Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity
Simple Summary
Abstract
1. Introduction
2. Results
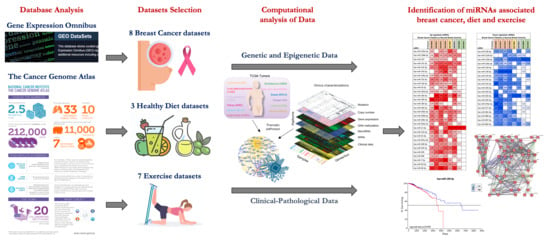
2.1. miRNA Microarray Expression Profiling Dataset Selection
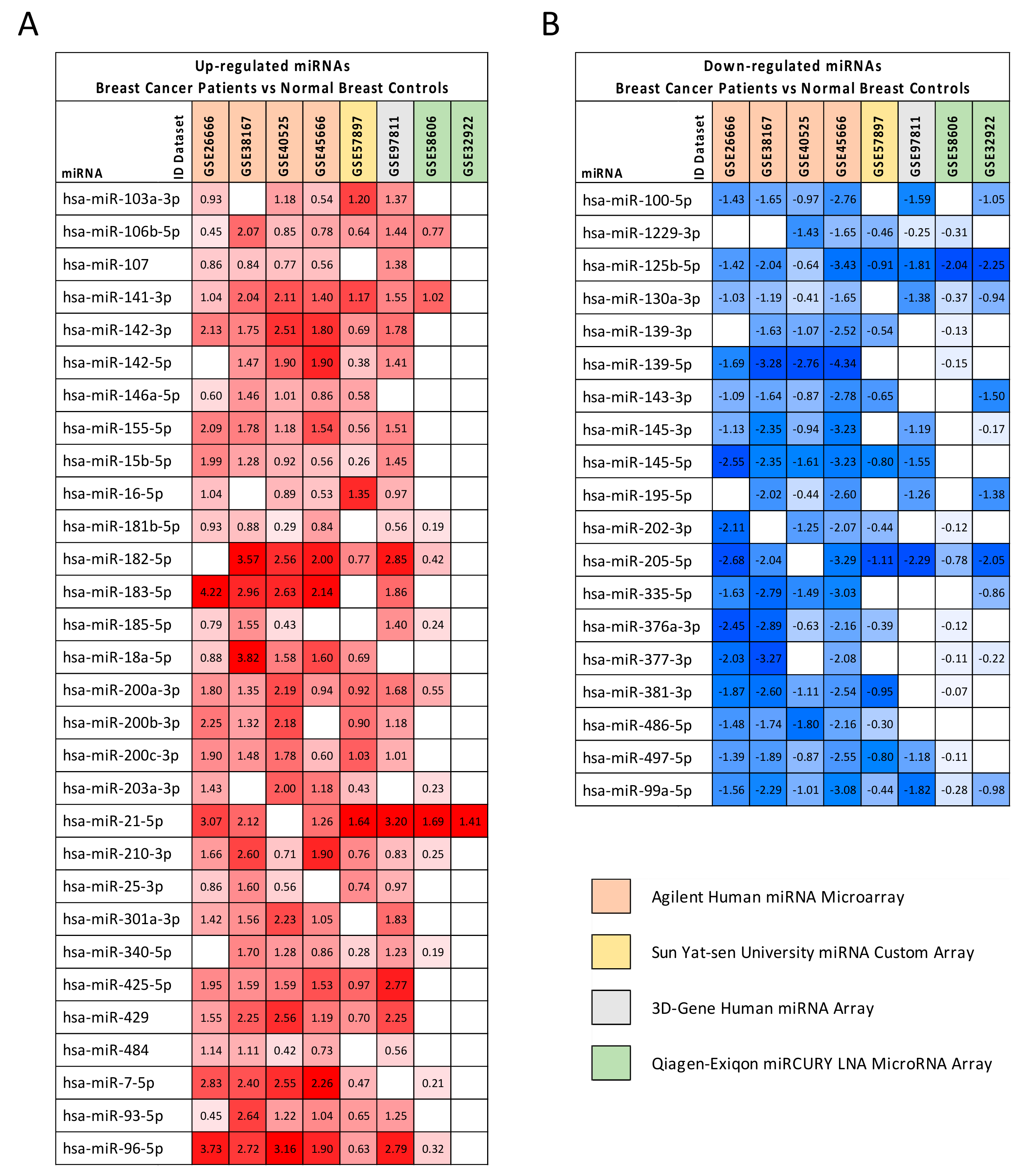
2.2. Computational Identification of microRNAs Involved in Breast Cancer
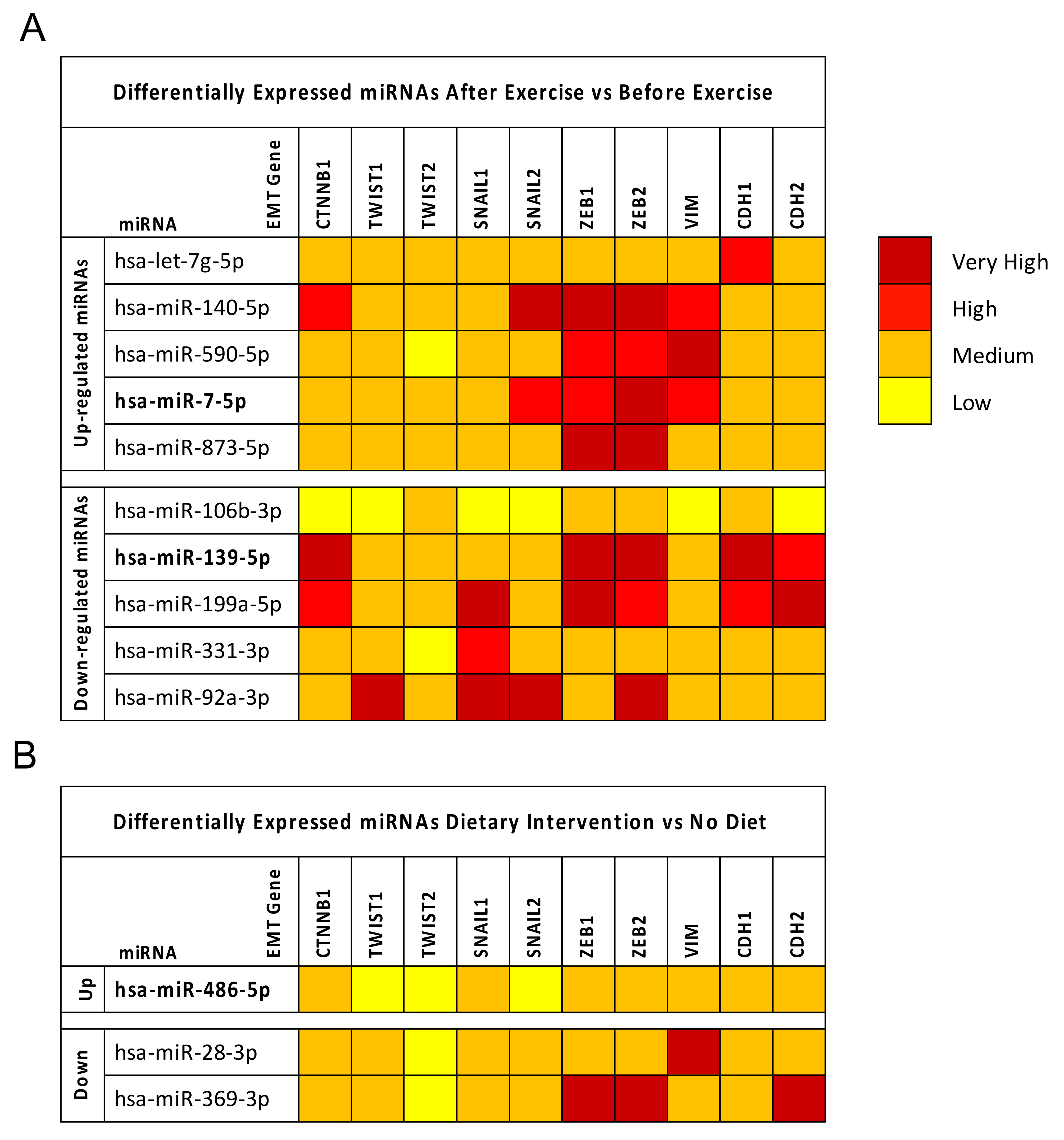
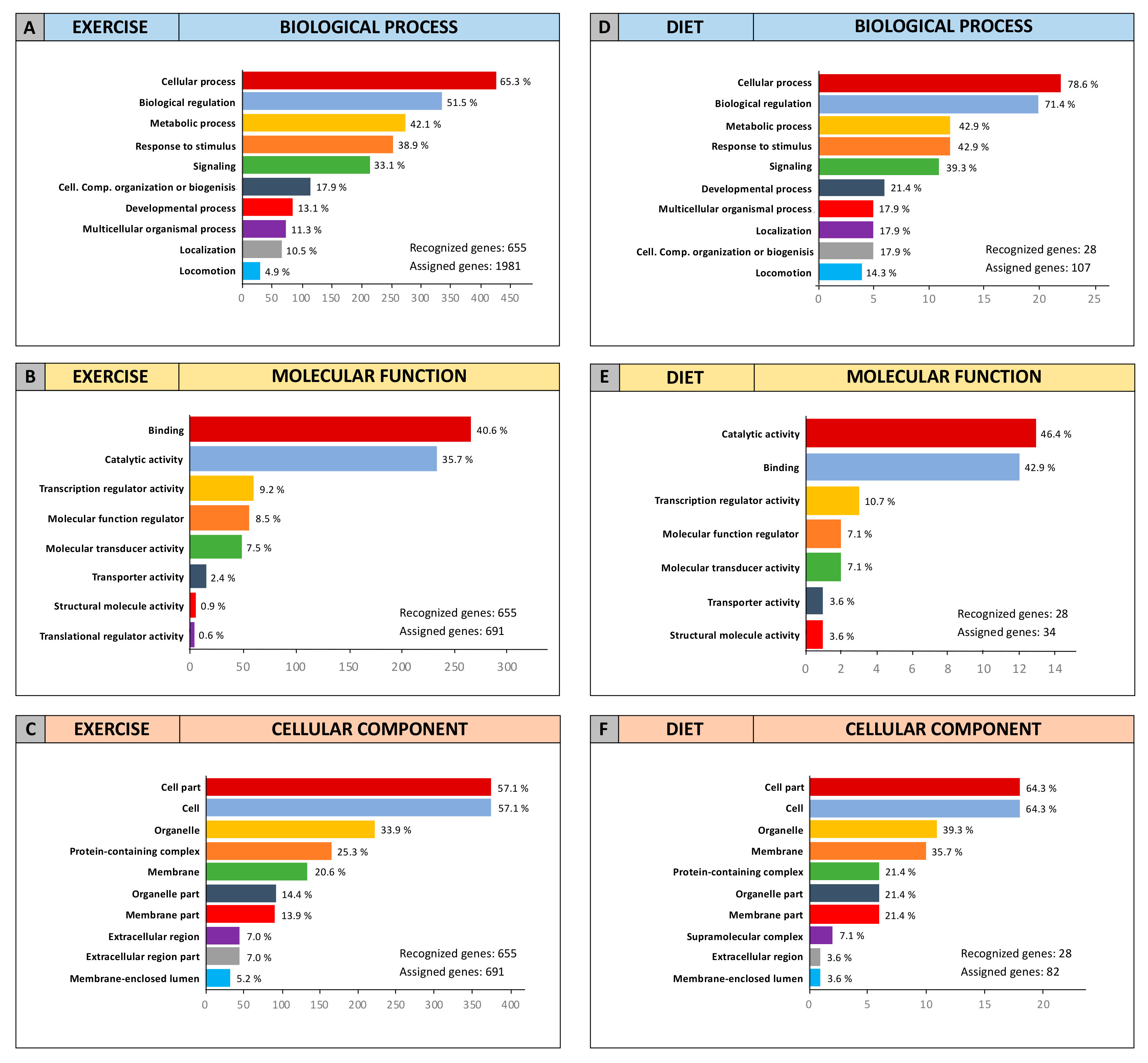
2.3. Computational Identification of microRNA Modulated by Diet and Exercise
2.4. Breast Cancer (BC) miRNA Modulation of Epithelial-Mesenchymal Transition (EMT) Genes
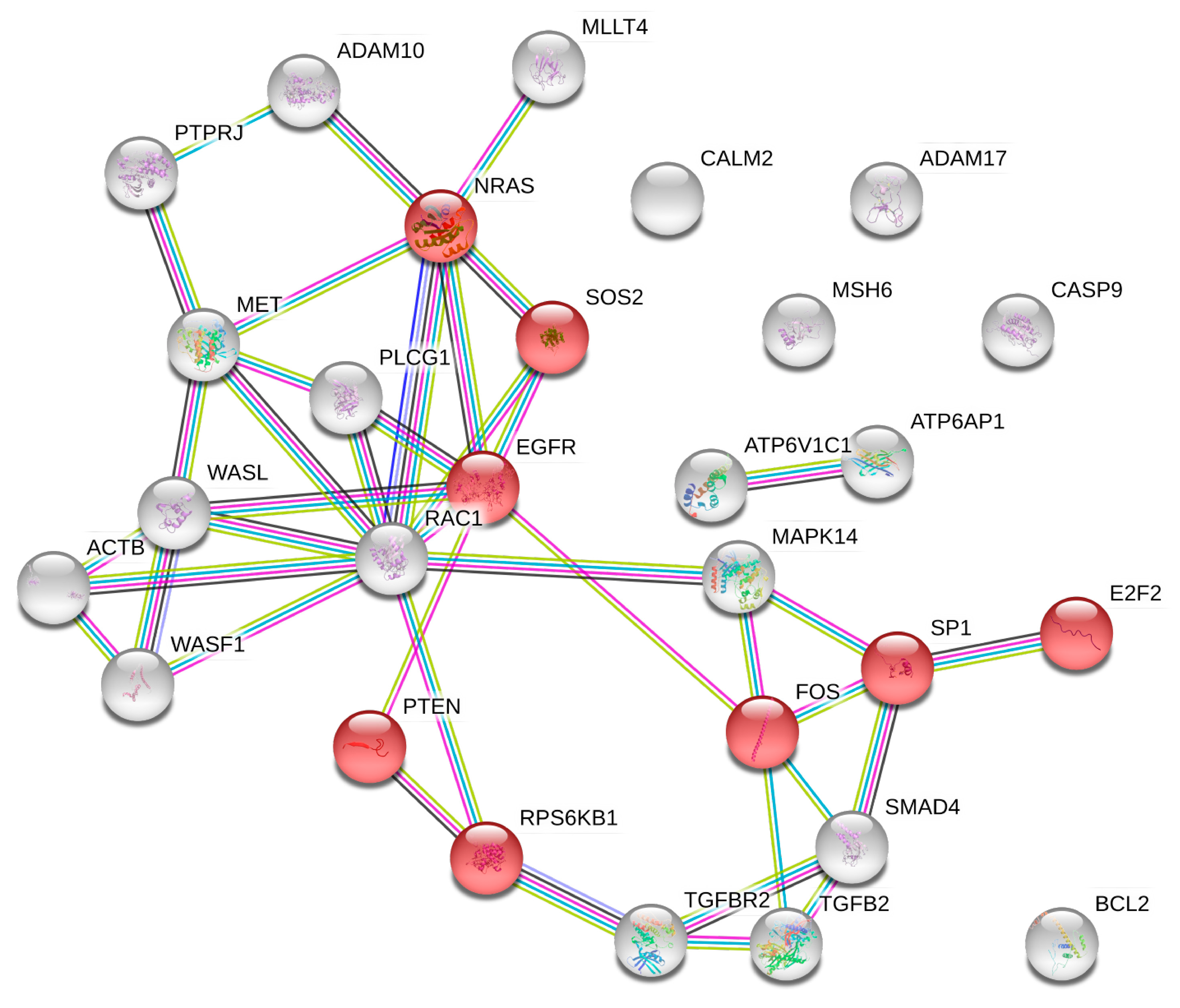
2.5. microRNA Pathway Prediction Analysis and miRNA-Targeted Genes Protein–Protein Interaction
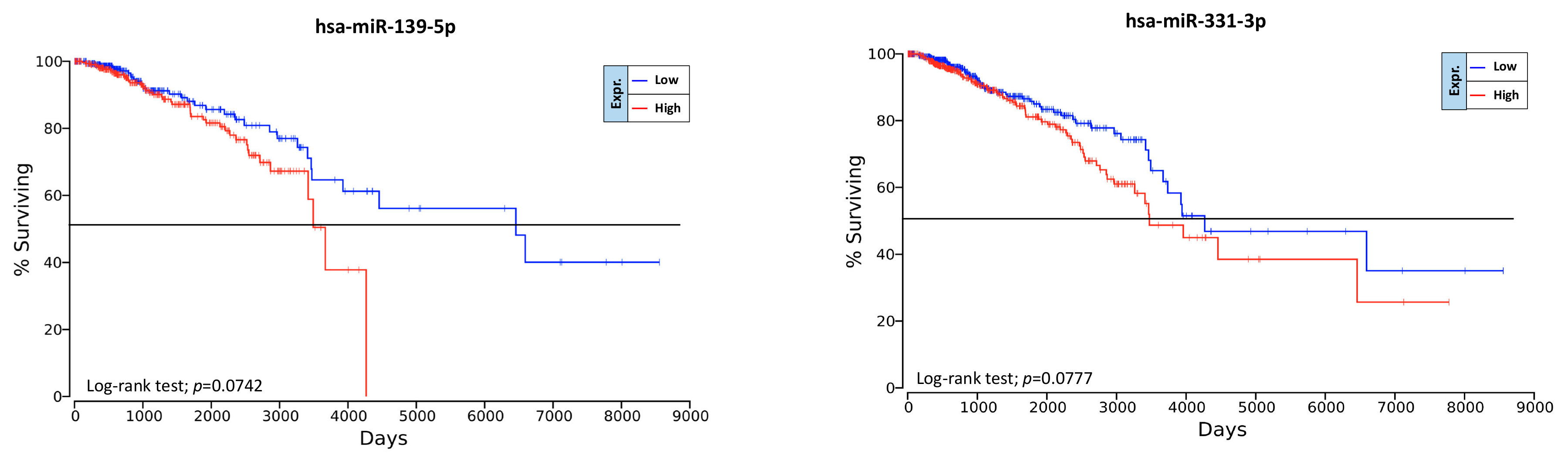
2.6. Overall Survival Predictive Value of BC, Exercise-Modulated, and Diet-Modulated microRNAs
3. Discussion
4. Materials and Methods
4.1. microRNA Expression Profiling Dataset Selection
4.2. Differential Analysis between Groups and microRNA Annotation
4.3. Identification of microRNAs Involved in Breast Cancer and Effectively Modulated by Diet and Exercise
4.4. Interaction between Selected microRNAs and Epithelial-Mesenchymal Transition (EMT) Genes
4.5. microRNA Pathway Prediction Analysis
4.6. miRNA-Targeted Genes Interaction and Gene Ontology (GO)
4.7. Prognostic Significance of Computationally Selected microRNAs
4.8. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front. Pharmacol. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, R.E.; Baker, J.A.; Helvie, M.A. Breast cancer deaths averted over 3 decades. Cancer 2019, 125, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Christofi, T.; Baritaki, S.; Falzone, L.; Libra, M.; Zaravinos, A. Current Perspectives in Cancer Immunotherapy. Cancers 2019, 11, 1472. [Google Scholar] [CrossRef] [PubMed]
- Carayol, M.; Ninot, G.; Senesse, P.; Bleuse, J.P.; Gourgou, S.; Sancho-Garnier, H.; Sari, C.; Romieu, I.; Romieu, G.; Jacot, W. Short- and long-term impact of adapted physical activity and diet counseling during adjuvant breast cancer therapy: The “APAD1” randomized controlled trial. BMC Cancer 2019, 19, 737. [Google Scholar] [CrossRef] [PubMed]
- Lyman, G.H.; Greenlee, H.; Bohlke, K.; Bao, T.; DeMichele, A.M.; Deng, G.E.; Fouladbakhsh, J.M.; Gil, B.; Hershman, D.L.; Mansfield, S.; et al. Integrative Therapies During and After Breast Cancer Treatment: ASCO Endorsement of the SIO Clinical Practice Guideline. J. Clin. Oncol. 2018, 36, 2647–2655. [Google Scholar] [CrossRef] [PubMed]
- Hack, C.C.; Hackl, J.; Hüttner, N.B.M.; Langemann, H.; Schwitulla, J.; Dietzel-Drentwett, S.; Fasching, P.A.; Beckmann, M.W.; Theuser, A.K. Self-reported Improvement in Side Effects and Quality of Life With Integrative Medicine in Breast Cancer Patients. Integr. Cancer Ther. 2018, 17, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Sudhakaran, M.; Sardesai, S.; Doseff, A.I. Flavonoids: New Frontier for Immuno-Regulation and Breast Cancer Control. Antioxidants 2019, 8, 103. [Google Scholar] [CrossRef]
- Lopes, C.M.; Dourado, A.; Oliveira, R. Phytotherapy and Nutritional Supplements on Breast Cancer. Biomed. Res. Int. 2017, 2017, 7207983. [Google Scholar] [CrossRef]
- Mendoza, L. Potential effect of probiotics in the treatment of breast cancer. Oncol. Rev. 2019, 13, 422. [Google Scholar] [CrossRef]
- Vivarelli, S.; Salemi, R.; Candido, S.; Falzone, L.; Santagati, M.; Stefani, S.; Torino, F.; Banna, G.L.; Tonini, G.; Libra, M. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Lopez, G.; Eddy, C.; Liu, W.; Li, Y.; Chen, M.; Bruera, E.; Cohen, L. Physical Therapist-Led Exercise Assessment and Counseling in Integrative Cancer Care: Effects on Patient Self-reported Symptoms and Quality of Life. Integr. Cancer Ther. 2019, 18, 1534735419832360. [Google Scholar] [CrossRef] [PubMed]
- Browall, M.; Mijwel, S.; Rundqvist, H.; Wengström, Y. Physical Activity During and After Adjuvant Treatment for Breast Cancer: An Integrative Review of Women’s Experiences. Integr. Cancer Ther. 2018, 17, 16–30. [Google Scholar] [CrossRef] [PubMed]
