Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Water Source Description
2.2. Plant Materials, Experimental Design, and Agronomic Practices
2.3. Measurements
2.3.1. Physio-Biochemical Parameters
2.3.2. Agronomical Parameters
2.4. Statistical Analysis
3. Results
3.1. Grouping Genotypes Based in Their Salt Tolerance Level
3.2. Physio-Biochemical Attributes
3.3. Agronomical Attributes
3.4. Additive Main Effect and Multiplicative Interaction Model (AMMI)
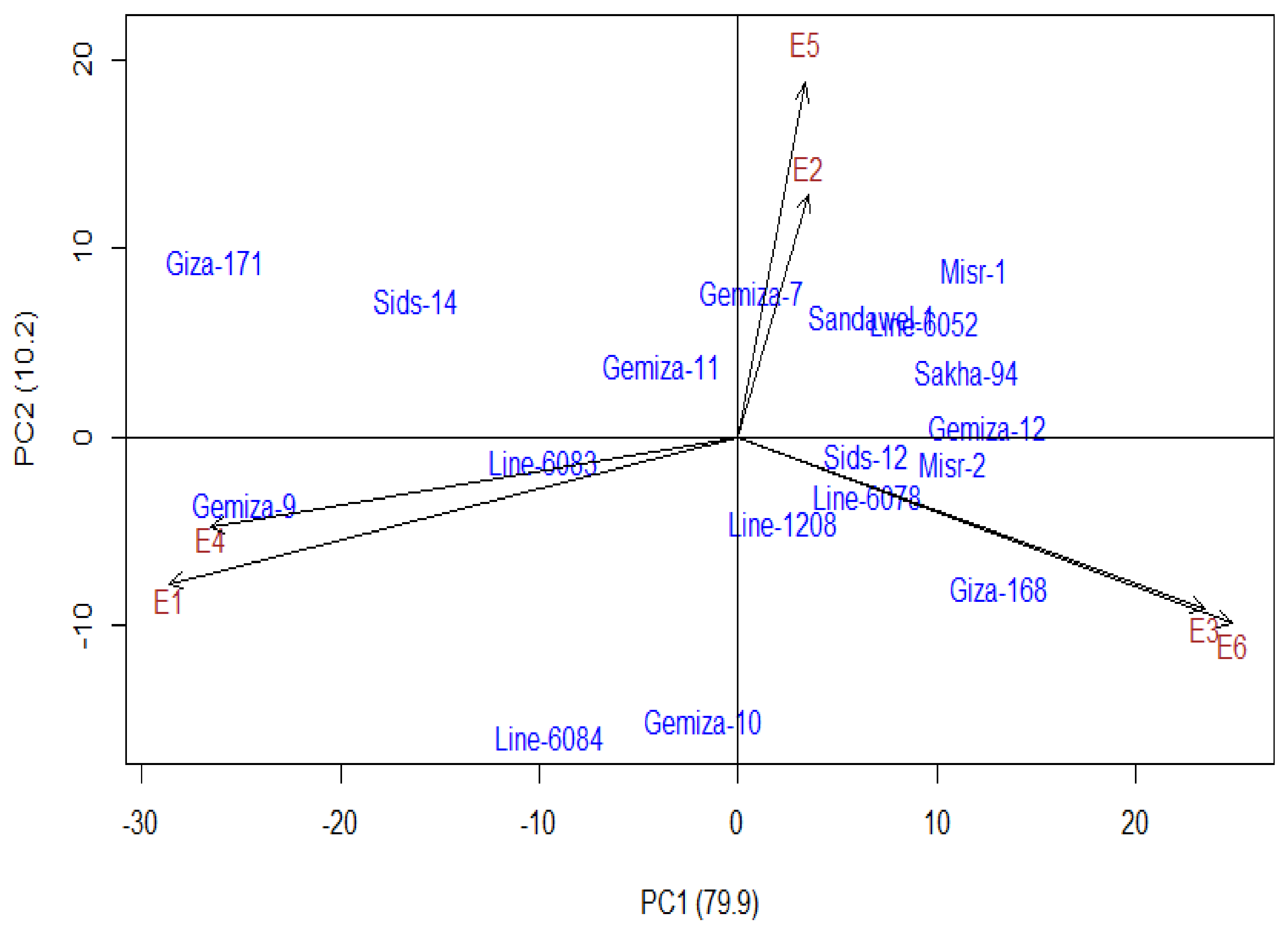
3.5. Inter-Relationship Between All Measured Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Yadava, R.; Gollen, B.; Kumar, S.; Verma, R.K.; Yadav, S. Nutritional contents and medicinal properties of wheat: A review. Life Sci. Med. Res. 2011, 22, 1–10. [Google Scholar]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Sec. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Database. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 21 June 2020).
