Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions
Abstract
:1. A Brief Historical Perspective
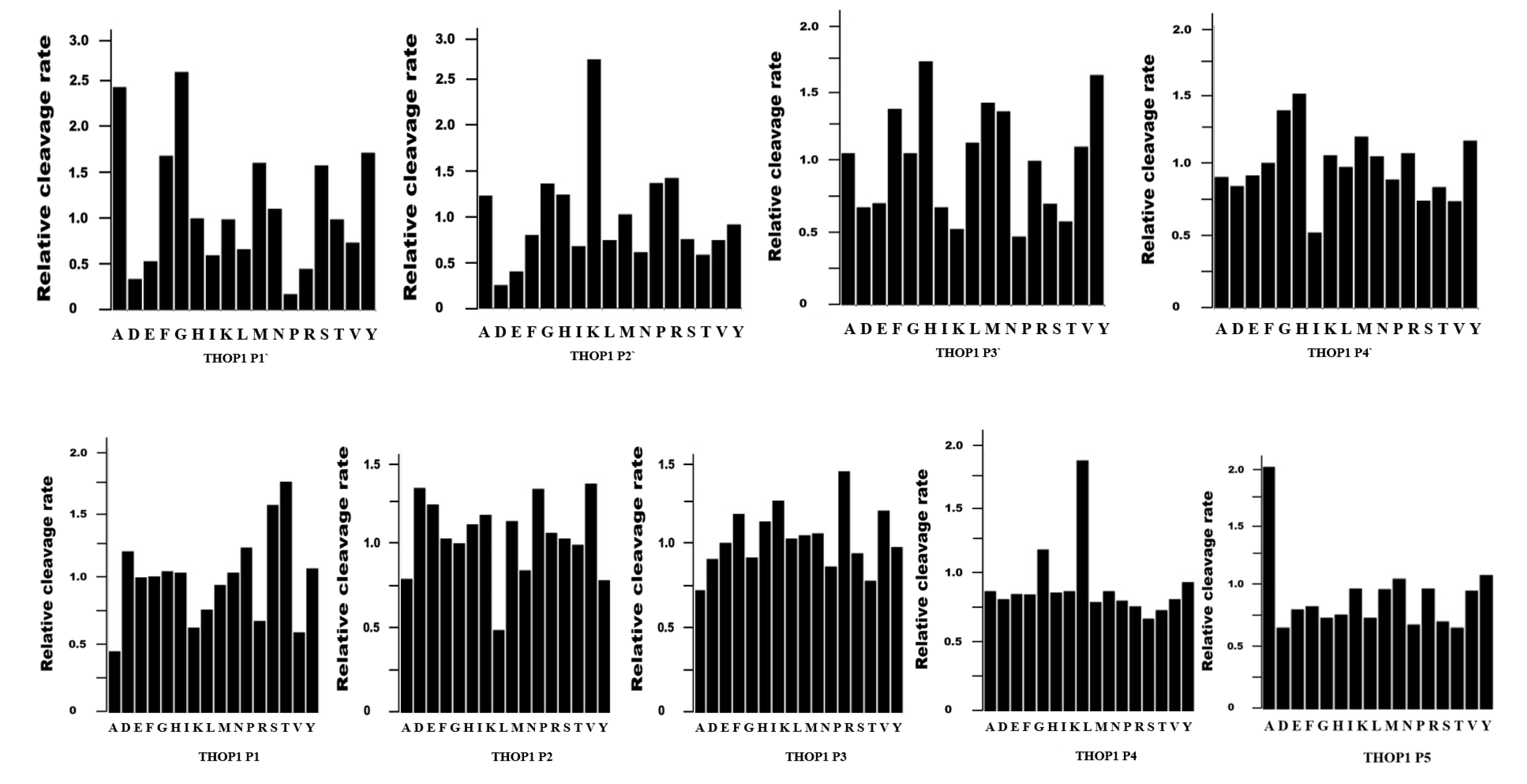
2. THOP1 Substrates: Only Size Matters
3. THOP1 Provides a Major Contribution to Major Histocompatibility Complex of Class I (MHC-I) Antigen Presentation
4. Seminal Experiments Evidencing that Intracellular Peptides (InPeps) Generated by Proteasome Activity, were Naturally Present in Cells and Tissues
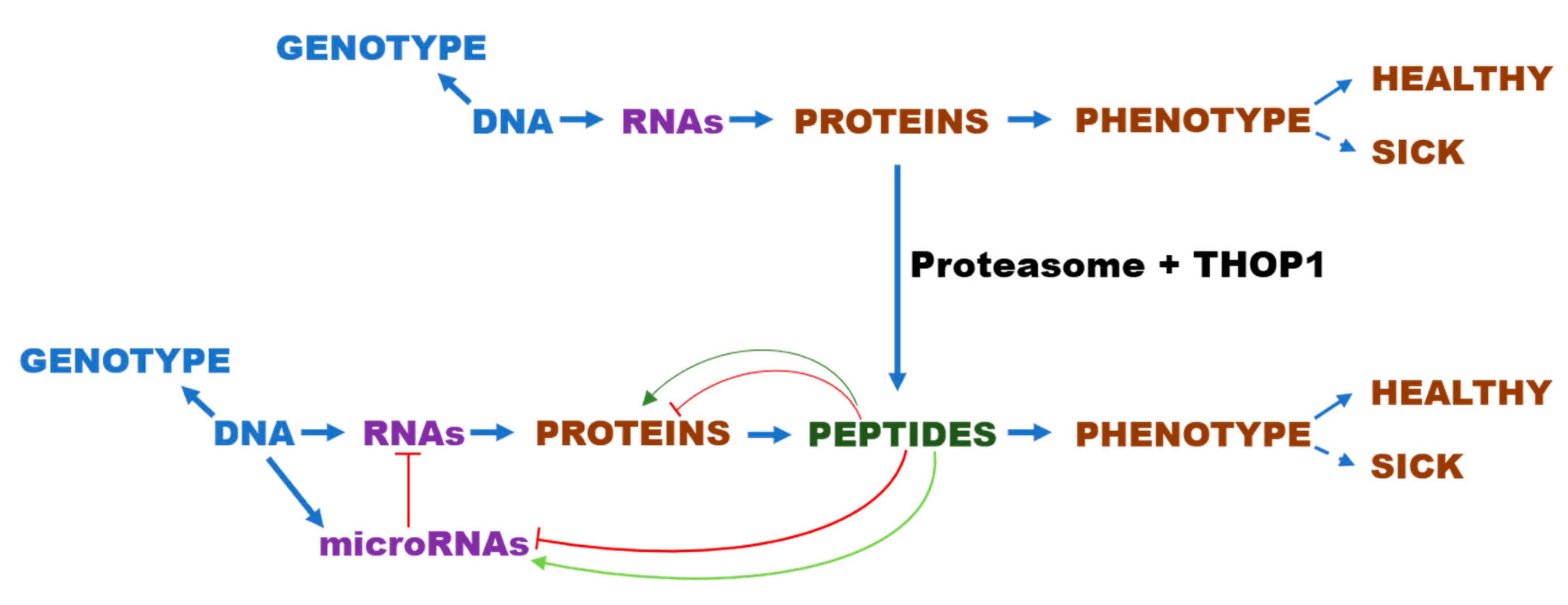
5. Identification of Functional InPeps Suggests that Protein Degradation by Proteasome Is Not the End, But Rather a Next Step in Protein Function
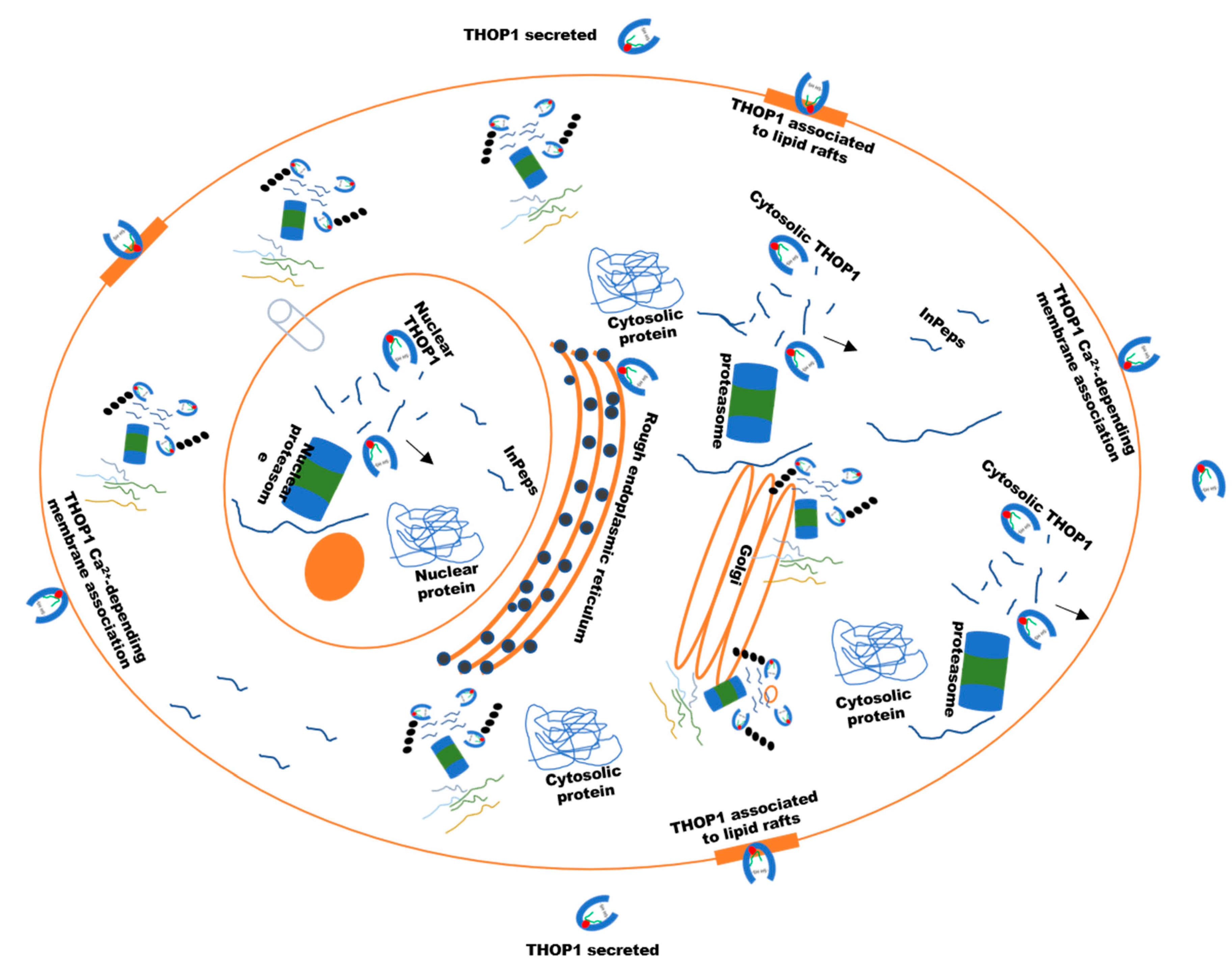
6. The Biological Significance of THOP1 Represented through both Neuropeptides and InPeps
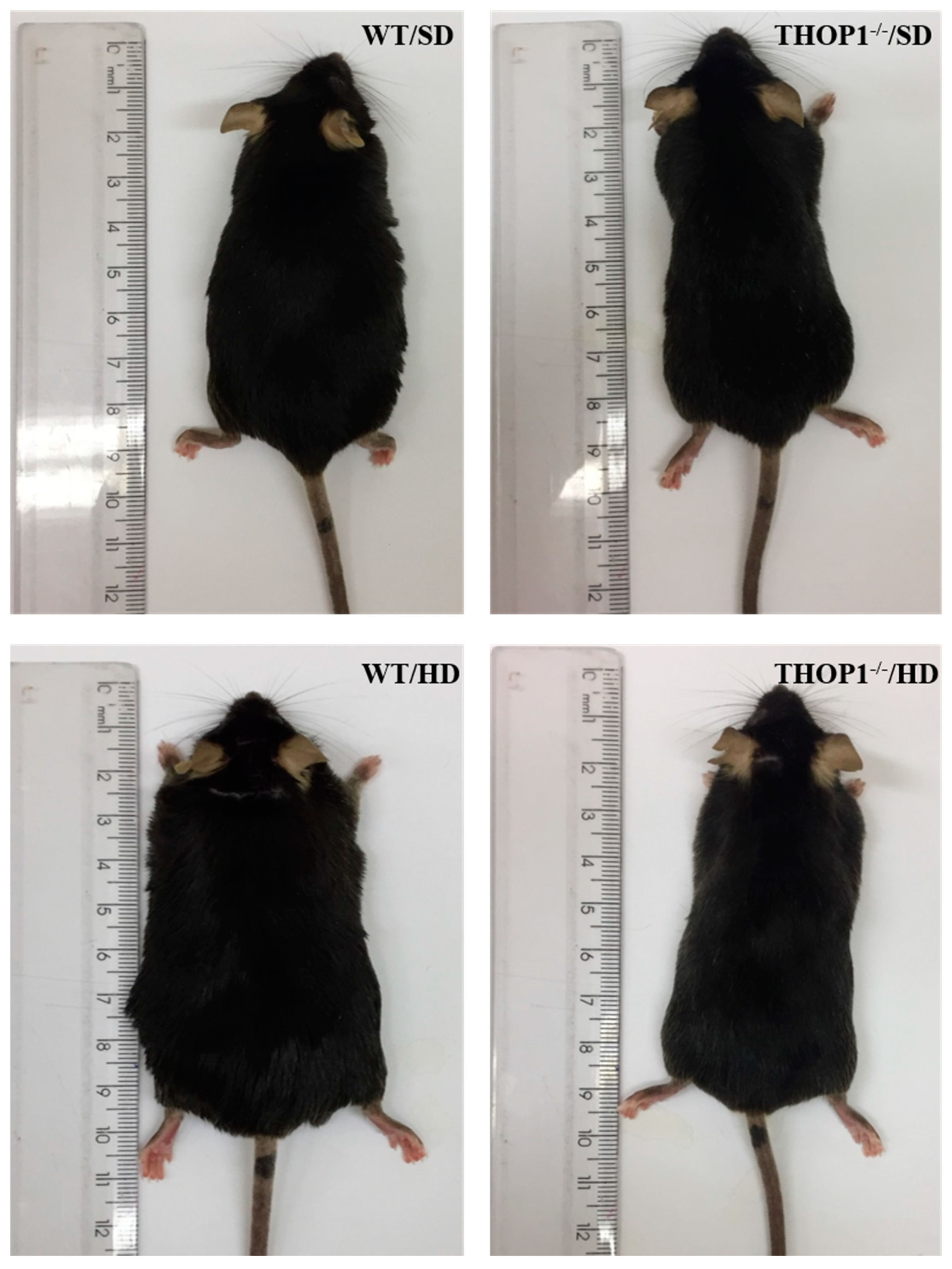
7. THOP1 Plays Key Unanticipated Physiological Functions on Energy Metabolism Regulation
8. THOP1 Relevance in Human Diseases and Diagnostics
9. Chemical Inhibitors of THOP1