- De Cicco, P.; Catani, M.V.; Gasperi, V.; Sibilano, M.; Quaglietta, M.; Savini, I. Nutrition and Breast Cancer: A Literature Review on Prevention, Treatment and Recurrence. Nutrients 2019, 11, 1514. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.A.; Tigu, A.B.; Muntean, M.; Cenariu, D.; Slaby, O.; Berindan-Neagoe, I. Molecular Links between Central Obesity and Breast Cancer. Int. J. Mol. Sci. 2019, 20, 5364. [Google Scholar] [CrossRef] [PubMed]
- Kotsopoulos, J. Menopausal hormones: Definitive evidence for breast cancer. Lancet 2019, 394, 1116–1118. [Google Scholar] [CrossRef]
- Key, T.J.; Balkwill, A.; Bradbury, K.E.; Reeves, G.K.; Kuan, A.S.; Simpson, R.F.; Green, J.; Beral, V. Foods, macronutrients and breast cancer risk in postmenopausal women: A large UK cohort. Int. J. Epidemiol. 2019, 48, 489–500. [Google Scholar] [CrossRef]
- Banna, G.L.; Torino, F.; Marletta, F.; Santagati, M.; Salemi, R.; Cannarozzo, E.; Falzone, L.; Ferraù, F.; Libra, M. Lactobacillus rhamnosus GG: An Overview to Explore the Rationale of Its Use in Cancer. Front. Pharmacol. 2017, 8, 603. [Google Scholar] [CrossRef]
- Ferroni, P.; Riondino, S.; Buonomo, O.; Palmirotta, R.; Guadagni, F.; Roselli, M. Type 2 Diabetes and Breast Cancer: The Interplay between Impaired Glucose Metabolism and Oxidant Stress. Oxid. Med. Cell Longev. 2015, 2015, 183928. [Google Scholar] [CrossRef]
- Alves, B.C.; Silva, T.R.; Spritzer, P.M. Sedentary Lifestyle and High-Carbohydrate Intake are Associated with Low-Grade Chronic Inflammation in Post-Menopause: A Cross-sectional Study. Rev. Bras. Ginecol. Obs. 2016, 38, 317–324. [Google Scholar] [CrossRef]
- Jang, H.; Chung, M.S.; Kang, S.S.; Park, Y. Association between the Dietary Inflammatory Index and Risk for Cancer Recurrence and Mortality among Patients with Breast Cancer. Nutrients 2018, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Romieu, I.; Ferrari, P.; Rinaldi, S.; Slimani, N.; Jenab, M.; Olsen, A.; Tjonneland, A.; Overvad, K.; Boutron-Ruault, M.C.; Lajous, M.; et al. Dietary glycemic index and glycemic load and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Am. J. Clin. Nutr. 2012, 96, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Bodén, S.; Myte, R.; Wennberg, M.; Harlid, S.; Johansson, I.; Shivappa, N.; Hébert, J.R.; Van Guelpen, B.; Nilsson, L.M. The inflammatory potential of diet in determining cancer risk; A prospective investigation of two dietary pattern scores. PLoS ONE 2019, 14, e0214551. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Pietrocola, F.; Kroemer, G. Nutrition, inflammation and cancer. Nat. Immunol. 2017, 18, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Ultimo, S.; Zauli, G.; Martelli, A.M.; Vitale, M.; McCubrey, J.A.; Capitani, S.; Neri, L.M. Influence of physical exercise on microRNAs in skeletal muscle regeneration, aging and diseases. Oncotarget 2018, 9, 17220–17237. [Google Scholar] [CrossRef] [PubMed]
- Quintanilha, B.J.; Reis, B.Z.; Duarte, G.B.S.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef]
- Giardina, S.; Hernández-Alonso, P.; Díaz-López, A.; Salas-Huetos, A.; Salas-Salvadó, J.; Bulló, M. Changes in circulating miRNAs in healthy overweight and obese subjects: Effect of diet composition and weight loss. Clin. Nutr. 2019, 38, 438–443. [Google Scholar] [CrossRef]
- Falzone, L.; Salemi, R.; Travali, S.; Scalisi, A.; McCubrey, J.A.; Candido, S.; Libra, M. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging 2016, 8, 933–944. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef]
- Gillman, A.S.; Gardiner, C.K.; Koljack, C.E.; Bryan, A.D. Body mass index, diet, and exercise: Testing possible linkages to breast cancer risk via DNA methylation. Breast Cancer Res. Treat. 2018, 168, 241–248. [Google Scholar] [CrossRef]
- Toledo, E.; Salas-Salvadó, J.; Donat-Vargas, C.; Buil-Cosiales, P.; Estruch, R.; Ros, E.; Corella, D.; Fitó, M.; Hu, F.B.; Arós, F.; et al. Mediterranean Diet and Invasive Breast Cancer Risk Among Women at High Cardiovascular Risk in the PREDIMED Trial: A Randomized Clinical Trial. JAMA Intern. Med. 2015, 175, 1752–1760. [Google Scholar] [CrossRef] [PubMed]