- Newell, N. Effects of soil salinity on plant growth. Plant Physiol. 2013, 1, 1–4. [Google Scholar]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef] [Green Version]
- Allel, D.; Ben-Amar, A.; Abdelly, C. Leaf photosynthesis, chlorophyll fluorescence and ion content of barley (Hordeum vulgare) in response to salinity. J. Plant Nutr. 2018, 41, 497–508. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Kotb, T.H.; Watanabe, T.; Ogino, Y.; Tanji, K.K. Soil salinization in the Nile Delta and related policy issues in Egypt. Agric. Water Manag. 2000, 43, 239–261. [Google Scholar] [CrossRef]
- Chen, J.; Mueller, V. Coastal climate change, soil salinity and human migration in Bangladesh. Nat. Clim. Chang. 2018, 8, 981–985. [Google Scholar] [CrossRef]
- Baker, N.R.; Rosenqvist, E. Applications of chlorophyll fluorescence can improve crop production strategies: An examination of future possibilities. J. Exp. Bot. 2004, 55, 1607–1621. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Qiu, L.; Xu, L.; Ye, L.; Chen, M.; Sun, D.; Chen, Z.; Zhang, H.; Jin, X.; Dai, F. Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS ONE 2011, 6, e22938. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.H.; Murata, N. Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 2011, 34, 1–20. [Google Scholar] [CrossRef] [PubMed]
- El-Hendawy, S.E.; Hassan, W.M.; Al-Suhaibani, N.A.; Refay, Y.; Abdella, K.A. Comparative performance of multivariable agro-physiological parameters for detecting salt tolerance of wheat cultivars under simulated saline field growing conditions. Front. Plant Sci. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desoky, E.-S.M.; Elrys, A.S.; Rady, M.M. Integrative moringa and licorice extracts application improves Capsicum annuum fruit yield and declines its contaminant contents on a heavy metals-contaminated saline soil. Ecotoxicol. Environ. Saf. 2019, 169, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Rady, M.; Desoky, E.-S.; Elrys, A.; Boghdady, M. Can licorice root extract be used as an effective natural biostimulant for salt-stressed common bean plants? S. Afr. J. Bot. 2019, 121, 294–305. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H.; Noctor, G. Oxygen processing in photosynthesis: Regulation and signalling. New Phytol. 2000, 146, 359–388. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Hoque, M.A.; Banu, M.N.A.; Okuma, E.; Amako, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Exogenous proline and glycinebetaine increase NaCl-induced ascorbate–glutathione cycle enzyme activities, and proline improves salt tolerance more than glycinebetaine in tobacco Bright Yellow-2 suspension-cultured cells. J. Plant Physiol. 2007, 164, 1457–1468. [Google Scholar] [CrossRef]
- Sudhir, P.; Murthy, S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Kahrizi, S.; Sedighi, M.; Sofalian, O. Effect of salt stress on proline and activity of antioxidant enzymes in ten durum wheat cultivars. Ann. Biol. Res. 2012, 3, 3870–3874. [Google Scholar]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Rady, M.M. Effect of 24-epibrassinolide on growth, yield, antioxidant system and cadmium content of bean (Phaseolus vulgaris L.) plants under salinity and cadmium stress. Sci. Hortic. 2011, 129, 232–237. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhao, X. Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS ONE 2014, 9, e112807. [Google Scholar] [CrossRef] [Green Version]
- Rios, J.J.; Martínez-Ballesta, M.C.; Ruiz, J.M.; Blasco, B.; Carvajal, M. Silicon-mediated improvement in plant salinity tolerance: The role of aquaporins. Front. Plant Sci. 2017, 8, 948. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+ permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2015; pp. 77–153. [Google Scholar]
- Schutzendubel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef]
