Effects of Inulin-Based Prebiotics Alone or in Combination with Probiotics on Human Gut Microbiota and Markers of Immune System: A Randomized, Double-Blind, Placebo-Controlled Study in Healthy Subjects
Abstract
:1. Introduction
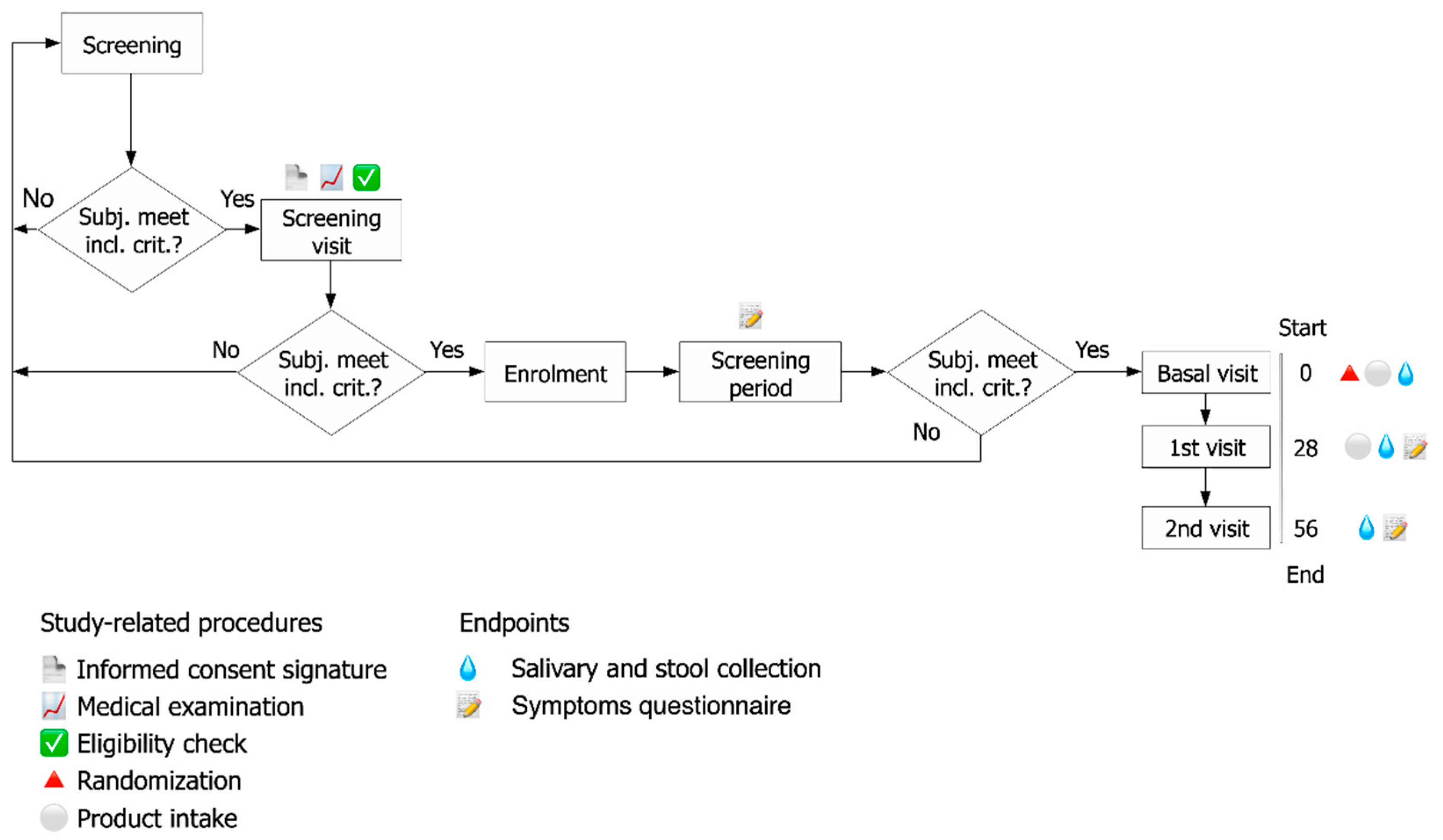
2. Materials and Methods
2.1. Study Design
- A.
- Prebiotic formulation (Group 1): 50 mg FOS (fructo-oligosaccharides with a degree of polymerization between 3–5) and 50 mg inulin (with a degree of polymerization of 10) (for a total of 100 mg of fibers), and common excipients used in formulations
- B.
- Probiotic and prebiotic (synbiotic) formulation (Group 2): 1 × 109 CFU L. acidophilus PBS066 (30 mg as lyophilized), 1 × 109 CFU L. plantarum PBS067 (12 mg