- Shapira, N. The potential contribution of dietary factors to breast cancer prevention. Eur. J. Cancer Prev. 2017, 26, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, P.; Baumann, F.T. Physical Activity, Exercise and Breast Cancer—What Is the Evidence for Rehabilitation, Aftercare, and Survival? A Review. Breast Care 2018, 13, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Arem, H.; Hubal, M.J.; Cartmel, B.; Li, F.; Harrigan, M.; Sanft, T.; Cheng, C.J.; Pusztai, L.; Irwin, M.L. Exercise and weight loss interventions and miRNA expression in women with breast cancer. Breast Cancer Res. Treat. 2018, 170, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Ntanasis-Stathopoulos, J.; Tzanninis, J.G.; Philippou, A.; Koutsilieris, M. Epigenetic regulation on gene expression induced by physical exercise. J. Musculoskelet. Neuronal Interact. 2013, 13, 133–146. [Google Scholar]
- Lope, V.; Martín, M.; Castelló, A.; Ruiz, A.; Casas, A.M.; Baena-Cañada, J.M.; Antolín, S.; Ramos-Vázquez, M.; García-Sáenz, J.Á.; Muñoz, M.; et al. Overeating, caloric restriction and breast cancer risk by pathologic subtype: The EPIGEICAM study. Sci. Rep. 2019, 9, 3904. [Google Scholar] [CrossRef]
- Augustin, L.S.; Libra, M.; Crispo, A.; Grimaldi, M.; De Laurentiis, M.; Rinaldo, M.; D’Aiuto, M.; Catalano, F.; Banna, G.; Ferrau’, F.; et al. Low glycemic index diet, exercise and vitamin D to reduce breast cancer recurrence (DEDiCa): Design of a clinical trial. BMC Cancer 2017, 17, 69. [Google Scholar] [CrossRef]
- Zhao, X.G.; Hu, J.Y.; Tang, J.; Yi, W.; Zhang, M.Y.; Deng, R.; Mai, S.J.; Weng, N.Q.; Wang, R.Q.; Liu, J.; et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019, 10, 479. [Google Scholar] [CrossRef]
- Hironaka-Mitsuhashi, A.; Matsuzaki, J.; Takahashi, R.U.; Yoshida, M.; Nezu, Y.; Yamamoto, Y.; Shiino, S.; Kinoshita, T.; Ushijima, T.; Hiraoka, N.; et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS ONE 2017, 12, e0187638. [Google Scholar] [CrossRef]
- Matamala, N.; Vargas, M.T.; González-Cámpora, R.; Miñambres, R.; Arias, J.I.; Menéndez, P.; Andrés-León, E.; Gómez-López, G.; Yanowsky, K.; Calvete-Candenas, J.; et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin. Chem. 2015, 61, 1098–1106. [Google Scholar] [CrossRef]
- Avery-Kiejda, K.A.; Braye, S.G.; Mathe, A.; Forbes, J.F.; Scott, R.J. Decreased expression of key tumour suppressor microRNAs is associated with lymph node metastases in triple negative breast cancer. BMC Cancer 2014, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Tanić, M.; Yanowski, K.; Andrés, E.; Gómez-López, G.; Socorro, M.R.; Pisano, D.G.; Martinez-Delgado, B.; Benítez, J. miRNA expression profiling of formalin-fixed paraffin-embedded (FFPE) hereditary breast tumors. Genom. Data 2014, 3, 75–79. [Google Scholar] [CrossRef]
- Lee, C.H.; Kuo, W.H.; Lin, C.C.; Oyang, Y.J.; Huang, H.C.; Juan, H.F. MicroRNA-regulated protein-protein interaction networks and their functions in breast cancer. Int. J. Mol. Sci. 2013, 14, 11560–11606. [Google Scholar] [CrossRef] [PubMed]
- Cimino, D.; De Pittà, C.; Orso, F.; Zampini, M.; Casara, S.; Penna, E.; Quaglino, E.; Forni, M.; Damasco, C.; Pinatel, E.; et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013, 27, 1223–1235. [Google Scholar] [CrossRef]
- Biagioni, F.; Bossel Ben-Moshe, N.; Fontemaggi, G.; Canu, V.; Mori, F.; Antoniani, B.; Di Benedetto, A.; Santoro, R.; Germoni, S.; De Angelis, F.; et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol. Med. 2012, 4, 1214–1229. [Google Scholar] [CrossRef] [PubMed]
- McCann, S.E.; Liu, S.; Wang, D.; Shen, J.; Hu, Q.; Hong, C.C.; Newman, V.A.; Zhao, H. Reduction of dietary glycaemic load modifies the expression of microRNA potentially associated with energy balance and cancer pathways in pre-menopausal women. Br. J. Nutr. 2013, 109, 585–592. [Google Scholar] [CrossRef]
- Kristensen, M.M.; Davidsen, P.K.; Vigelsø, A.; Hansen, C.N.; Jensen, L.J.; Jessen, N.; Bruun, J.M.; Dela, F.; Helge, J.W. miRNAs in human subcutaneous adipose tissue: Effects of weight loss induced by hypocaloric diet and exercise. Obesity 2017, 25, 572–580. [Google Scholar] [CrossRef]