- Desoky, E.-S.M.; El Sayed, A.I.; Merwad, A.-R.M.; Rady, M.M. Stimulating antioxidant defenses, antioxidant gene expression, and salt tolerance in Pisum sativum seedling by pretreatment using licorice root extract (LRE) as an organic biostimulant. Plant Physiol. Biochem. 2019, 142, 292–302. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; de Almeida Viégas, R.; da Rocha, I.M.A.; Moreira, A.C.d.O.M.; de Azevedo Moreira, R.; Oliveira, J.T.A. Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J. Plant Physiol. 2003, 160, 115–123. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotech. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadshani, S.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Multi-dimensional evaluation of response to salt stress in wheat. PLoS ONE 2019, 14, e0222659. [Google Scholar] [CrossRef]
- El Hendawy, S.; Ruan, Y.; Hu, Y.; Schmidhalter, U. A comparison of screening criteria for salt tolerance in wheat under field and controlled environmental conditions. J. Agron. Crop Sci. 2009, 195, 356–367. [Google Scholar] [CrossRef]
- Hasan, A.; Hafiz, H.R.; Siddiqui, N.; Khatun, M.; Islam, R.; Mamun, A.-A. Evaluation of wheat genotypes for salt tolerance based on some physiological traits. J. Crop Sci. Biotechnol. 2015, 18, 333–340. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.; Shen, J.; Baum, M.; Ogbonnaya, F.; Léon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Ellis, R.; Forster, B.; Gordon, D.; Handley, L.; Keith, R.; Lawrence, P.; Meyer, R.; Powell, W.; Robinson, D.; Scrimgeour, C. Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J. Exp. Bot. 2002, 53, 1163–1176. [Google Scholar] [CrossRef] [Green Version]
- Bağci, S.A.; Ekiz, H.; Yilmaz, A. Salt tolerance of sixteen wheat genotypes during seedling growth. Turk. J. Agric. For. 2007, 31, 363–372. [Google Scholar]
- Khan, M.; Yasmin, S.; Ansari, R.; Shirazi, M.; Ashraf, M. Screening for salt tolerance in wheat genotypes at an early seedling stage. Pak. J. Bot. 2007, 39, 2501–2509. [Google Scholar]
- El-Hendawy, S.; Hu, Y.; Sakagami, J.-I.; Schmidhalter, U. Screening Egyptian wheat genotypes for salt tolerance at early growth stages. Int. J. Plant Prod. 2011, 5, 283–298. [Google Scholar]
- Muhammad, Z.; Hussain, F. Effect of NaCl salinity on the germination and seedling growth of seven wheat genotypes. Pak. J. Bot. 2012, 44, 1845–1850. [Google Scholar]
- Chahine, K.; Sourour, A.; Youssef, T.; Hajer, S.-A. Salinity effect on plant growth at the seedling stage of durum wheat (Triticum durum Desf.). J. Plant Breed. Crop Sci. 2013, 5, 20–25. [Google Scholar] [CrossRef]
- Hussain, B.; Khan, A.S.; Ali, Z. Genetic variation in wheat germplasm for salinity tolerance atseedling stage: Improved statistical inference. Turk. J. Agric. For. 2015, 39, 182–192. [Google Scholar] [CrossRef]
- Jovović, M.; Tunguz, V.; Mirosavljević, M.; Pržulj, N. Effect of salinity and drought stress on germination and early seedlings growth of bread wheat (Triticum aestivum L.). Genetika 2018, 50, 285–298. [Google Scholar] [CrossRef]
- Guellim, A.; Catterou, M.; Chabrerie, O.; Tetu, T.; Hirel, B.; Dubois, F.; Ben Ahmed, H.; Kichey, T. Identification of phenotypic and physiological markers of salt stress tolerance in durum wheat (Triticum durum Desf.) through integrated server analyses. Agronomy 2019, 9, 844. [Google Scholar] [CrossRef] [Green Version]
- Al-Ashkar, I.; Alderfasi, A.; Ben Romdhane, W.; Seleiman, M.F.; El-Said, R.A.; Al-Doss, A. Morphological and genetic diversity within salt tolerance detection in eighteen wheat genotypes. Plants 2020, 9, 287. [Google Scholar] [CrossRef] [Green Version]
- Igartua, E.; Gracia, M.; Lasa, J. Field responses of grain sorghum to a salinity gradient. Field Crops Res. 1995, 42, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Allel, D.; BenAmar, A.; Badri, M.; Abdelly, C. Evaluation of salinity tolerance indices in North African barley accessions at reproductive stage. Czech J. Genet. Plant Breed. 2019, 55, 61–69. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Mansour, E.; Moustafa, E.S.; Qabil, N.; Abdelsalam, A.; Wafa, H.A.; El Kenawy, A.; Casas, A.M.; Igartua, E. Assessing different barley growth habits under Egyptian conditions for enhancing resilience to climate change. Field Crops Res. 2018, 224, 67–75. [Google Scholar] [CrossRef]
