eur. J. Pharmacol. 2011, 650, 364–370. [Google Scholar] [CrossRef]
- Ortiz-Andrade, R.R.; Sánchez-Salgado, J.C.; Navarrete-Vázquez, G.; Webster, S.P.; Binnie, M.; García-Jiménez, S.; León-Rivera, I.; Cigarroa-Vázquez, P.; Villalobos-Molina, R.; Estrada-Soto, S. Antidiabetic and toxicological evaluations of naringenin in normoglycaemic and NIDDM rat models and its implications on extra-pancreatic glucose regulation. Diabetes Obes. Metab. 2008, 10, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Duncan, B.B.; Schmidt, M.I.; Pankow, J.S.; Ballantyne, C.M.; Couper, D.; Vigo, A.; Hoogeveen, R.; Folsom, A.R.; Heiss, G. Low-grade systemic inflammation and the development of type 2 diabetes: The atherosclerosis risk in communities study. Diabetes 2003, 52, 1799–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahadoran, Z.; Mirmiran, P.; Azizi, F. Dietary polyphenols as potential nutraceuticals in management of diabetes: A review. J. Diabetes Metab. Disord. 2013, 12, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabha, B.; Neethu, S.; Krishnan, S.L.; Sherin, D.R.; Madhukrishnan, M.; Ananthakrishnan, R.; Rameshkumar, K.B.; Manojkumar, T.K.; Jayamurthy, P.; Radhakrishnan, K.V. Antidiabetic potential of phytochemicals isolated from the stem bark of Myristica fatua Houtt. var. magnifica (Bedd.) Sinclair. Bioorg. Med. Chem. 2018, 26, 3461–3467. [Google Scholar] [CrossRef] [PubMed]
- Kakarala, K.K.; Jamil, K. Protease activated receptor-2 (PAR2): Possible target of phytochemicals. J. Biomol. Str. Dyn. 2015, 33, 2003–2022. [Google Scholar] [CrossRef]
- Emran, T.B.; Rahman, M.A.; Uddin, M.M.N.; Rahman, M.M.; Uddin, M.Z.; Dash, R.; Layzu, C. Effects of organic extracts and their different fractions of five Bangladeshi plants on in vitro thrombolysis. BMC Complement. Altern. Med. 2015, 15, 128. [Google Scholar] [CrossRef] [Green Version]
- Ragazzi, E.; Veronese, G. Quantitative analysis of phenolic compounds after thin-layer chromatographic separation. J. Chromatogr. A 1973, 77, 369–375. [Google Scholar] [CrossRef]
- Olech, M.; Komsta, Ł.; Nowak, R.; Cieśla, Ł.; Waksmundzka-Hajnos, M. Investigation of antiradical activity of plant material by thin-layer chromatography with image processing. Food Chem. 2012, 132, 549–553. [Google Scholar] [CrossRef]
- Olaokun, O.O.; McGaw, L.J.; Eloff, J.N.; Naidoo, V. Evaluation of the inhibition of carbohydrate hydrolysing enzymes, antioxidant activity and polyphenolic content of extracts of ten African Ficus species (Moraceae) used traditionally to treat diabetes. BMC Complement. Altern. Med. Ther. 2013, 13, 94. [Google Scholar] [CrossRef] [Green Version]
- Sim, L.; Quezada-Calvillo, R.; Sterchi, E.E.; Nichols, B.L.; Rose, D.R. Human intestinal maltase–glucoamylase: Crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008, 375, 782–792. [Google Scholar] [CrossRef] [PubMed]



| Extract/Fractions | DPPH Scavenging Expressed as TAE (Trolox Antioxidant Equivalent) µg/mL | Total Phenolic Content (mg GAE/g Dry Extract) |
|---|---|---|
| Crude methanol | 1.66 ± 0.63 a | 49.78 ± 0.40 a |