as lyophilized), 1 × 109 CFU B. animalis spp. lactis BL050 (30 mg as lyophilized) (all the strains are from the private collection of Roelmi HPC, Origgio, Italy), with 50 mg FOS (fructo-oligosaccharides with a degree of polymerization between 3–5) and 50 mg inulin (with a degree of polymerization of 10) (for a total of 100 mg of fibers), and common excipients used in formulations
- D.
- Placebo formulation (Group 3): the same formulation without prebiotics and probiotics included in the other two formulations.
2.2. Subjects of the Study
2.3. Randomization
2.4. Assessment of Clinical Effects
2.4.1. Endpoints
2.4.2. Symptoms Questionnaire
2.4.3. Assessment of Fecal Calprotectin
2.4.4. Assessment of Fecal β-Defensin2
2.4.5. Assessment of Salivary IgA
2.4.6. Assessment of Total Antioxidant Capacity (TAC) in Salivary Samples through FRAP
2.5. Probiotic Strains and DNA Extraction
2.6. Fecal Microbiology Analysis by Quantitative PCR
2.7. Gut Microbiome Community Characterization by Next Generation Sequencing
2.8. Microbial Composition and Community Structure Analysis
2.9. Statistical Methods
3. Results
3.1. Subjects of the Study
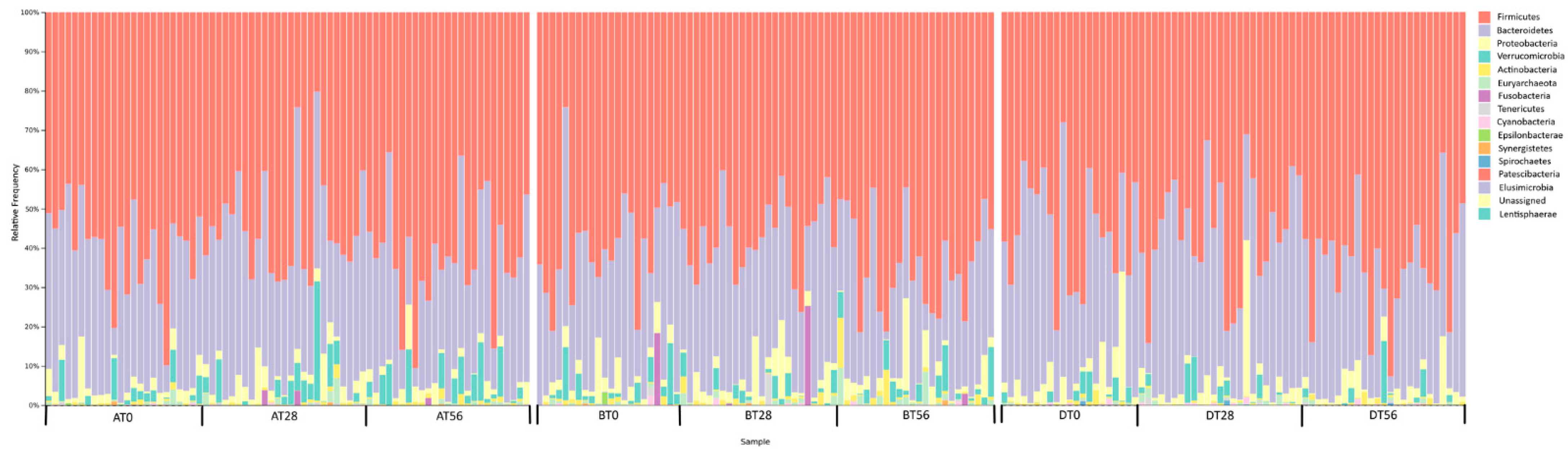
3.2. Gut Microbiota Profile of Recruited Participants during the Treatment
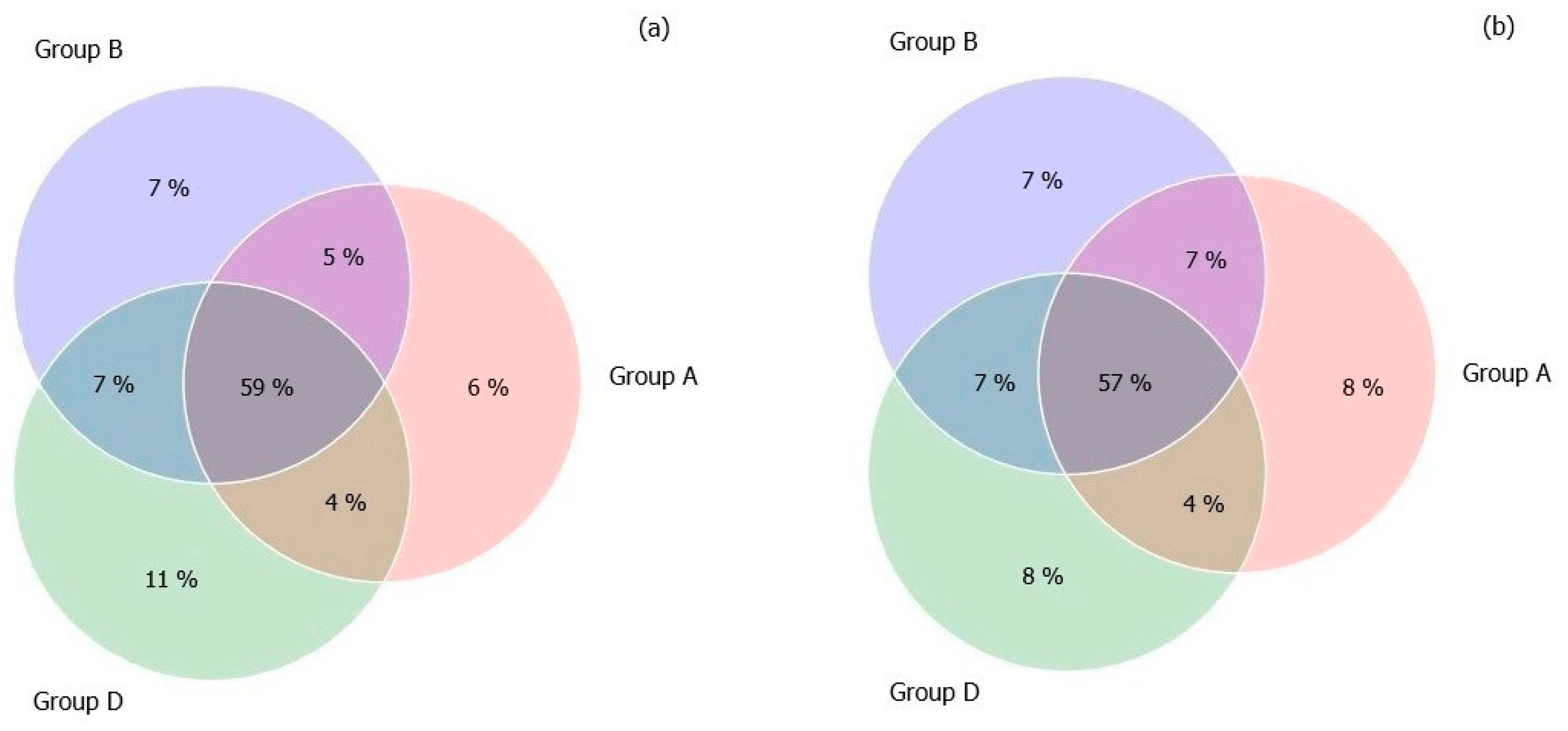
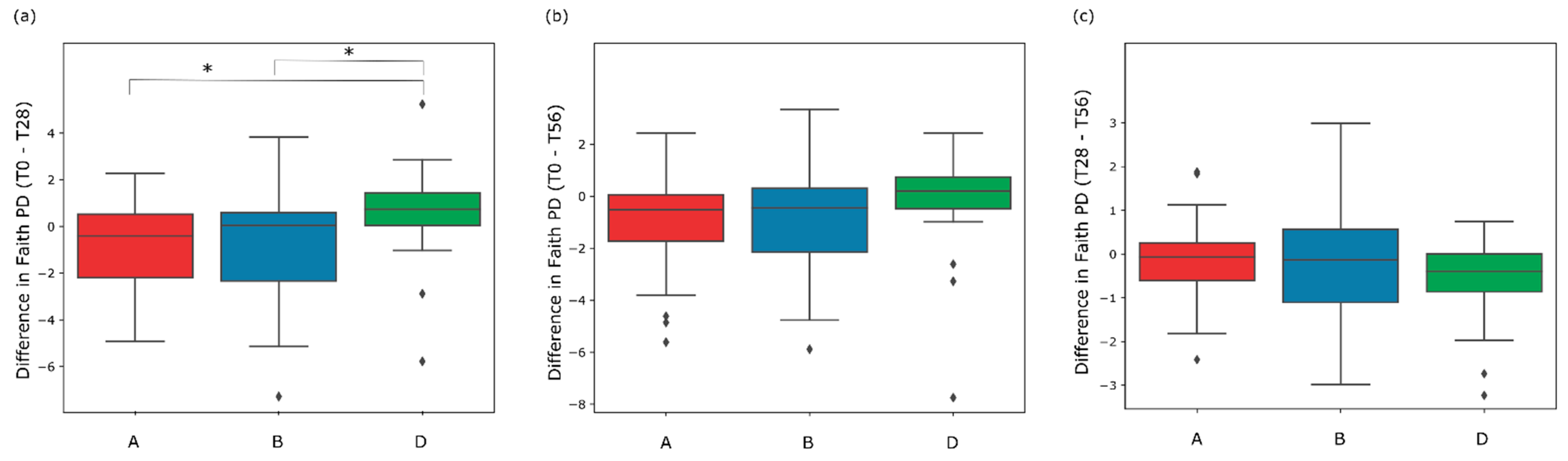
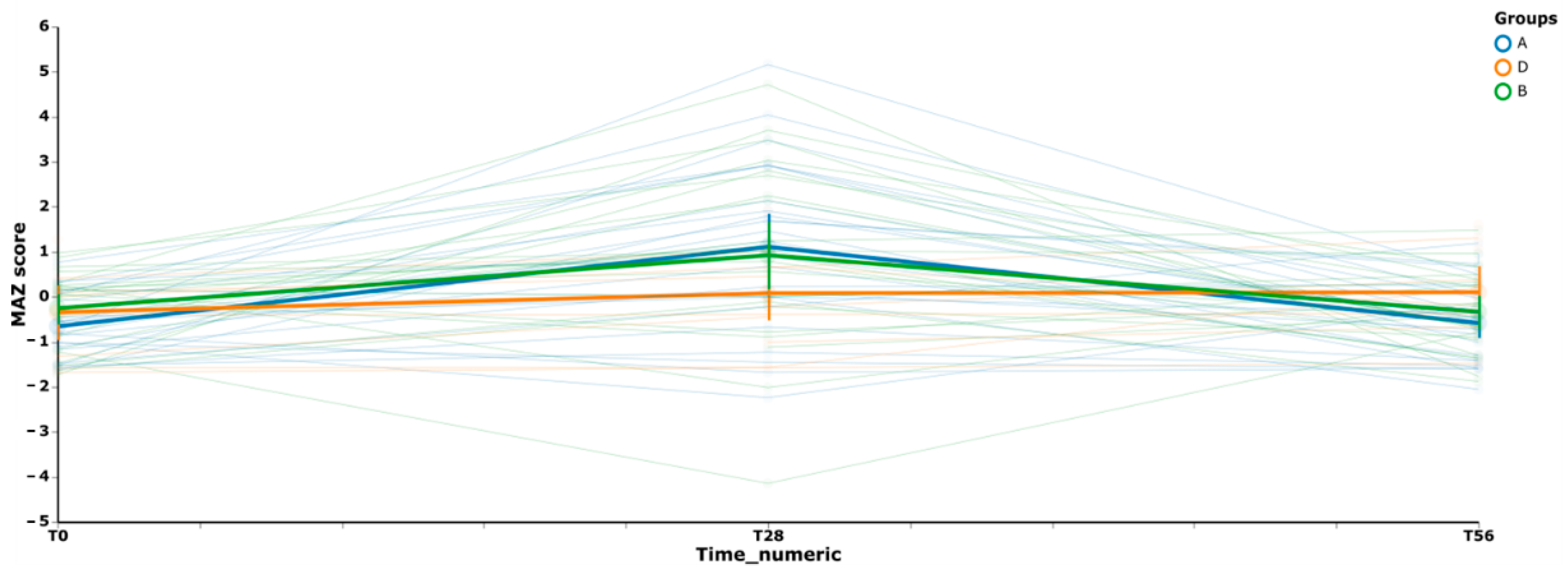
3.3. Gut Microbiome Diversity in Elderly Subjects
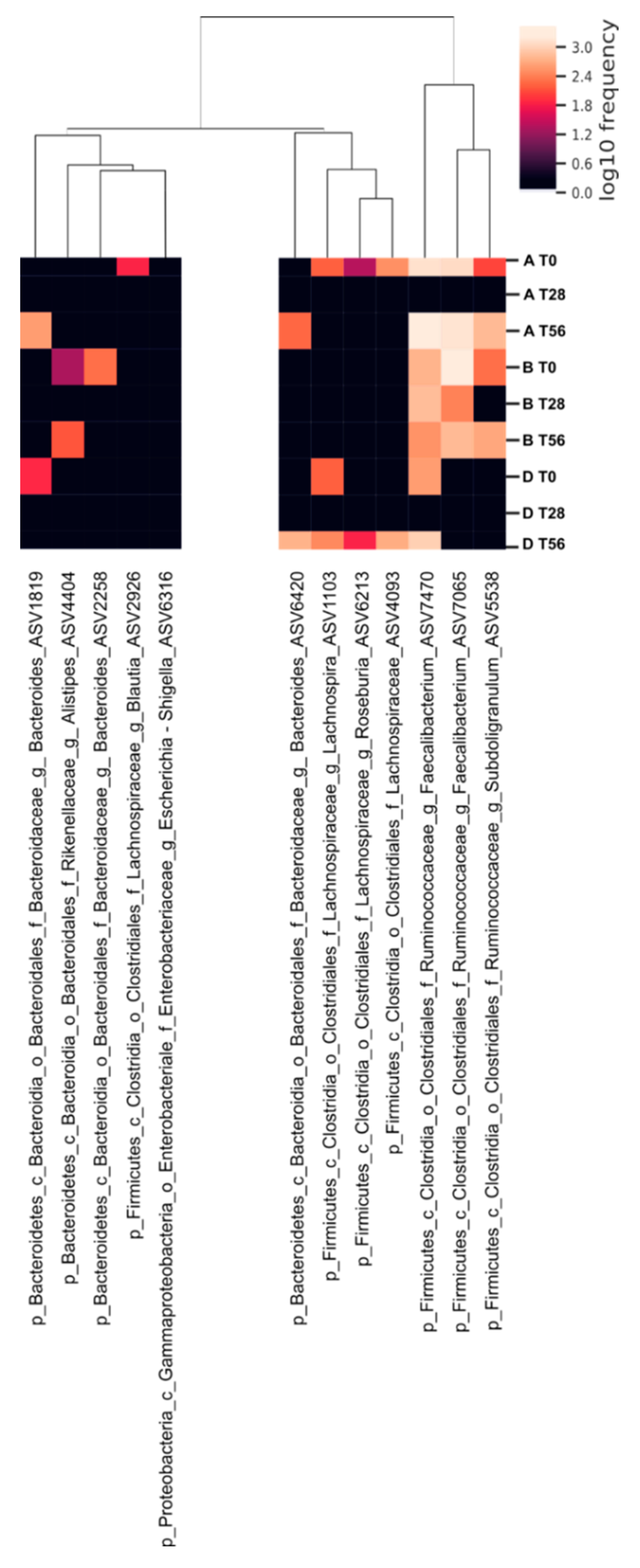