- Mansour, E.; Abdul-Hamid, M.I.; Yasin, M.T.; Qabil, N.; Attia, A. Identifying drought-tolerant genotypes of barley and their responses to various irrigation levels in a Mediterranean environment. Agric. Water Manag. 2017, 194, 58–67. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weatherley, P. Studies in the water relations of the cotton plant. I. The field measurement of water deficits in leaves. New Phytol. 1950, 87, 81–97. [Google Scholar] [CrossRef]
- Premachandra, G.; Saneoka, H.; Ogata, S. Cell membrane stability, an indicator of drought tolerance as affected by applied nitrogen in soybean. J. Agric. Sci. 1990, 115, 63–66. [Google Scholar] [CrossRef]
- Sullivan, C.Y. Selection for Drought and Heat Tolerance in Grain Sorghum; John Wiley & Sons: New York, NY, USA, 1979; pp. 263–281. [Google Scholar]
- Zhang, Z.L.; Qu, W. Experimental Guidance of Plant Physiology; High Education: Beijing, China, 2004. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Irigoyen, J.; Einerich, D.; Sánchez-Díaz, M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol. Plant. 1992, 84, 55–60. [Google Scholar] [CrossRef]
- Mukherjee, S.; Choudhuri, M. Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 1983, 58, 166–170. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Aebi, H. Catalase In Vitro; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Maechlay, A.; Chance, B. The Assay of Catalase and Peroxidase; John Wiley & Sons, Inc.: New York, NY, USA, 1954; pp. 357–424. [Google Scholar]
- Klapheck, S.; Zimmer, I.; Cosse, H. Scavenging of hydrogen peroxide in the endosperm of Ricinus communis by ascorbate peroxidase. Plant Cell Physiol. 1990, 31, 1005–1013. [Google Scholar]
- Wolf, B. A comprehensive system of leaf analyses and its use for diagnosing crop nutrient status. Commun. Soil Sci. Plant Anal. 1982, 13, 1035–1059. [Google Scholar] [CrossRef]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.; Ricciardi, G.; Borghi, B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Gauch, H.G., Jr. Statistical analysis of yield trials by AMMI and GGE. Crop Sci. 2006, 46, 1488–1500. [Google Scholar] [CrossRef]
- Aragüés, R.; Urdanoz, V.; Çetin, M.; Kirda, C.; Daghari, H.; Ltifi, W.; Lahlou, M.; Douaik, A. Soil salinity related to physical soil characteristics and irrigation management in four Mediterranean irrigation districts. Agric. Water Manag. 2011, 98, 959–966. [Google Scholar] [CrossRef] [Green Version]
- El-Hendawy, S.E.; Hu, Y.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Eur. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Genc, Y.; Tester, M.; McDonald, G. Calcium requirement of wheat in saline and non-saline conditions. Plant Soil 2010, 327, 331–345. [Google Scholar] [CrossRef]
- Tavakkoli, E.; Rengasamy, P.; McDonald, G.K. The response of barley to salinity stress differs between hydroponic and soil systems. Funct. Plant Biol. 2010, 37, 621–633. [Google Scholar] [CrossRef]
- Houshmand, S.; Arzani, A.; Maibody, S.A.M.; Feizi, M. Evaluation of salt-tolerant genotypes of durum wheat derived from in vitro and field experiments. Field Crops Res. 2005, 91, 345–354. [Google Scholar] [CrossRef]
- Shafi, M.; Bakhat, J.; Khan, M.J.; Khan, M.A.; Anwar, S. Effect of salinity on yield and ion accumulation of wheat genotypes. Pak. J. Bot. 2010, 42, 4113–4121. [Google Scholar]
- Sheng, M.; Tang, M.; Chen, H.; Yang, B.; Zhang, F.; Huang, Y. Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 2008, 18, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Srinieng, K.; Saisavoey, T.; Karnchanatat, A. Effect of salinity stress on antioxidative enzyme activities in tomato cultured in vitro. Pak. J. Bot. 2015, 47, 1–10. [Google Scholar]
- Rezende, R.A.L.S.; Rodrigues, F.A.; Soares, J.D.R.; Silveira, H.R.d.O.; Pasqual, M.; Dias, G.d.M.G. Salt stress and exogenous silicon influence physiological and anatomical features of in vitro-grown cape gooseberry. Cienc. Rural 2018, 48, e20170176. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Kao, C.H. Cell wall peroxidase activity, hydrogen peroxide level and NaCl-inhibited root growth of rice seedlings. Plant Soil 2001, 230, 135–143. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Hong, C.Y.; Liu, L.F.; Kao, C.H. Relative importance of Na+ and Cl– in NaCl-induced antioxidant systems in roots of rice seedlings. Physiol. Plant. 2004, 122, 86–94. [Google Scholar] [CrossRef]