- D’Amore, S.; Vacca, M.; Cariello, M.; Graziano, G.; D’Orazio, A.; Salvia, R.; Sasso, R.C.; Sabbà, C.; Palasciano, G.; Moschetta, A. Genes and miRNA expression signatures in peripheral blood mononuclear cells in healthy subjects and patients with metabolic syndrome after acute intake of extra virgin olive oil. Biochim. Biophys. Acta 2016, 1861, 1671–1680. [Google Scholar] [CrossRef]
- Kern, F.; Ludwig, N.; Backes, C.; Maldener, E.; Fehlmann, T.; Suleymanov, A.; Meese, E.; Hecksteden, A.; Keller, A.; Meyer, T. Systematic Assessment of Blood-Borne MicroRNAs Highlights Molecular Profiles of Endurance Sport and Carbohydrate Uptake. Cells 2019, 8, 1045. [Google Scholar] [CrossRef]
- Chilton, W.L.; Marques, F.Z.; West, J.; Kannourakis, G.; Berzins, S.P.; O’Brien, B.J.; Charchar, F.J. Acute exercise leads to regulation of telomere-associated genes and microRNA expression in immune cells. PLoS ONE 2014, 9, e92088. [Google Scholar] [CrossRef]
- Radom-Aizik, S.; Zaldivar, F.P., Jr.; Haddad, F.; Cooper, D.M. Impact of brief exercise on circulating monocyte gene and microRNA expression: Implications for atherosclerotic vascular disease. Brain Behav. Immun. 2014, 39, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F.; Haddad, F.; Cooper, D.M. Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J. Appl. Physiol. 2013, 114, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F., Jr.; Leu, S.Y.; Adams, G.R.; Oliver, S.; Cooper, D.M. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Version 2. Clin. Transl. Sci. 2012, 5, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F., Jr.; Oliver, S.; Galassetti, P.; Cooper, D.M. Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. J. Appl. Physiol. 2010, 109, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Witard, O.C.; Ball, D. The interaction between nutrition and exercise for promoting health and performance. Proc. Nutr. Soc. 2018, 77, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Baguley, B.J.; Bolam, K.A.; Wright, O.R.L.; Skinner, T.L. The Effect of Nutrition Therapy and Exercise on Cancer-Related Fatigue and Quality of Life in Men with Prostate Cancer: A Systematic Review. Nutrients 2017, 9, 1003. [Google Scholar] [CrossRef]
- Kouvari, M.; Panagiotakos, D.B.; Naumovski, N.; Chrysohoou, C.; Georgousopoulou, E.N.; Yannakoulia, M.; Tousoulis, D.; Pitsavos, C.; ATTICA study Investigators. Dietary anti-inflammatory index, metabolic syndrome and transition in metabolic status; a gender-specific analysis of ATTICA prospective study. Diabetes Res. Clin. Pract. 2020, 161, 108031. [Google Scholar] [CrossRef]
- Zabłocka-Słowińska, K.A.; Skórska, K.; Płaczkowska, S.; Prescha, A.; Pawełczyk, K.; Kosacka, M.; Porębska, I.; Grajeta, H. The relationships between glycemic index and glycemic load of diets and nutritional status and antioxidant/oxidant status in the serum of patients with lung cancer. Adv. Clin. Exp. Med. 2019, 28, 1027–1036. [Google Scholar] [CrossRef]
- Saha, S.K.; Lee, S.B.; Won, J.; Choi, H.Y.; Kim, K.; Yang, G.M.; Dayem, A.A.; Cho, S.G. Correlation between Oxidative Stress, Nutrition, and Cancer Initiation. Int. J. Mol. Sci. 2017, 18, 1544. [Google Scholar] [CrossRef]
- Mentella, M.C.; Scaldaferri, F.; Ricci, C.; Gasbarrini, A.; Miggiano, G.A.D. Cancer and Mediterranean Diet: A Review. Nutrients 2019, 11, 2059. [Google Scholar] [CrossRef]
- Hardman, W.E. Diet components can suppress inflammation and reduce cancer risk. Nutr. Res. Pract. 2014, 8, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Candido, S.; Lupo, G.; Pennisi, M.; Basile, M.S.; Anfuso, C.D.; Petralia, M.C.; Gattuso, G.; Vivarelli, S.; Spandidos, D.A.; Libra, M.; et al. The analysis of miRNA expression profiling datasets reveals inverse microRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019, 42, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Lupo, G.; La Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.D.; Salinas, E.A.; Newtson, A.M.; Sharma, D.; Keeney, M.E.; Warrier, A.; Smith, B.J.; Bender, D.P.; Goodheart, M.J.; Thiel, K.W.; et al. An integrated prediction model of recurrence in endometrial endometrioid cancers. Cancer Manag. Res. 2019, 11, 5301–5315. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Candido, S.; Salemi, R.; Basile, M.S.; Scalisi, A.; McCubrey, J.A.; Torino, F.; Signorelli, S.S.; Montella, M.; Libra, M. Computational identification of microRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget 2016, 7, 72758–72766. [Google Scholar] [CrossRef]