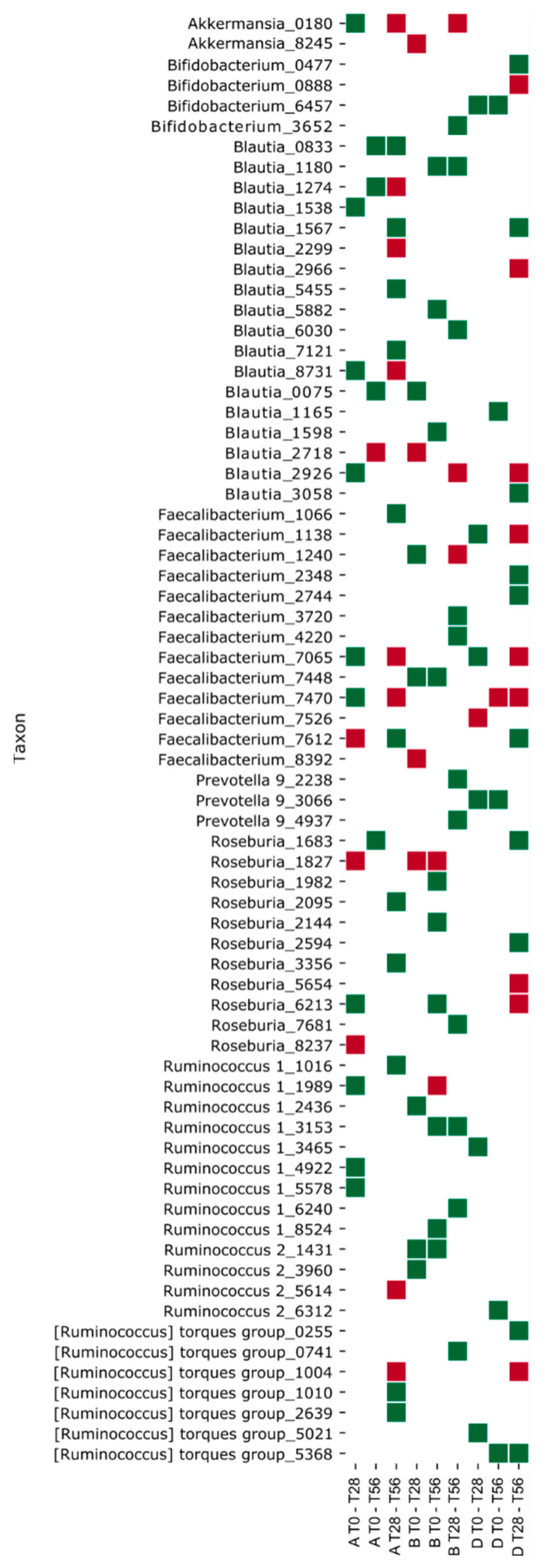
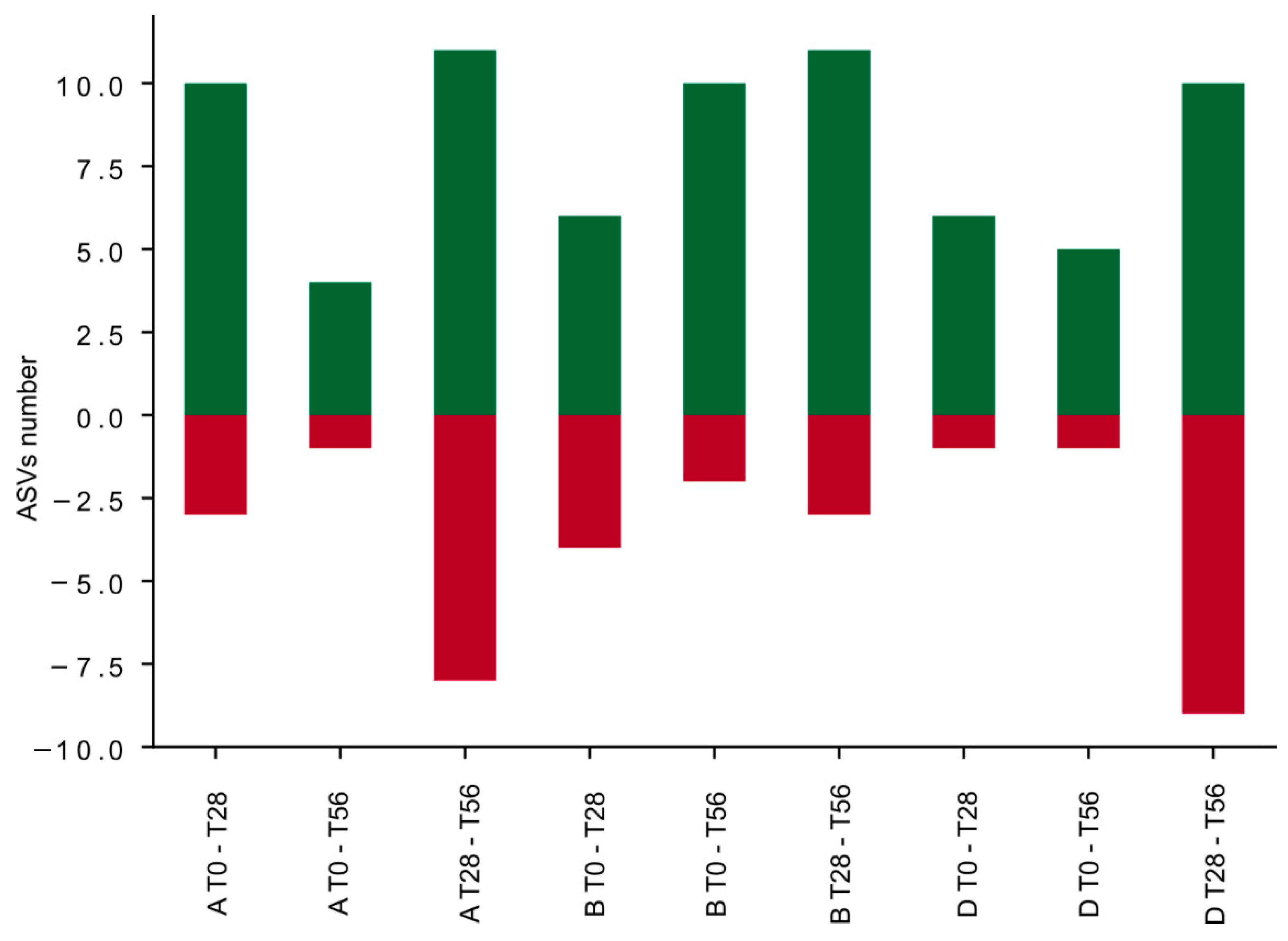
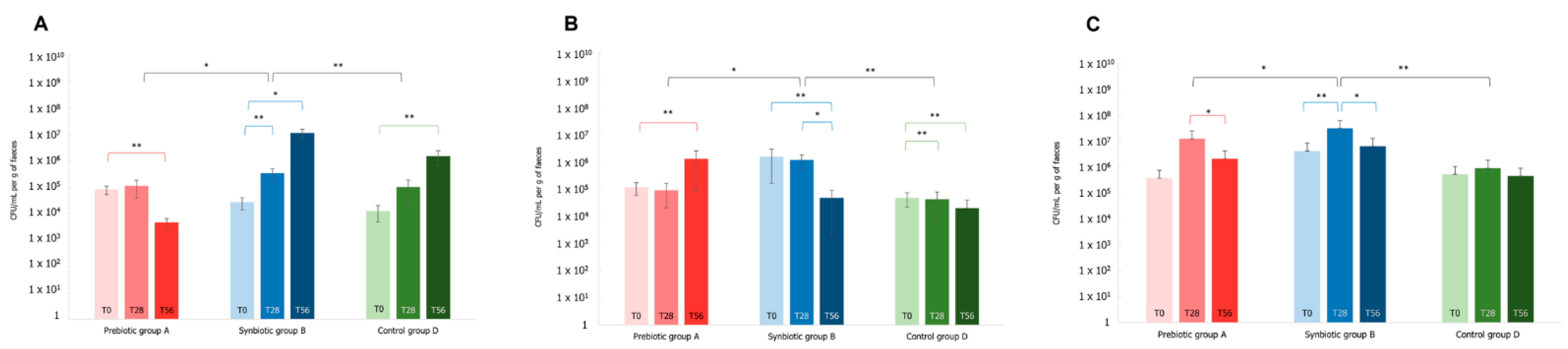
3.4. Effects of the Synbiotic Intake on the Most Relevant Taxa
3.5. Effect of the Synbiotic on Lactobacillus spp. and Bifidobacterium spp.
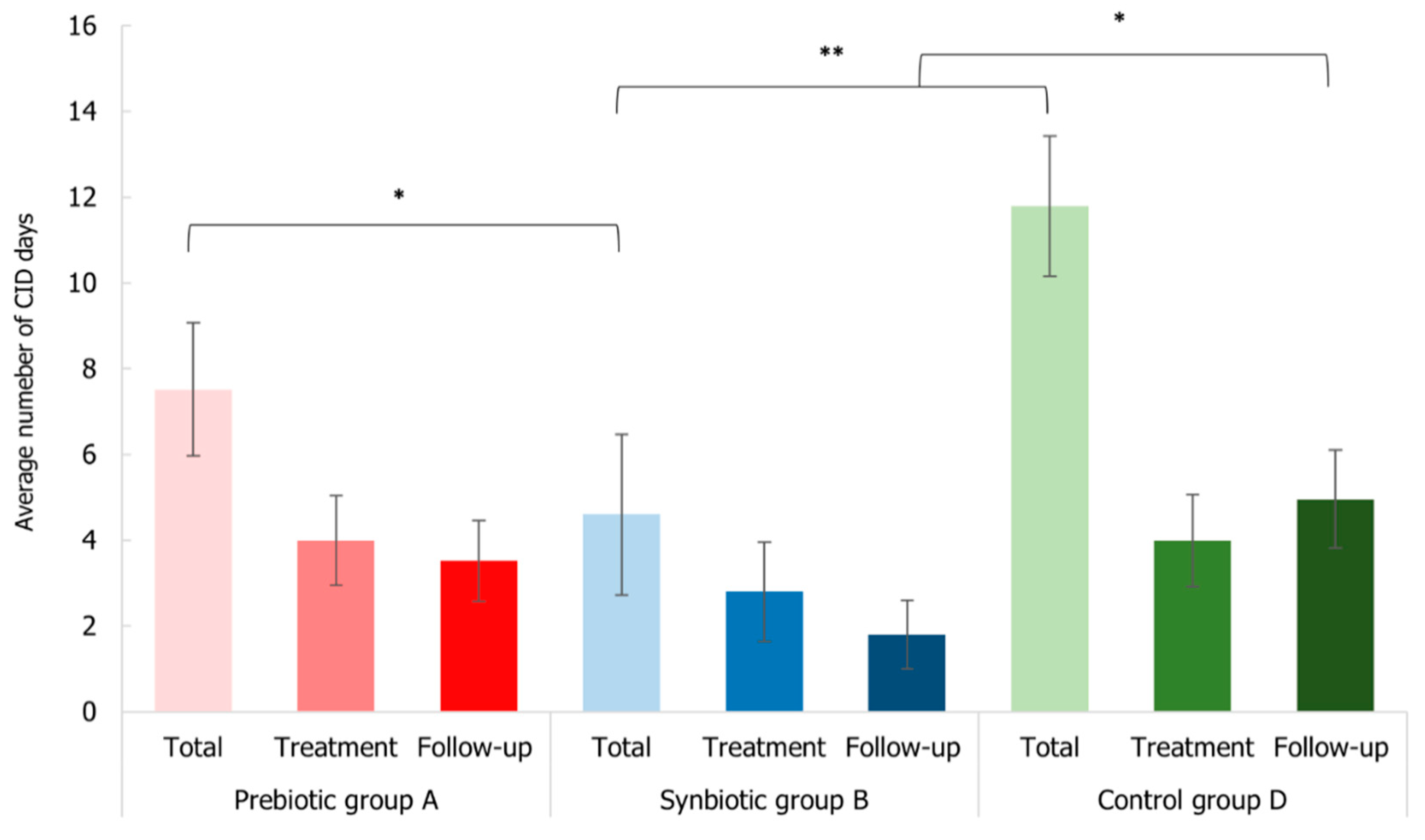
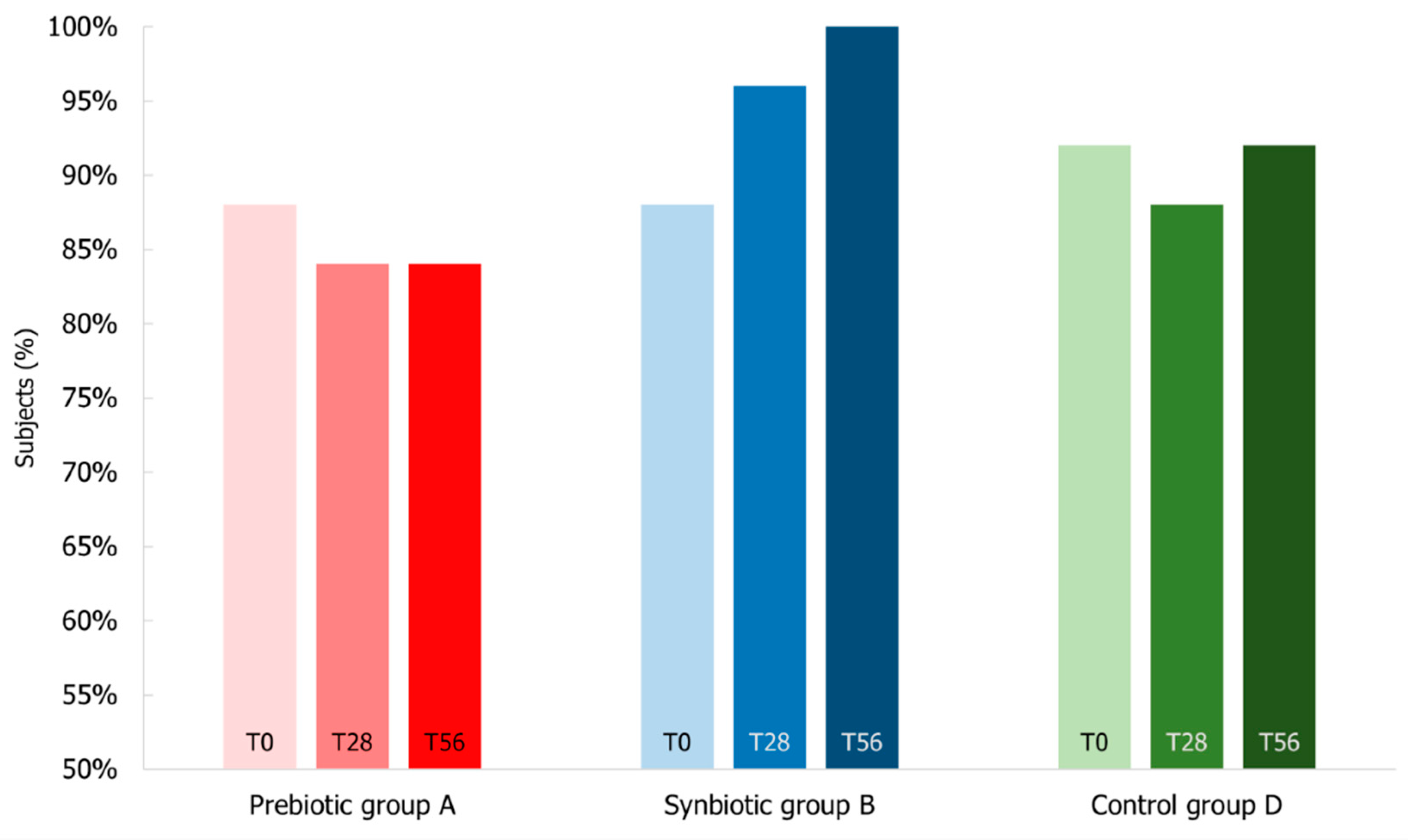
3.6. Impact of Synbiotic Intake on Common Infectious Disease Incidence
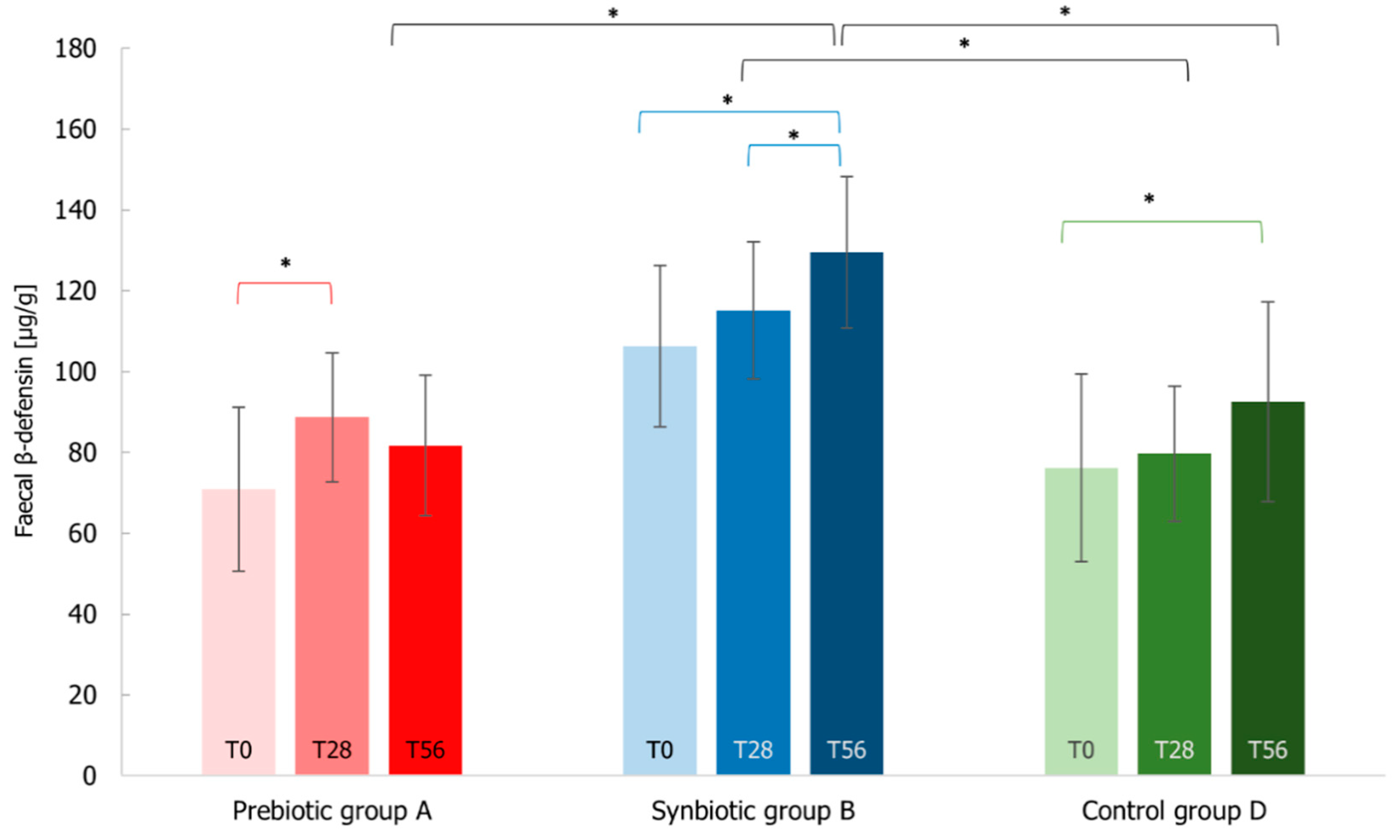
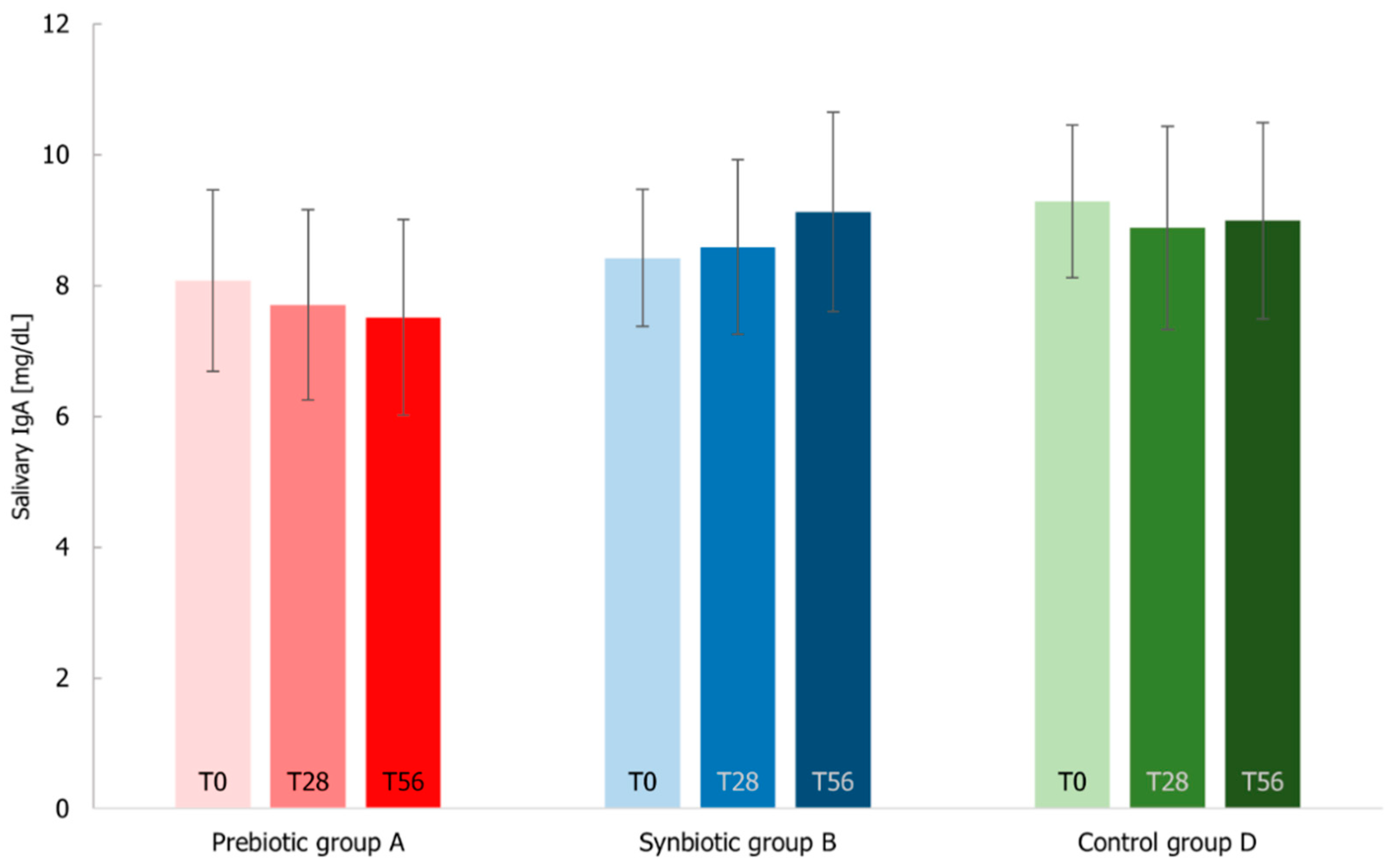
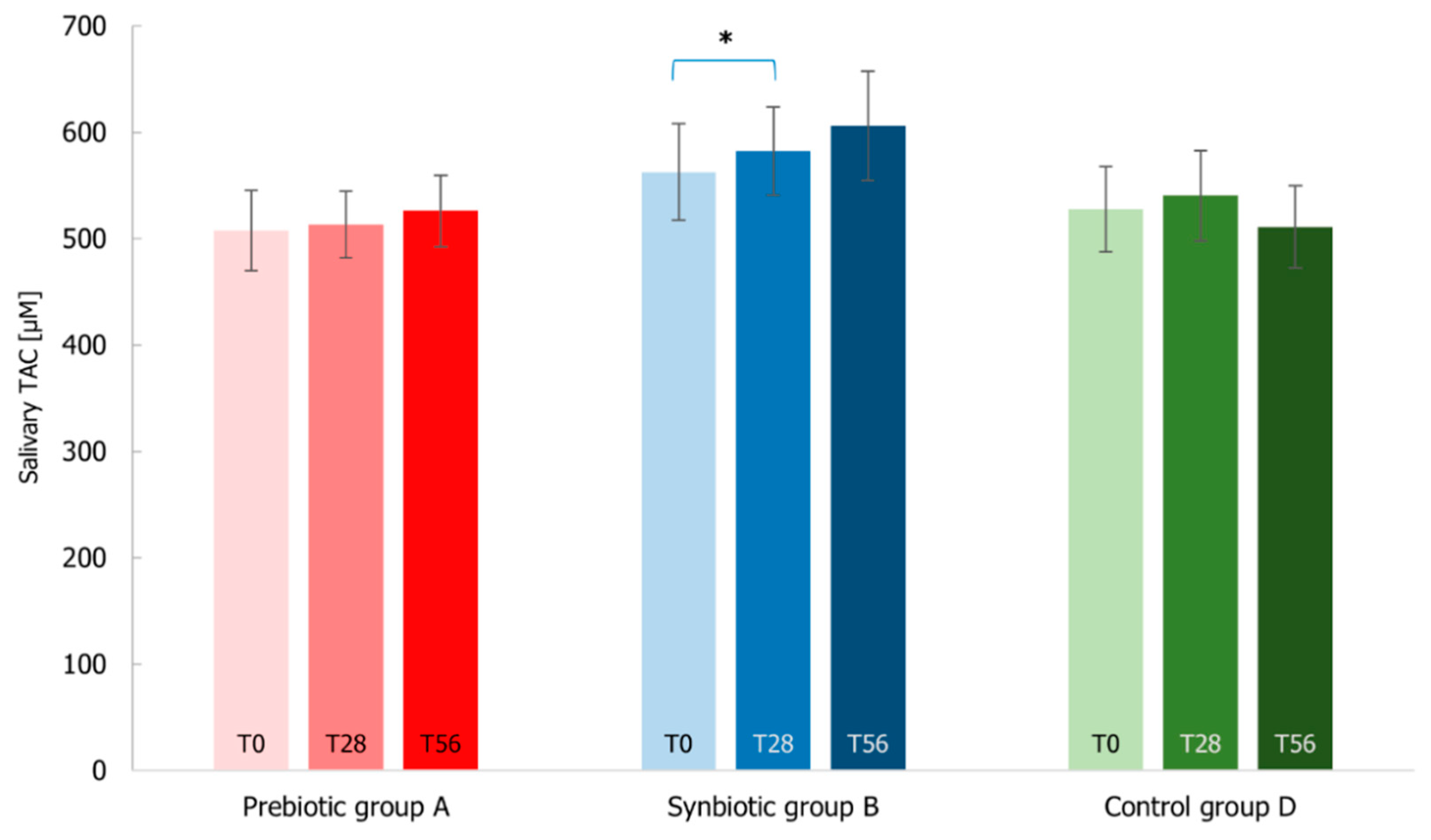
3.7. Immunological Markers Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soenen, S.; Rayner, C.K.; Jones, K.L.; Horowitz, M. The ageing gastrointestinal tract. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2018, 16, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Stower, H. Gut–brain communication. Nat. Med. 2019, 25, 1799. [Google Scholar] [CrossRef]
- Tandon, D.; Haque, M.M.; Gote, M.; Jain, M.; Bhaduri, A.; Dubey, A.K.; Mande, S.S. A prospective randomized, double-blind, placebo-controlled, dose-response relationship study to investigate efficacy of fructo-oligosaccharides (FOS) on human gut microflora. Sci. Rep. 2019, 9, 5473. [Google Scholar] [CrossRef] [Green Version]
- Bartosch, S.; Woodmansey, E.J.; Paterson, J.C.M.; McMurdo, M.E.T.; Macfarlane, G.T. Microbiological Effects of Consuming a Synbiotic Containing Bifidobacterium bifidum, Bifidobacterium lactis, and Oligofructose in Elderly Persons, Determined by Real-Time Polymerase Chain Reaction and Counting of Viable Bacteria. Clin. Infect. Dis. 2005, 40, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Wu, S.; Zeng, Z.; Fu, Z. Effects of environmental pollutants on gut microbiota. Environ. Pollut. 2017, 222, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Woodmansey, E.J. Intestinal bacteria and ageing. J. Appl. Microbiol. 2007, 102, 1178–1186. [Google Scholar] [CrossRef]
- United Nations. World Population Ageing 2015. Department of Economic and Social Affairs, Population Division. (ST/ESA/SER.A/390) 2015. Available online: https://www.un.org/en/development/desa/population/publications/pdf/ageing/WPA2015_Report.pdf (accessed on 5 March 2020).