- Zhao, H.; Liang, H.; Chu, Y.; Sun, C.; Wei, N.; Yang, M.; Zheng, C. Effects of salt stress on chlorophyll fluorescence and the antioxidant system in Ginkgo biloba L. seedlings. HortScience 2019, 54, 2125–2133. [Google Scholar] [CrossRef] [Green Version]
- Stępień, P.; Kłbus, G. Water relations and photosynthesis in Cucumis sativus L. leaves under salt stress. Biol. Plant. 2006, 50, 610–616. [Google Scholar] [CrossRef]
- Trapp, S.; Feificova, D.; Rasmussen, N.F.; Bauer-Gottwein, P. Plant uptake of NaCl in relation to enzyme kinetics and toxic effects. Environ. Exp. Bot. 2008, 64, 1–7. [Google Scholar] [CrossRef]
- Saqib, M.; Akhtar, J.; Abbas, G.; Nasim, M. Salinity and drought interaction in wheat (Triticum aestivum L.) is affected by the genotype and plant growth stage. Acta Physiol. Plant. 2013, 35, 2761–2768. [Google Scholar] [CrossRef]
- Valero, E.; Macià, H.; Ildefonso, M.; Hernández, J.-A.; González-Sánchez, M.-I.; García-Carmona, F. Modeling the ascorbate-glutathione cycle in chloroplasts under light/dark conditions. BMC Syst. Biol. 2015, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sairam, R.K.; Rao, K.V.; Srivastava, G. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Rady, M.M.; Kuşvuran, A.; Alharby, H.F.; Alzahrani, Y.; Kuşvuran, S. Pretreatment with proline or an organic bio-stimulant induces salt tolerance in wheat plants by improving antioxidant redox state and enzymatic activities and reducing the oxidative stress. J. Plant Growth Regul. 2019, 38, 449–462. [Google Scholar] [CrossRef]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Alzahrani, Y.; Rady, M.M. Compared to antioxidants and polyamines, the role of maize grain-derived organic biostimulants in improving cadmium tolerance in wheat plants. Ecotoxicol. Environ. Saf. 2019, 182, 109378. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Torabian, S. Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by biochar under salt stress. Ecotoxicol. Environ. Saf. 2017, 137, 64–70. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Howladar, S.M. A novel Moringa oleifera leaf extract can mitigate the stress effects of salinity and cadmium in bean (Phaseolus vulgaris L.) plants. Ecotoxicol. Environ. Saf. 2014, 100, 69–75. [Google Scholar] [CrossRef]
- Semida, W.M.; Rady, M.M. Presoaking application of propolis and maize grain extracts alleviates salinity stress in common bean (Phaseolus vulgaris L.). Sci. Hortic. 2014, 168, 210–217. [Google Scholar] [CrossRef]
- Elrys, A.S.; Abdo, A.I.; Abdel-Hamed, E.M.; Desoky, E.-S.M. Integrative application of licorice root extract or lipoic acid with fulvic acid improves wheat production and defenses under salt stress conditions. Ecotoxicol. Environ. Saf. 2020, 190, 110144. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Chow, W.; Sun, L.; Chen, J.; Chen, Y.; Peng, C. The influence of low temperature on photosynthesis and antioxidant enzymes in sensitive banana and tolerant plantain (Musa sp.) cultivars. Photosynthetica 2011, 49, 201–208. [Google Scholar] [CrossRef]
- Xu, R.; Yamada, M.; Fujiyama, H. Lipid peroxidation and antioxidative enzymes of two turfgrass species under salinity stress. Pedosphere 2013, 23, 213–222. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Insights into the significance of antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Exogenous silicon attenuates cadmium-induced oxidative stress in Brassica napus L. by modulating AsA-GSH pathway and glyoxalase system. Front. Plant Sci. 2017, 8, 1061. [Google Scholar] [CrossRef]
- Zhou, Y.; Wen, Z.; Zhang, J.; Chen, X.; Cui, J.; Xu, W.; Liu, H.-Y. Exogenous glutathione alleviates salt-induced oxidative stress in tomato seedlings by regulating glutathione metabolism, redox status, and the antioxidant system. Sci. Hortic. 2017, 220, 90–101. [Google Scholar] [CrossRef]
- Hernández, J.A.; Almansa, M.S. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol. Plant. 2002, 115, 251–257. [Google Scholar] [CrossRef]
- Yadavi, A.; Aboueshaghi, R.; Dehnavi, M.; Balouchi, H. Effect of micronutrients foliar application on grain qualitative characteristics and some physiological traits of bean (Phaseolus vulgaris L.) under drought stress. Indian J. Fundam. Appl. Life Sci. 2014, 4, 124–131. [Google Scholar]