- Hafsi, S.; Candido, S.; Maestro, R.; Falzone, L.; Soua, Z.; Bonavida, B.; Spandidos, D.A.; Libra, M. Correlation between the overexpression of Yin Yang 1 and the expression levels of miRNAs in Burkitt’s lymphoma: A computational study. Oncol. Lett. 2016, 11, 1021–1025. [Google Scholar] [CrossRef]
- Huang, G.L.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G.A. MiR-200 family and cancer: From a meta-analysis view. Mol. Asp. Med. 2019, 70, 57–71. [Google Scholar] [CrossRef]
- Wang, H.; Tan, Z.; Hu, H.; Liu, H.; Wu, T.; Zheng, C.; Wang, X.; Luo, Z.; Wang, J.; Liu, S.; et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer 2019, 19, 738. [Google Scholar] [CrossRef]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Coker-Gurkan, A.; Bulut, D.; Genc, R.; Arisan, E.D.; Obakan-Yerlikaya, P.; Palavan-Unsal, N. Curcumin prevented human autocrine growth hormone (GH) signaling mediated NF-κB activation and miR-183-96-182 cluster stimulated epithelial mesenchymal transition in T47D breast cancer cells. Mol. Biol. Rep. 2019, 46, 355–369. [Google Scholar] [CrossRef]
- Zhang, Q.; Ren, W.; Huang, B.; Yi, L.; Zhu, H. MicroRNA-183/182/96 cooperatively regulates the proliferation of colon cancer cells. Mol. Med. Rep. 2015, 12, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sheng, C.; Huang, L.; Zhang, H.; Huang, L.; Cheng, Z.; Zhu, Q. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014, 16, 473. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mou, Q.; Li, P.; Yang, Z.; Wang, Z.; Niu, J.; Liu, Y.; Sun, Z.; Lv, S.; Zhang, B.; et al. MiR-486-5p inhibits IL-22-induced epithelial-mesenchymal transition of breast cancer cell by repressing Dock1. J. Cancer 2019, 10, 4695–4706. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Humphries, B.; Yang, C.; Wang, Z. MiR-205 Dysregulations in Breast Cancer: The Complexity and Opportunities. Noncoding RNA 2019, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Pajic, M.; Froio, D.; Daly, S.; Doculara, L.; Millar, E.; Graham, P.H.; Drury, A.; Steinmann, A.; de Bock, C.E.; Boulghourjian, A.; et al. miR-139-5p Modulates Radiotherapy Resistance in Breast Cancer by Repressing Multiple Gene Networks of DNA Repair and ROS Defense. Cancer Res. 2018, 78, 501–515. [Google Scholar] [CrossRef]
- Xia, M.; Li, H.; Wang, J.J.; Zeng, H.J.; Wang, S.H. MiR-99a suppress proliferation, migration and invasion through regulating insulin-like growth factor 1 receptor in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1755–1763. [Google Scholar]
- Gao, L.; Guo, Q.; Li, X.; Yang, X.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; Xing, Y.; Xi, T.; et al. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine 2019, 41, 395–407. [Google Scholar] [CrossRef]
- Chen, J.; Shin, V.Y.; Siu, M.T.; Ho, J.C.; Cheuk, I.; Kwong, A. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer 2016, 16, 887. [Google Scholar] [CrossRef]
- Fedele, M.; Cerchia, L.; Chiappetta, G. The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas. Cancers 2017, 9, 134. [Google Scholar] [CrossRef]
- Rakha, E.A.; Green, A.R. Molecular classification of breast cancer: What the pathologist needs to know. Pathology 2017, 49, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, F.; Ferns, G.A.; Talebian, S.; Nourbakhsh, M.; Avan, A.; Shahidsales, S. Role of regulatory miRNAs of the PI3K/AKT signaling pathway in the pathogenesis of breast cancer. Gene 2020, 737, 144459. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, T.; Mao, J.; Zhang, J.; Fan, S.; Lu, Y.; Sun, Z.; Zhang, Q.; Song, B.; Li, L. PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2019, 38, 256. [Google Scholar] [CrossRef] [PubMed]
- Kjaer, I.M.; Bechmann, T.; Brandslund, I.; Madsen, J.S. Prognostic and predictive value of EGFR and EGFR-ligands in blood of breast cancer patients: A systematic review. Clin. Chem. Lab. Med. 2018, 56, 688–701. [Google Scholar] [CrossRef]