10. Concluding Remarks and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
Non-Standard Abbreviations
| thimet oligopeptidase | THOP1 |
| angiotensin converting enzyme 1 | ACE1 |
| neprilysin | NEP |
| dipeptidyl peptidase IV | DPP4 |
| prolyl oligopeptidase | POP |
| insulin degrading enzyme | IDE |
| neurolysin | Nln |
| cluster of differentiation 36/fatty acid translocase | CD36/FAT |
| lipoprotein lipase | LPL |
| macrophage mannose receptor 1 | CD206 |
| integrin alpha-X | CD11C |
| murine macrophage F4/80 | glycoprotein (F4/80) |
| peroxisome proliferator-activated receptor gamma | PPAR-γ |
| fatty acid synthase | FAS |
References
- Orlowski, M.; Michaud, C.; Chu, T.G. A soluble metalloendopeptidase from rat brain. Purification of the enzyme and determination of specificity with synthetic and natural peptides. Eur. J. Biochem. 1983, 135, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.J.; Brown, M.A.; Dando, P.M.; Knight, C.G.; McKie, N.; Rawlings, N.D.; Serizawa, A. Thimet oligopeptidase and oligopeptidase M or neurolysin. Methods Enzym. 1995, 248, 529–556. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of peptidases. Biochem. J. 1993, 290, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Barrett, A.J. Thimet oligopeptidase (EC 3.4.24.15): The same by any name? Biochem. J. 1991, 277, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Tisljar, U.; Barrett, A.J. Thiol-dependent metallo-endopeptidase characteristics of Pz-peptidase in rat and rabbit. Biochem. J. 1990, 267, 531–533. [Google Scholar] [CrossRef] [Green Version]
- Tisljar, U.; de Camargo, A.C.; da Costa, C.A.; Barrett, A.J. Activity of Pz-peptidase and endo-oligopeptidase are due to the same enzyme. Biochem. Biophys. Res. Commun. 1989, 162, 1460–1464. [Google Scholar] [CrossRef]
- Barrett, A.J.; Brown, M.A. Chicken liver Pz-peptidase, a thiol-dependent metallo-endopeptidase. Biochem. J. 1990, 271, 701–706. [Google Scholar] [CrossRef] [Green Version]