- Allen, D.; McKee, I.; Farage, P.; Baker, N. Analysis of limitations to CO2 assimilation on exposure of leaves of two Brassica napus cultivars to UV-B. Plant Cell Environ. 1997, 20, 633–640. [Google Scholar] [CrossRef]
- Sitohy, M.Z.; Desoky, E.-S.M.; Osman, A.; Rady, M.M. Pumpkin seed protein hydrolysate treatment alleviates salt stress effects on Phaseolus vulgaris by elevating antioxidant capacity and recovering ion homeostasis. Sci. Hortic. 2020, 271, 109495. [Google Scholar] [CrossRef]
- Borsani, O.; Valpuesta, V.; Botella, M. Developing salt tolerant plants in a new century: A molecular biology approach. Plant Cell Tiss. Org. 2003, 73, 101–115. [Google Scholar] [CrossRef]
- Wahome, P.; Jesch, H.; Grittner, I. Mechanisms of salt stress tolerance in two rose rootstocks: Rosa chinensis ‘Major’and R. rubiginosa. Sci. Hortic. 2001, 87, 207–216. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Meloni, D.A.; Martínez, C.A. Glycinebetaine improves salt tolerance in vinal (Prosopis ruscifolia Griesbach) seedlings. Braz. J. Plant Physiol. 2009, 21, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Vysotskaya, L.; Hedley, P.E.; Sharipova, G.; Veselov, D.; Kudoyarova, G.; Morris, J.; Jones, H.G. Effect of salinity on water relations of wild barley plants differing in salt tolerance. AoB Plant. 2010, 2010, 1–8. [Google Scholar] [CrossRef]
- Mansour, E.; Merwad, A.; Yasin, M.; Abdul-Hamid, M.; El-Sobky, E.; Oraby, H. Nitrogen use efficiency in spring wheat: Genotypic variation and grain yield response under sandy soil conditions. J. Agric. Sci. 2017, 155, 1407–1423. [Google Scholar] [CrossRef]
- Zeng, L.; Shannon, M.; Grieve, C. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 2002, 127, 235–245. [Google Scholar] [CrossRef]
- Hammami, Z.; Sbei, H.; Kadri, K.; Jmel, Z.; Sahli, A.; Belhaj Fraj, M.; Naser, H.; Teixeira da Silva, J.; Trifa, Y. Evaluation of performance of different barley genotypes irrigated with saline water in South Tunisian Saharan conditions. Environ. Exp. Bot. 2016, 14, 15–21. [Google Scholar] [CrossRef]
- Mansour, E.; Moustafa, E.S.; El-Naggar, N.Z.; Abdelsalam, A.; Igartua, E. Grain yield stability of high-yielding barley genotypes under Egyptian conditions for enhancing resilience to climate change. Crop Pasture Sci. 2018, 69, 681–690. [Google Scholar] [CrossRef]

| Genotypes | Grain Yield (kg ha−1) | Yield Index (YI) | Cluster Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | ||
| Gemiza-11 | 4960 | 3973 | 2992 | 3975 | 1.30 | 1.40 | 1.47 | 1.39 | A |
| Gemiza-9 | 5392 | 3549 | 2359 | 3767 | 1.42 | 1.25 | 1.16 | 1.27 | B |
| Gemiza-10 | 4263 | 2849 | 2460 | 3191 | 1.12 | 1.00 | 1.20 | 1.11 | B |
| Giza-171 | 5209 | 3680 | 2050 | 3646 | 1.37 | 1.29 | 1.00 | 1.22 | B |
| Sids-14 | 4602 | 3339 | 1976 | 3306 | 1.21 | 1.17 | 0.97 | 1.12 | B |
| Line-6084 | 4718 | 3080 | 2525 | 3441 | 1.24 | 1.08 | 1.24 | 1.19 | B |
| Line-6083 | 4210 | 2892 | 1937 | 3013 | 1.11 | 1.02 | 0.95 | 1.02 | B |
| Giza-168 | 3182 | 2442 | 2130 | 2585 | 0.84 | 0.86 | 1.04 | 0.91 | C |
| Gemiza-7 | 3560 | 2768 | 1809 | 2712 | 0.94 | 0.97 | 0.89 | 0.93 | C |
| Sakha-94 | 3185 | 2681 | 2007 | 2624 | 0.84 | 0.94 | 0.98 | 0.92 | C |
| Sids-12 | 3613 | 2854 | 2193 | 2887 | 0.95 | 1.00 | 1.07 | 1.01 | C |
| Misr-1 | 3254 | 2840 | 2075 | 2723 | 0.86 | 1.00 | 1.02 | 0.96 | C |
| Line-6052 | 3185 | 2677 | 1884 | 2582 | 0.84 | 0.94 | 0.92 | 0.90 | C |
| Line-6078 | 3360 | 2551 | 1947 | 2619 | 0.88 | 0.90 | 0.95 | 0.91 | C |
| Line-1208 | 3669 | 2690 | 2041 | 2800 | 0.96 | 0.95 | 1.00 | 0.97 | C |
| Gemiza-12 | 2786 | 2225 | 1668 | 2226 | 0.73 | 0.78 | 0.82 | 0.78 | D |
| Shandawel-1 | 2680 | 2097 | 1246 | 2008 | 0.70 | 0.74 | 0.61 | 0.68 | D |
| Misr-2 | 2659 | 1983 | 1462 | 2034 | 0.70 | 0.70 | 0.72 | 0.70 | D |
| Genotypes | Total Chlorophyll (mg g−1 Fresh Weight) | Pn (µmol CO2 m−2 s−1) | E (mmol H2O m−2 s−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 2.96 | 2.22 | 1.66 | 2.28 a† | 12.72 | 10.14 | 6.89 | 9.92 a | 7.05 | 4.73 | 3.55 | 5.11 a |