- Huang, J.; Luo, Q.; Xiao, Y.; Li, H.; Kong, L.; Ren, G. The implication from RAS/RAF/ERK signaling pathway increased activation in epirubicin treated triple negative breast cancer. Oncotarget 2017, 8, 108249–108260. [Google Scholar] [CrossRef]
- Tobin, N.P.; Bergh, J. Analysis of Cyclin D1 in Breast Cancer: A Call to Arms. Curr. Breast Cancer Rep. 2012, 4, 171–173. [Google Scholar] [CrossRef]
- Kim, I.; Choi, S.; Kim, S. BRCA-Pathway: A structural integration and visualization system of TCGA breast cancer data on KEGG pathways. BMC Bioinform. 2018, 19, 42. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Q.; Jiang, X.Q.; Li, D.Q.; Jiang, X.C.; Wu, X.B.; Cao, Y.L. miR-146a promoted breast cancer proliferation and invasion by regulating NM23-H1. J. Biochem. 2020, 167, 41–48. [Google Scholar] [CrossRef]
- Marques, M.M.; Evangelista, A.F.; Macedo, T.; Vieira, R.A.D.C.; Scapulatempo-Neto, C.; Reis, R.M.; Carvalho, A.L.; Silva, I.D.C.G.D. Expression of tumor suppressors miR-195 and let-7a as potential biomarkers of invasive breast cancer. Clinics 2018, 73, e184. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, G.; Sun, R.; Pan, X.; Wang, X.; Li, H.; Sun, Y. miR-185-5p inhibits F-actin polymerization and reverses epithelial mesenchymal transition of human breast cancer cells by modulating RAGE. Mol. Med. Rep. 2018, 18, 2621–2630. [Google Scholar] [CrossRef]
- Maskey, N.; Li, D.; Xu, H.; Song, H.; Wu, C.; Hua, K.; Song, J.; Fang, L. MicroRNA-340 inhibits invasion and metastasis by downregulating ROCK1 in breast cancer cells. Oncol. Lett. 2017, 14, 2261–2267. [Google Scholar] [CrossRef]
- Zearo, S.; Kim, E.; Zhu, Y.; Zhao, J.T.; Sidhu, S.B.; Robinson, B.G.; Soon, P.S. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer 2014, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- McAnena, P.; Tanriverdi, K.; Curran, C.; Gilligan, K.; Freedman, J.E.; Brown, J.A.L.; Kerin, M.J. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 2019, 19, 436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Sun, D.W.; Mao, L.; Zhang, J.; Jiang, L.H.; Li, J.; Wu, Y.; Ji, H.; Chen, W.; Wang, J.; et al. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem. Biophys. Res. Commun. 2015, 465, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Tuaeva, N.O.; Falzone, L.; Porozov, Y.B.; Nosyrev, A.E.; Trukhan, V.M.; Kovatsi, L.; Spandidos, D.A.; Drakoulis, N.; Kalogeraki, A.; Mamoulakis, C.; et al. Translational Application of Circulating DNA in Oncology: Review of the Last Decades Achievements. Cells 2019, 8, 1251. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef]
- Filetti, V.; Falzone, L.; Rapisarda, V.; Caltabiano, R.; Eleonora Graziano, A.C.; Ledda, C.; Loreto, C. Modulation of microRNA expression levels after naturally occurring asbestiform fibers exposure as a diagnostic biomarker of mesothelial neoplastic transformation. Ecotoxicol. Environ. Saf. 2020, 198, 110640. [Google Scholar] [CrossRef]
- Battaglia, R.; Palini, S.; Vento, M.E.; La Ferlita, A.; Lo Faro, M.J.; Caroppo, E.; Borzì, P.; Falzone, L.; Barbagallo, D.; Ragusa, M.; et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci. Rep. 2019, 9, 84. [Google Scholar] [CrossRef]
- Salemi, R.; Falzone, L.; Madonna, G.; Polesel, J.; Cinà, D.; Mallardo, D.; Ascierto, P.A.; Libra, M.; Candido, S. MMP-9 as a Candidate Marker of Response to BRAF Inhibitors in Melanoma Patients With BRAFV600E Mutation Detected in Circulating-Free DNA. Front. Pharmacol. 2018, 9, 856. [Google Scholar] [CrossRef]
- Falzone, L.; Scola, L.; Zanghì, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging 2018, 10, 1000–1014. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Romano, G.L.; Salemi, R.; Bucolo, C.; Tomasello, B.; Lupo, G.; Anfuso, C.D.; Spandidos, D.A.; Libra, M.; Candido, S. Prognostic significance of deregulated microRNAs in uveal melanomas. Mol. Med. Rep. 2019, 19, 2599–2610. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]





| Series Accession | Number of Control | Number of BC Tissue | Total Number | Samples | Platform | Ref. |
|---|---|---|---|---|---|---|
| Breast Cancer Datasets | ||||||