| Hexane (HF) | 3.06 ± 0.77 a | 42.12 ± 1.53 b |
| Chloroform (CF) | 2.99 ± 2.78 b | 53.08 ± 1.98 b |
| Ethyl acetate (EF) | 2.24 ± 1.69 b | 54.41 ± 1.22 b |
| Methanol (MF) | 1.51 ± 0.66 a | 56.53 ± 1.94 b |
| Ascorbic acid | 0.56 ± 0.88 | ND |
| Extract | 50% Inhibitory Concentration (IC50) (µg/mL) | |
|---|---|---|
| α-Amylase Inhibition | α-Glucosidase Inhibition | |
| Crude methanol | 16.16 ± 2.23 a,c | 12.25 ± 1.03 a,b |
| Fraction | ||
| Hexane | 50.93 ± 0.25 b | 39.28 ± 1.02 a |
| Chloroform | 60.85 ± 0.72 b | 52.24 ± 1.02 a |
| Ethyl acetate | 29.18 ± 1.14 a | 25.20 ± 0.76 a |
| Methanol | 10.76 ± 1.33 a,c | 12.27 ± 1.55 a,b |
| MF sub-fraction | ||
| MF1 | 26.84 ± 1.67 c | 58.75 ± 1.48 b |
| MF2 | 36.93 ± 1.01 c | 28.63 ± 1.21 b |
| MF3 | 12.49 ± 0.96 c,d | 10.19 ± 1.04 b |
| MF4 | 45.11 ± 1.36 c | 27.63 ± 2.02 b,d |
| MF3 sub-fraction | ||
| MF3-1 | 8.79 ± 2.23 d | 6.88 ± 0.79 c |
| MF3-2 | 8.71 ± 1.42 d | 7.84 ± 2.61 d |
| Naringenin | 5.81 ± 2.14 d | 4.77 ± 2.99 d |
| Acarbose (positive control) | 1.24 ± 1.64 | 1.92 ± 0.73 |
| NMR | Spectra Signal Correlations |
|---|---|
| 1H | δ 7.37 and 6.83 ppm, (J = 8.6 Hz, H-2′, 6′ and H-3′, 5); δ 5.87 and 5.89 ppm (J = 2.2 Hz, H-6 and H-8). 5-OH δ 5.44 (1H, dd, J = 13.0 and 3.0 Hz), δ 2.70 (1H, dd, J = 17.2 and 3.0 Hz) and δ 3.15 (1H, dd, J = 17.2 and 13.0 Hz) |
| 13C | δ 98.20 (C-6), δ 97.50 (C-8), δ 115.22 (C-3′ and C-5′) and δ 128.10 (C-2′ and C-6′) |
| HMBC | 2JC-H and 3JC-H, δ 196.28 (C-4), δ 160.10 (C-4′), δ 166.59 (C-7), δ 164.11 (C-5), δ 163.50 (C-8a), δ 129.49 (C-1′) and δ 102.19 (C-4a). |
| Compound | Binding Energy (kcal/mol) |
|---|---|
| Acarbose | −7.9 |
| Naringenin | −7.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olaokun, O.O.; Manonga, S.A.; Zubair, M.S.; Maulana, S.; Mkolo, N.M. Molecular Docking and Molecular Dynamics Studies of Antidiabetic Phenolic Compound Isolated from Leaf Extract of Englerophytum magalismontanum (Sond.) T.D.Penn. Molecules 2022, 27, 3175. https://doi.org/10.3390/molecules27103175
Olaokun OO, Manonga SA, Zubair MS, Maulana S, Mkolo NM. Molecular Docking and Molecular Dynamics Studies of Antidiabetic Phenolic Compound Isolated from Leaf Extract of Englerophytum magalismontanum (Sond.) T.D.Penn. Molecules. 2022; 27(10):3175. https://doi.org/10.3390/molecules27103175
Chicago/Turabian StyleOlaokun, Oyinlola Oluwunmi, Sizakele Annousca Manonga, Muhammad Sulaiman Zubair, Saipul Maulana, and Nqobile Monate Mkolo. 2022. "Molecular Docking and Molecular Dynamics Studies of Antidiabetic Phenolic Compound Isolated from Leaf Extract of Englerophytum magalismontanum (Sond.) T.D.Penn." Molecules 27, no. 10: 3175. https://doi.org/10.3390/molecules27103175
APA StyleOlaokun, O. O., Manonga, S. A., Zubair, M. S., Maulana, S., & Mkolo, N. M. (2022). Molecular Docking and Molecular Dynamics Studies of Antidiabetic Phenolic Compound Isolated from Leaf Extract of Englerophytum magalismontanum (Sond.) T.D.Penn. Molecules, 27(10), 3175. https://doi.org/10.3390/molecules27103175