| Group B n = 6 | 2.92 | 2.09 | 1.39 | 2.13 b | 12.50 | 9.91 | 6.42 | 9.61 b | 7.01 | 4.52 | 3.22 | 4.92 b |
| Group C n = 8 | 2.67 | 1.90 | 1.30 | 1.95 c | 11.24 | 9.18 | 6.07 | 8.83 c | 6.16 | 4.17 | 3.05 | 4.46 c |
| Group D n = 3 | 2.50 | 1.81 | 1.15 | 1.82 d | 10.60 | 8.39 | 5.24 | 8.07 d | 5.37 | 3.72 | 2.80 | 3.96 d |
| Mean | 2.76 A | 2.00 B | 1.37 C | 11.77 A | 9.40 B | 6.16 C | 6.40 A | 4.29 B | 3.15 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.011 | < 0.001 | 0.07 | < 0.001 | 0.15 | |||||
| Group (G) | 3 | < 0.001 | 0.029 | < 0.001 | 0.11 | < 0.001 | 0.16 | |||||
| S × G | 6 | < 0.001 | 0.047 | < 0.001 | 0.20 | 0.003 | 0.29 | |||||
| Genotypes | Gs (mmol H2O m−2 s−1) | RWC (%) | MSI (%) | |||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 0.59 | 0.40 | 0.30 | 0.43 a | 65.81 | 51.56 | 42.04 | 53.13 a | 60.78 | 49.01 | 36.22 | 48.67 a |
| Group B n = 6 | 0.56 | 0.39 | 0.27 | 0.41 b | 63.15 | 50.44 | 38.71 | 50.77 b | 59.96 | 48.07 | 33.90 | 47.31 b |
| Group C n = 8 | 0.49 | 0.36 | 0.26 | 0.37 c | 56.99 | 47.89 | 39.48 | 48.12 c | 55.45 | 44.35 | 32.60 | 44.13 c |
| Group D n = 3 | 0.44 | 0.33 | 0.21 | 0.33 d | 53.93 | 46.39 | 36.36 | 45.56 d | 51.18 | 40.71 | 26.90 | 39.60 d |
| Mean | 0.52 A | 0.37 B | 0.26 C | 59.97 A | 49.07 B | 39.14 C | 56.84 A | 45.54 B | 32.40 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.003 | < 0.001 | 1.71 | < 0.001 | 0.16 | |||||
| Group (G) | 3 | < 0.001 | 0.006 | < 0.001 | 1.42 | < 0.001 | 0.32 | |||||
| S × G | 6 | < 0.001 | 0.010 | 0.002 | 2.77 | < 0.001 | 0.55 | |||||
| Genotypes | MDA (nmol g−1 Fresh Weight) | EL (%) | Soluble Sugars (mg g−1 Dry Weight) | Free Proline (µg g−1 Dry Weight) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 49.33 | 60.18 | 69.00 | 59.50 d† | 2.80 | 6.60 | 10.46 | 6.62 d | 25.95 | 40.53 | 56.80 | 41.09 a | 127.7 | 192.5 | 242.8 | 187.7 a |
| Group B n = 6 | 50.07 | 61.55 | 72.64 | 61.42 c | 3.05 | 6.93 | 12.51 | 7.49 c | 25.89 | 39.57 | 53.26 | 39.57 b | 123.5 | 183.7 | 230.5 | 179.2 b |
| Group C n = 8 | 53.65 | 64.26 | 74.94 | 64.28 b | 4.20 | 7.63 | 13.12 | 8.32 b | 24.11 | 35.23 | 50.03 | 36.46 c | 98.6 | 164.1 | 223.7 | 162.2 c |
| Group D n = 3 | 57.61 | 67.92 | 82.56 | 69.36 a | 5.36 | 8.69 | 15.07 | 9.71 a | 21.79 | 30.62 | 44.66 | 32.36 d | 84.7 | 154.7 | 207.5 | 148.9 d |
| Mean | 52.67 C | 63.48 B | 74.78 A | 3.85 C | 7.46 B | 12.79 A | 24.43 C | 36.48 B | 51.19 A | 108.6 C | 173.8 B | 226.1 A | ||||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | p-value | LSD | |||||||
| Salinity (S) | 2 | < 0.001 | 0.57 | < 0.001 | 0.20 | < 0.001 | 1.44 | < 0.001 | 1.64 | |||||||
| Group (G) | 3 | < 0.001 | 0.62 | < 0.001 | 0.28 | < 0.001 | 1.29 | < 0.001 | 1.82 | |||||||
| S × G | 6 | < 0.001 | 1.04 | < 0.001 | 0.49 | < 0.001 | 2.93 | < 0.001 | 2.31 | |||||||
| Genotypes | AsA (µmol g−1 dry weight) | SOD (U µg−1 protein) | CAT (U mg−1 min−1) | POD (µg g−1 fresh weight min−1) | ||||||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 1.74 | 2.53 | 3.03 | 2.43 a | 52.41 | 69.93 | 87.46 | 69.93 a | 0.50 | 0.70 | 0.96 | 0.72 a | 90.33 | 131.57 | 177.93 | 133.28 a |
| Group B n = 6 | 1.72 | 2.46 | 2.86 | 2.35 b | 51.92 | 67.42 | 82.62 | 67.32 b | 0.50 | 0.65 | 0.85 | 0.67 b | 88.94 | 126.67 | 167.34 | 127.65 b |
| Group C n = 8 | 1.61 | 2.25 | 2.77 | 2.21 c | 47.43 | 63.49 | 79.78 | 63.57 c | 0.46 | 0.60 | 0.81 | 0.62 c | 82.22 | 113.58 | 161.78 | 119.19 c |
| Group D n = 3 | 1.53 | 2.08 | 2.56 | 2.06 d | 42.27 | 58.10 | 73.71 | 58.03 d | 0.42 | 0.57 | 0.73 | 0.57 d | 77.53 | 102.46 | 143.91 | 107.97 d |
| Mean | 1.65 C | 2.33 B | 2.80 A | 48.51 C | 64.74 B | 80.89 A | 0.47 C | 0.63 B | 0.84 A | 84.76 C | 118.57 B | 162.74 A | ||||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | p-value | LSD | |||||||
| Salinity (S) | 2 | < 0.001 | 0.009 | < 0.001 | 0.27 | < 0.001 | 0.009 | < 0.001 | 0.94 | |||||||
| Group (G) | 3 | < 0.001 | 0.013 | < 0.001 | 0.56 | < 0.001 | 0.012 | < 0.001 | 1.41 | |||||||
| S × G | 6 | < 0.001 | 0.023 | < 0.001 | 0.95 | < 0.001 | 0.015 | < 0.001 | 2.45 | |||||||
| Genotypes | Cl− (%) | Na+ (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 1.55 | 2.05 | 3.30 | 2.30 c† | 4.35 | 8.21 | 11.15 | 7.90 d |