| GSE57897 | 31 | 422 | 453 | Normal and BC tissues | GPL18722 Homo sapiens microRNA array | [38] |
| GSE97811 | 16 | 45 | 61 | Normal and BC FFPE tissues | GPL21263 3D-Gene Human miRNA V21_1.0.0 | [39] |
| GSE58606 | 11 | 122 | 133 | Normal and BC FFPE tissues | GPL18838 miRCURY LNA microRNA Array 7th generation | [40] |
| GSE38167 | 23 | 31 | 54 | Normal and BC FFPE tissues | GPL14943 Agilent-029297 Human miRNA Microarray | [41] |
| GSE32922 | 15 | 22 | 37 | Normal and BC tissues | GPL7723 miRCURY LNA microRNA Array, v.11.0 | [42] |
| GSE45666 | 15 | 101 | 116 | Normal and BC tissues | GPL14767 Agilent-021827 Human miRNA Microarray | [43] |
| GSE26666 | 17 | 77 | 94 | Frozen normal and BC tissues | GPL8227 Agilent-019118 Human miRNA Microarray 2.0 | [44] |
| GSE40525 | 59 | 61 | 120 | Normal and BC FFPE tissues | GPL8227 Agilent-019118 Human miRNA Microarray 2.0 | [45] |
| Samples before Diet | Samples after Diet | Diet Datasets | ||||
| GSE27474 | 14 | 14 | 28 | Serum Samples | GPL8179 Illumina Human v2 MicroRNA expression beadchip | [46] |
| GSE87103 | 12 | 12 | 24 | Subcutaneous adipose tissues | GPL11434 miRCURY LNA microRNA Array, 6th generation | [47] |
| GSE75026 | 12 | 12 | 24 | PBMCs samples | GPL8179 Illumina Human v2 MicroRNA expression beadchip | [48] |
| Samples before Exercise | Samples after Exercise | Exercise Datasets | ||||
| GSE133910 | 46 | 44 | 90 | Peripheral blood samples | GPL25134 Agilent-070156 Human_miRNA_V21.0_Microarray | [49] |
| GSE87103 | 12 | 12 | 24 | Subcutaneous adipose tissues | GPL11434 miRCURY LNA microRNA Array, 6th generation | [47] |
| GSE45041 | 10 | 10 | 20 | White blood cells | GPL16770 Agilent-031181 Unrestricted Human miRNA V16.0 | [50] |
| GSE51837 | 12 | 12 | 24 | Monocytes | GPL10850 Agilent-021827 Human miRNA Microarray (V3) | [51] |
| GSE41915 | 11 | 11 | 22 | NK cells | GPL10850 Agilent-021827 Human miRNA Microarray (V3) | [52] |
| GSE28745 | 12 | 12 | 24 | PBMCs | GPL8227 Agilent-019118 Human miRNA Microarray 2.0 | [53] |
| GSE18999 | 11 | 11 | 22 | Neutrophils | GPL7731 Agilent-019118 Human miRNA Microarray 2.0 | [54] |
| miRNA | GEO Datasets log2FC Average | TCGA BRCA | |
|---|---|---|---|
| p-Value | |||
| Upregulated miRNAs | |||
| hsa-miR-103a-3p | 1.044 | 1.532 | 1.031 × 10−19 |
| hsa-miR-106b-5p | 1.000 | 2.423 | 7.270 × 10−28 |
| hsa-miR-107 | 0.882 | 1.676 | 3.424 × 10−17 |
| hsa-miR-141-3p | 1.476 | 5.343 | 2.723 × 10−29 |
| hsa-miR-142-3p | 1.775 | 3.040 | 2.214 × 10−18 |
| hsa-miR-142-5p | 1.413 | 2.283 | 1.313 × 10−13 |
| hsa-miR-146a-5p * | 0.902 | #N/D | #N/D |
| hsa-miR-155-5p | 1.443 | 1.961 | 4.674 × 10−18 |
| hsa-miR-15b-5p | 1.076 | 1.908 | 1.745 × 10−11 |
| hsa-miR-16-5p | 0.955 | 1.591 | 2.960 × 10−8 |
| hsa-miR-181b-5p | 0.616 | 2.219 | 1.238 × 10−22 |
| hsa-miR-182-5p | 2.027 | 4.799 | 2.421 × 10−40 |
| hsa-miR-183-5p | 2.759 | 7.102 | 3.388 × 10−47 |
| hsa-miR-185-5p | 0.883 | 1.548 | 2.932 × 10−9 |
| hsa-miR-18a-5p | 1.714 | 1.905 | 5.766 × 10−14 |
| hsa-miR-200a-3p | 1.346 | 3.948 | 2.352 × 10−26 |
| hsa-miR-200b-3p | 1.564 | 2.846 | 4.468 × 10−20 |
| hsa-miR-200c-3p | 1.298 | 2.691 | 2.049 × 10−17 |
| hsa-miR-203a-3p | 1.055 | 2.287 | 2.680 × 10−14 |
| hsa-miR-21-5p | 2.054 | 4.448 | 5.680 × 10−41 |
| hsa-miR-210-3p | 1.244 | 5.632 | 1.111 × 10−31 |
| hsa-miR-25-3p * | 0.946 | #N/D | #N/D |
| hsa-miR-301a-3p | 1.620 | 2.314 | 7.058 × 10−24 |
| hsa-miR-340-5p | 0.924 | 2.001 | 8.384 × 10−25 |
| hsa-miR-425-5p | 1.733 | 1.740 | 8.393 × 10−8 |
| hsa-miR-429 | 1.751 | 4.884 | 6.208 × 10−33 |
| hsa-miR-484 | 0.792 | 1.399 | 2.801 × 10−6 |
| hsa-miR-7-5p | 1.786 | 1.850 | 7.483 × 10−21 |
| hsa-miR-93-5p | 1.209 | 2.110 | 5.223 × 10−26 |
| hsa-miR-96-5p | 2.178 | 6.970 | 7.515 × 10−47 |
| Downregulated miRNAs | |||
| hsa-miR-100-5p | −1.574 | −4.066 | 1.191 × 10−52 |
| hsa-miR-1229-3p | −0.819 | 1.274 | 2.105 × 10−6 |
| hsa-miR-125b-5p | −1.819 | −4.107 | 6.680 × 10−48 |
| hsa-miR-130a-3p | −0.996 | −2.371 | 2.085 × 10−30 |
| hsa-miR-139-3p | −1.178 | −8.512 | 2.131 × 10−52 |
| hsa-miR-139-5p | −2.444 | −8.699 | 6.482 × 10−58 |
| hsa-miR-143-3p | −1.422 | −2.288 | 1.458 × 10−28 |