- Finlay, B.B.; Pettersson, S.; Melby, M.; Bosch, T.C.G. The Microbiome Mediates Environmental Effects on Aging. BioEssays 2019, 41, e1800257. [Google Scholar] [CrossRef]
- Clements, S.J.; Carding, S. Diet, the intestinal microbiota, and immune health in aging. Crit. Rev. Food Sci. Nutr. 2017, 58, 651–661. [Google Scholar] [CrossRef] [Green Version]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimaraes, V.D.; Sokol, H.; Dore, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Salazar, N.; Valdés-Varela, L.; González, S.; Gueimonde, M.; de Los Reyes-Gavilán, C.G. Nutrition and the gut microbiome in the elderly. Gut Microbes 2016, 8, 82–97. [Google Scholar] [CrossRef] [Green Version]
- Kotani, Y.; Shinkai, S.; Okamatsu, H.; Toba, M.; Ogawa, K.; Yoshida, H.; Fukaya, T.; Fujiwara, Y.; Chaves, P.H.; Kakumoto, K.; et al. Oral intake of Lactobacillus pentosus strain b240 accelerates salivary immunoglobulin A secretion in the elderly: A randomized, placebo-controlled, double-blind trial. Immun. Ageing 2010, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guigoz, Y.; Doré, J.; Schiffrin, E.J. The inflammatory status of old age can be nurtured from the intestinal environment. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 13–20. [Google Scholar] [CrossRef]
- Kolibab, K.; Smithson, S.L.; Shriner, A.K.; Khuder, S.; Romero-Steiner, S.; Carlone, G.M.; Westerink, M.J. Immune response to pneumococcal polysaccharides 4 and 14 in elderly and young adults. I Antibody concentrations, avidity and functional activity. Immun. Ageing 2005, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Maciejczyk, M.; Zalewska, A.; Ładny, J.R. Salivary Antioxidant Barrier, Redox Status, and Oxidative Damage to Proteins and Lipids in Healthy Children, Adults, and the Elderly. Oxidative Med. Cell. Longev. 2019, 2019, 4393460. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Fukuda, S.; Fujiwara, A.; Suda, W.; Hattori, M.; Kikuchi, J.; Ohno, H. Multiple Omics Uncovers Host–Gut Microbial Mutualism During Prebiotic Fructooligosaccharide Supplementation. DNA Res. 2014, 21, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Ouwehand, A.C.; Tiihonen, K.; Saarinen, M.; Putaala, H.; Rautonen, N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: Intestinal and immune parameters. Br. J. Nutr. 2008, 101, 367–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlane, S.; Cleary, S.; Bahrami, B.; Reynolds, N.; Macfarlane, G.T. Synbiotic consumption changes the metabolism and composition of the gut microbiota in older people and modifies inflammatory processes: A randomised, double-blind, placebo-controlled crossover study. Aliment. Pharmacol. Ther. 2013, 38, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.S.; Kong, C.; Gao, R.Y.; Yan, X.; Yu, H.J.; Wen, B.; Zhu, Q.; Shen, T.Y.; Sun, Z.L.; Qin, H.L. Analysis of fecal microbiota in patients with functional constipation undergoing treatment with synbiotics. Eur. J. Clin. Microbiol. 2018, 37, 555–563. [Google Scholar] [CrossRef]
- NCI Drug Dictionary. Synbiotic Supplement. NCI Thesaurus. 2020. Available online: https://ncit.nci.nih.gov/ncitbrowser/ConceptReport.jsp?dictionary=NCI_Thesaurus&ns=ncit&code=C99230 (accessed on 5 March 2020).
- Krumbeck, J.A.; Rasmussen, H.E.; Hutkins, R.W.; Clarke, J.; Shawron, K.; Keshavarzian, A.; Walter, J. Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics. Microbiome 2018, 6, 121. [Google Scholar] [CrossRef]
- Krumbeck, J.A.; Walter, J.; Hutkins, R.W. Synbiotics for Improved Human Health: Recent Developments, Challenges, and Opportunities. Annu. Rev. Food Sci. Technol. 2018, 9, 451–479. [Google Scholar] [CrossRef]
- World Health Organization. Guidelines for Good Clinical Practice (GCP) for Trials on Pharmaceutical Products. 1995. Available online: https://apps.who.int/medicinedocs/pdf/whozip13e/whozip13e.pdf (accessed on 5 March 2020).
- Presti, I.; D’Orazio, G.; Labra, M.; La Ferla, B.; Mezzasalma, V.; Bizzaro, G.; Giardina, S.; Michelotti, A.; Tursi, F.; Vassallo, M.; et al. Evaluation of the probiotic properties of new Lactobacillus and Bifidobacterium strains and their in vitro effect. Appl. Microbiol. Biotechnol. 2015, 99, 5613–5626. [Google Scholar] [CrossRef]
- De Giani, A.; Bovio, F.; Forcella, M.; Fusi, P.; Sello, G.; Di Gennaro, P. Identification of a bacteriocin-like compound from Lactobacillus plantarum with antimicrobial activity and effects on normal and cancerogenic human intestinal cells. AMB Express 2019, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Mezzasalma, V.; Manfrini, E.; Ferri, E.; Sandionigi, A.; La Ferla, B.; Schiano, I.; Michelotti, A.; Nobile, V.; Labra, M.; Di Gennaro, P. A Randomized, Double-Blind, Placebo-Controlled Trial: The Efficacy of Multispecies Probiotic Supplementation in Alleviating Symptoms of Irritable Bowel Syndrome Associated with Constipation. BioMed Res. Int. 2016, 2016, 4740907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefevre, M.; Racedo, S.M.; Ripert, G.; Housez, B.; Cazaubiel, M.; Maudet, C.; Jüsten, P.; Marteau, P.; Urdaci, M.C. Probiotic strain Bacillus subtilis CU1 stimulates immune system of elderly during common infectious disease period: A randomized, double-blind placebo-controlled study. Immun. Ageing 2015, 12, 12–24. [Google Scholar] [CrossRef] [Green Version]
- Klingberg, E.; Magnusson, M.K.; Strid, H.; Deminger, A.; Ståhl, A.; Sundin, J.; Simrén, M.; Carlsten, H.; Öhman, L.; Forsblad-D’Elia, H. A distinct gut microbiota composition in patients with ankylosing spondylitis is associated with increased levels of fecal calprotectin. Arthritis Res. Ther. 2019, 21, 248. [Google Scholar] [CrossRef] [Green Version]
- Schwiertz, A.; Huber, H.; Rusch, K. Human beta-defensin-2 levels in healthy individuals. Am. J. Gastroenterol. 2009, 104, 2110–2111. [Google Scholar] [CrossRef]
- Kabeerdoss, J.; Devi, R.S.; Mary, R.R.; Prabhavathi, D.; Vidya, R.; Mechenro, J.; Mahendri, N.V.; Pugazhendhi, S.; Ramakrishna, B. Effect of yoghurt containing Bifidobacterium lactis Bb12®on faecal excretion of secretory immunoglobulin A and human beta-defensin 2 in healthy adult volunteers. Nutr. J. 2011, 10, 138. [Google Scholar] [CrossRef] [Green Version]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Karlík, M.; Valkovič, P.; Hančinová, V.; Krížová, L.; Tóthová, L.; Celec, P. Markers of oxidative stress in plasma and saliva in patients with multiple sclerosis. Clin. Biochem. 2015, 48, 24–28. [Google Scholar] [CrossRef]
- Kim, W.S.; Elmogy, M.; Earle, M.; Haj-Ahmad, Z.; Bak, R.; Haj-Ahmad, Y. Application Note 91 Stool DNA Preservation; APP91-v3; Norgen Biotek Corp.: Thorold, ON, Canada, 2019. [Google Scholar]