| Group B n = 6 | 1.57 | 2.21 | 3.18 | 2.32 c | 4.56 | 8.77 | 12.18 | 8.50 c |
| Group C n = 8 | 1.75 | 2.61 | 3.51 | 2.62 b | 5.84 | 9.76 | 12.87 | 9.49 b |
| Group D n = 3 | 1.87 | 2.87 | 3.38 | 2.71 a | 6.78 | 10.24 | 15.24 | 10.75 a |
| Mean | 1.69 C | 2.43 B | 3.34 A | 5.38 C | 9.24 B | 12.86 A | ||
| ANOVA | df | p-value | LSD | p-value | LSD | |||
| Salinity (S) | 2 | < 0.001 | 0.02 | < 0.001 | 0.15 | |||
| Group (G) | 3 | < 0.001 | 0.05 | < 0.001 | 0.14 | |||
| S × G | 6 | < 0.001 | 0.10 | < 0.001 | 0.27 | |||
| Genotypes | K+ (%) | K+/Na+ ratio | ||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 19.97 | 16.50 | 11.32 | 15.93 a | 4.59 | 2.01 | 1.01 | 2.54 a |
| Group B n = 6 | 19.77 | 15.09 | 10.00 | 14.95 b | 4.37 | 1.73 | 0.83 | 2.31 b |
| Group C n = 8 | 18.30 | 13.41 | 9.41 | 13.71 c | 3.15 | 1.38 | 0.74 | 1.75 c |
| Group D n = 3 | 17.27 | 12.36 | 8.29 | 12.64 d | 2.55 | 1.21 | 0.54 | 1.43 d |
| Mean | 18.83 A | 14.34 B | 9.75 C | 3.66 A | 1.58 B | 0.78 C | ||
| ANOVA | df | p-value | LSD | p-value | LSD | |||
| Salinity (S) | 2 | < 0.001 | 0.04 | < 0.001 | 0.14 | |||
| Group (G) | 3 | < 0.001 | 0.17 | < 0.001 | 0.07 | |||
| S × G | 6 | < 0.001 | 0.28 | < 0.001 | 0.17 | |||
| Genotypes | Plant Height (cm) | Spike Length (cm) | Number of Spikes m−2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 92.50 | 88.67 | 74.00 | 85.06 a† | 12.31 | 10.85 | 9.89 | 11.02 a | 182.9 | 161.9 | 151.0 | 162.2 a |
| Group B n = 6 | 87.54 | 76.36 | 67.91 | 77.27 b | 11.87 | 10.72 | 9.68 | 10.76 b | 173.6 | 154.3 | 133.3 | 156.9 b |
| Group C n = 8 | 83.40 | 77.02 | 70.23 | 76.88 bc | 11.70 | 10.63 | 9.60 | 10.64 b | 144.9 | 130.0 | 117.8 | 130.9 c |
| Group D n = 3 | 81.89 | 76.72 | 69.61 | 76.07 c | 11.13 | 10.28 | 9.13 | 10.18 c | 126.3 | 110.1 | 98.6 | 111.7 d |
| Mean | 86.33 A | 79.69 B | 70.44 C | 11.75 A | 10.62 B | 9.58 C | 156.9 A | 139.1 B | 125.2 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.79 | < 0.001 | 0.13 | < 0.001 | 2.25 | |||||
| Group (G) | 3 | < 0.001 | 0.91 | < 0.001 | 0.17 | < 0.001 | 3.64 | |||||
| S × G | 6 | < 0.001 | 2.03 | < 0.001 | 0.29 | < 0.001 | 6.80 | |||||
| Genotypes | Number of grains per spike | 1000-grain weight (g) | Grain yield (kg ha-1) | |||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 59.17 | 54.00 | 44.17 | 52.44 a | 55.59 | 49.55 | 41.10 | 48.75 a | 4960.0 | 3973.0 | 2992.0 | 3975.0 a |
| Group B n = 6 | 55.31 | 47.48 | 41.32 | 48.03 b | 53.16 | 47.55 | 40.78 | 47.16 b | 4732.3 | 3231.5 | 2217.8 | 3393.9 b |
| Group C n = 8 | 51.60 | 44.65 | 40.04 | 45.43 c | 50.79 | 46.03 | 40.65 | 45.82 c | 3376.0 | 2687.9 | 2010.8 | 2691.5 c |
| Group D n = 3 | 51.00 | 44.06 | 38.39 | 44.48 d | 48.66 | 44.01 | 39.00 | 43.89 d | 2708.3 | 2101.7 | 1458.7 | 2089.6 d |
| Mean | 54.27 A | 47.54 B | 40.98 C | 52.05 A | 46.78 B | 40.38 C | 3944 A | 2999 B | 2170 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.52 | < 0.001 | 0.40 | < 0.001 | 74.58 | |||||
| Group (G) | 3 | < 0.001 | 0.77 | < 0.001 | 0.62 | < 0.001 | 124.3 | |||||
| S × G | 6 | 0.003 | 1.33 | < 0.001 | 1.03 | < 0.001 | 203.4 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansour, E.; Moustafa, E.S.A.; Desoky, E.-S.M.; Ali, M.M.A.; Yasin, M.A.T.; Attia, A.; Alsuhaibani, N.; Tahir, M.U.; El-Hendawy, S. Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants 2020, 9, 1324. https://doi.org/10.3390/plants9101324
Mansour E, Moustafa ESA, Desoky E-SM, Ali MMA, Yasin MAT, Attia A, Alsuhaibani N, Tahir MU, El-Hendawy S. Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants. 2020; 9(10):1324. https://doi.org/10.3390/plants9101324
Chicago/Turabian StyleMansour, Elsayed, Ehab S. A. Moustafa, El-Sayed M. Desoky, Mohamed M. A. Ali, Mohamed A. T. Yasin, Ahmed Attia, Nasser Alsuhaibani, Muhammad Usman Tahir, and Salah El-Hendawy. 2020. "Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions" Plants 9, no. 10: 1324. https://doi.org/10.3390/plants9101324
APA StyleMansour, E., Moustafa, E. S. A., Desoky, E.-S. M., Ali, M. M. A., Yasin, M. A. T., Attia, A., Alsuhaibani, N., Tahir, M. U., & El-Hendawy, S. (2020). Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants, 9(10), 1324. https://doi.org/10.3390/plants9101324