| hsa-miR-145-3p | −1.501 | −3.196 | 7.255 × 10−54 |
| hsa-miR-145-5p | −2.015 | −5.892 | 2.501 × 10−50 |
| hsa-miR-195-5p | −1.541 | −2.867 | 1.560 × 10−39 |
| hsa-miR-202-3p * | −1.198 | #N/D | #N/D |
| hsa-miR-205-5p | −2.033 | −5.921 | 4.198 × 10−35 |
| hsa-miR-335-5p | −1.961 | −4.846 | 4.980 × 10−27 |
| hsa-miR-376a-3p | −1.441 | −1.364 | 2.287 × 10−8 |
| hsa-miR-377-3p | −1.542 | −1.418 | 2.186 × 10−7 |
| hsa-miR-381-3p | −1.523 | −2.486 | 3.350 × 10−26 |
| hsa-miR-486-5p | −1.496 | −10.611 | 3.737 × 10−26 |
| hsa-miR-497-5p | −1.254 | −2.726 | 1.543 × 10−33 |
| hsa-miR-99a-5p | −1.433 | −5.165 | 6.270 × 10−63 |
| KEGG Pathway | p-Value | N. of Targeted Genes | Involved miRNAs |
|---|---|---|---|
| Pathways in cancer (hsa05200) | 4.05564 × 10−7 | 179 | 10 |
| PI3K-Akt signaling pathway (hsa04151) | 0.011123026 | 140 | 10 |
| Proteoglycans in cancer (hsa05205) | 2.21725 × 10−17 | 107 | 10 |
| MAPK signaling pathway (hsa04010) | 0.011123026 | 104 | 10 |
| Viral carcinogenesis (hsa05203) | 6.08511 × 10−8 | 95 | 10 |
| Transcriptional misregulation in cancer (hsa05202) | 0.002760238 | 80 | 10 |
| Hippo signaling pathway (hsa04390) | 1.31008 × 10−8 | 76 | 10 |
| FoxO signaling pathway (hsa04068) | 1.48491 × 10−6 | 71 | 10 |
| Cell cycle (hsa04110) | 3.14391 × 10−5 | 66 | 10 |
| Insulin signaling pathway (hsa04910) | 0.009388457 | 65 | 10 |
| AMPK signaling pathway (hsa04152) | 0.006097228 | 62 | 10 |
| Thyroid hormone signaling pathway (hsa04919) | 0.000182888 | 61 | 10 |
| HIF-1 signaling pathway (hsa04066) | 3.77879 × 10−5 | 57 | 10 |
| TNF signaling pathway (hsa04668) | 0.014299582 | 54 | 10 |
| Prostate cancer (hsa05215) | 2.74229 × 10−5 | 52 | 10 |
| Small cell lung cancer (hsa05222) | 2.1672 × 10−5 | 50 | 10 |
| Estrogen signaling pathway (hsa04915) | 0.00014031 | 47 | 10 |
| ErbB signaling pathway (hsa04012) | 0.001313884 | 45 | 9 |
| Chronic myeloid leukemia (hsa05220) | 1.45098 × 10−6 | 44 | 10 |
| Renal cell carcinoma (hsa05211) | 1.69642 × 10−7 | 42 | 10 |
| TGF-beta signaling pathway (hsa04350) | 4.62474 × 10−6 | 41 | 10 |
| Pancreatic cancer (hsa05212) | 9.61555 × 10−6 | 40 | 10 |
| Colorectal cancer (hsa05210) | 5.0244 × 10−7 | 39 | 10 |
| p53 signaling pathway (hsa04115) | 0.00014031 | 39 | 10 |
| Apoptosis (hsa04210) | 0.018765462 | 38 | 10 |
| Glioma (hsa05214) | 2.56338 × 10−6 | 37 | 10 |
| Central carbon metabolism in cancer (hsa05230) | 2.56338 × 10−6 | 36 | 10 |
| Non-small cell lung cancer (hsa05223) | 8.72499 × 10−6 | 34 | 10 |
| ECM-receptor interaction (hsa04512) | 1.96805 × 10−5 | 34 | 10 |
| mTOR signaling pathway (hsa04150) | 0.003253273 | 34 | 10 |
| Melanoma (hsa05218) | 0.009412459 | 34 | 10 |
| Endometrial cancer (hsa05213) | 3.18028 × 10−5 | 32 | 10 |
| Acute myeloid leukemia (hsa05221) | 0.019544711 | 29 | 10 |
| Bladder cancer (hsa05219) | 0.000423154 | 25 | 9 |
| Thyroid cancer (hsa05216) | 0.000804104 | 17 | 10 |
| KEGG Pathway | p-Value | N. of Targeted Genes | Involved miRNAs |
|---|---|---|---|
| Adherens junction (hsa04520) | 0.00127981 | 10 | 2 |
| Choline metabolism in cancer (hsa05231) | 0.0417592 | 10 | 2 |
| Epithelial cell signaling in Helicobacter pylori infection (hsa05120) | 0.04463459 | 9 | 3 |
| Colorectal cancer (hsa05210) | 0.02665458 | 8 | 2 |
| Glioma (hsa05214) | 0.02665458 | 7 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falzone, L.; Grimaldi, M.; Celentano, E.; Augustin, L.S.A.; Libra, M. Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers 2020, 12, 2555. https://doi.org/10.3390/cancers12092555
Falzone L, Grimaldi M, Celentano E, Augustin LSA, Libra M. Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers. 2020; 12(9):2555. https://doi.org/10.3390/cancers12092555
Chicago/Turabian StyleFalzone, Luca, Maria Grimaldi, Egidio Celentano, Livia S. A. Augustin, and Massimo Libra. 2020. "Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity" Cancers 12, no. 9: 2555. https://doi.org/10.3390/cancers12092555
APA StyleFalzone, L., Grimaldi, M., Celentano, E., Augustin, L. S. A., & Libra, M. (2020). Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers, 12(9), 2555. https://doi.org/10.3390/cancers12092555