- Panek, M.; Paljetak, H.; Barešić, A.; Perić, M.; Matijašić, M.; Lojkić, I.; Bender, D.V.; Krznarić, Z.; Verbanac, D. Methodology challenges in studying human gut microbiota—Effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018, 8, 5143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manfredi, E. Biological Sciences. Ph.D. Thesis, University of Milano-Bicocca, Milan, Italy, 2015. [Google Scholar]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. The PD Phylogenetic diversity framework: Linking evolutionary history to feature diversity for biodiversity conservation. In Biodiversity Conservation and Phylogenetic Systematics; Topics in Biodiversity and Conservation; Pellens, R., Grandcolas, P., Eds.; Springer: Cham, Switzerland, 2016; pp. 39–56. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac–An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef]
- Zhang, Y.; Hedo, R.; Rivera, A.; Rull, R.; Richardson, S.; Tu, X.M. Post hoc power analysis: Is it an informative and meaningful analysis? Gen. Psychiatry 2019, 32, e100069. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, S.; Cuthill, I.C. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. 2007, 82, 591–605. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van De Wiele, T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Sadeghi, M.; Karam, G.A.; Vazirinejad, R. Salivary IgA and IgE levels in healthy subjects: Relation to age and gender. Braz. Oral Res. 2010, 24, 21–27. [Google Scholar] [CrossRef]
- Costabile, A.; Bergillos-Meca, T.; Rasinkangas, P.; Korpela, K.; De Vos, W.M.; Gibson, G.R. Effects of Soluble Corn Fiber Alone or in Synbiotic Combination with Lactobacillus rhamnosus GG and the Pilus-Deficient Derivative GG-PB12 on Fecal Microbiota, Metabolism, and Markers of Immune Function: A Randomized, Double-Blind, Placebo-Controlled, Crossover Study in Healthy Elderly (Saimes Study). Front. Immunol. 2017, 8, 1443. [Google Scholar] [CrossRef]
- Tran, T.; Cousin, F.; Lynch, D.B.; Menon, R.; Brulc, J.; Brown, J.R.-M.; O’Herlihy, E.; Butto, L.F.; Power, K.; Jeffery, I.; et al. Prebiotic supplementation in frail older people affects specific gut microbiota taxa but not global diversity. Microbiome 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.-C.; Young, J.; Lin, C.-S.; Chang, C.-J.; Lu, C.-C.; Martel, J.; Ojcius, D.; Ko, Y.-F. Impact of the gut microbiota, prebiotics, and probiotics on human health and disease. Biomed. J. 2014, 37, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Zhang, X.; Huang, L.; Shen, R.; Qin, H. Gut Microbiota Alteration After Long-Term Consumption of Probiotics in the Elderly. Probiotics Antimicrob. Proteins 2018, 11, 655–666. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Xu, J.; Xue, Z.; Zhang, M.; Pang, X.; Zhang, X.; Zhao, L. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 2015, 5, 14405. [Google Scholar] [CrossRef] [Green Version]
- Bron, P.A.; Kleerebezem, M.; Brummer, R.-J.; Cani, P.D.; Mercenier, A.; Macdonald, T.T.; Garcia-Ródenas, C.L.; Wells, J.M. Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 2017, 117, 93–107. [Google Scholar] [CrossRef]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2015, 65, 415–425. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derrien, M.; Belzer, C.; de Vos, W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017, 106, 171–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Huang, Y.; Lu, L.; Yang, W.; Huang, T.; Lin, Z.; Lin, C.; Kwan, H.Y.; Wong, H.L.X.; Chen, Y.; et al. Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats. Microbiome 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Mujagic, Z.; Jonkers, D.M.A.E.; Ludidi, S.; Keszthelyi, D.; Hesselink, M.A.; Weerts, Z.Z.R.M.; Kievit, R.N.; Althof, J.F.; Leue, C.; Kruimel, J.W.; et al. Biomarkers for visceral hypersensitivity in patients with irritable bowel syndrome. Neurogastroenterol. Motil. 2017, 29, e13137. [Google Scholar] [CrossRef] [PubMed]
- Möndel, M.; Schroeder, B.; Zimmermann, K.; Huber, H.; Nuding, S.; Beisner, J.; Fellermann, K.; Stange, E.F.; Wehkamp, J. Probiotic E. coli treatment mediates antimicrobial human β-defensin synthesis and fecal excretion in humans. Mucosal Immunol. 2008, 2, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Ghadimi, D.; Hassan, M.; Njeru, P.N.; De Vrese, M.; Geis, A.; Shalabi, S.I.; Abdel-Razek, S.T.; Abdel-Khair, A.E.-A.A.; Heller, K.J.; Schrezenmeir, J. Suppression subtractive hybridization identifies bacterial genomic regions that are possibly involved in hBD-2 regulation by enterocytes. Mol. Nutr. Food Res. 2011, 55, 1533–1542. [Google Scholar] [CrossRef]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef]
| A Group | B Group | D Group | |
|---|---|---|---|
| N° of subjects (female/male) | 25 (20/5) | 25 (20/5) | 25 (17/8) |
| Age (years) | 68.76 ± 4.95 | 70.08 ± 4.61 | 69.76 ± 5.20 |
| BMI (kg/m2) | 22.77 ± 1.68 | 22.74 ± 1.38 | 23.25 ± 1.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Giani, A.; Sandionigi, A.; Zampolli, J.; Michelotti, A.; Tursi, F.; Labra, M.; Di Gennaro, P. Effects of Inulin-Based Prebiotics Alone or in Combination with Probiotics on Human Gut Microbiota and Markers of Immune System: A Randomized, Double-Blind, Placebo-Controlled Study in Healthy Subjects. Microorganisms 2022, 10, 1256. https://doi.org/10.3390/microorganisms10061256
De Giani A, Sandionigi A, Zampolli J, Michelotti A, Tursi F, Labra M, Di Gennaro P. Effects of Inulin-Based Prebiotics Alone or in Combination with Probiotics on Human Gut Microbiota and Markers of Immune System: A Randomized, Double-Blind, Placebo-Controlled Study in Healthy Subjects. Microorganisms. 2022; 10(6):1256. https://doi.org/10.3390/microorganisms10061256
Chicago/Turabian StyleDe Giani, Alessandra, Anna Sandionigi, Jessica Zampolli, Angela Michelotti, Francesco Tursi, Massimo Labra, and Patrizia Di Gennaro. 2022. "Effects of Inulin-Based Prebiotics Alone or in Combination with Probiotics on Human Gut Microbiota and Markers of Immune System: A Randomized, Double-Blind, Placebo-Controlled Study in Healthy Subjects" Microorganisms 10, no. 6: 1256. https://doi.org/10.3390/microorganisms10061256
APA StyleDe Giani, A., Sandionigi, A., Zampolli, J., Michelotti, A., Tursi, F., Labra, M., & Di Gennaro, P. (2022). Effects of Inulin-Based Prebiotics Alone or in Combination with Probiotics on Human Gut Microbiota and Markers of Immune System: A Randomized, Double-Blind, Placebo-Controlled Study in Healthy Subjects. Microorganisms, 10(6), 1256. https://doi.org/10.3390/microorganisms10061256