- Shrimpton, C.N.; Glucksman, M.J.; Lew, R.A.; Tullai, J.W.; Margulies, E.H.; Roberts, J.L.; Smith, A.I. Thiol activation of endopeptidase EC 3.4.24.15. A novel mechanism for the regulation of catalytic activity. J. Biol. Chem. 1997, 272, 17395–17399. [Google Scholar] [CrossRef] [Green Version]
- Demasi, M.; Piassa Filho, G.M.; Castro, L.M.; Ferreira, J.C.; Rioli, V.; Ferro, E.S. Oligomerization of the cysteinyl-rich oligopeptidase EP24.15 is triggered by S-glutathionylation. Free Radic. Biol. Med. 2008, 44, 1180–1190. [Google Scholar] [CrossRef]
- Pierotti, A.; Dong, K.W.; Glucksman, M.J.; Orlowski, M.; Roberts, J.L. Molecular cloning and primary structure of rat testes metalloendopeptidase EC 3.4.24.15. Biochemistry 1990, 29, 10323–10329. [Google Scholar] [CrossRef]
- McKie, N.; Dando, P.M.; Rawlings, N.D.; Barrett, A.J. Thimet oligopeptidase: Similarity to ‘soluble angiotensin II-binding protein’ and some corrections to the published amino acid sequence of the rat testis enzyme. Biochem. J. 1993, 295, 57–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummins, P.M.; Pabon, A.; Margulies, E.H.; Glucksman, M.J. Zinc coordination and substrate catalysis within the neuropeptide processing enzyme endopeptidase EC 3.4.24.15. Identification of active site histidine and glutamate residues. J. Biol. Chem. 1999, 274, 16003–16009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rioli, V.; Gozzo, F.C.; Heimann, A.S.; Linardi, A.; Krieger, J.E.; Shida, C.S.; Almeida, P.C.; Hyslop, S.; Eberlin, M.N.; Ferro, E.S. Novel natural peptide substrates for endopeptidase 24.15, neurolysin, and angiotensin-converting enzyme. J. Biol. Chem. 2003, 278, 8547–8555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, K.; Hines, C.S.; Coll-Rodriguez, J.; Rodgers, D.W. Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization. J. Biol. Chem. 2004, 279, 20480–20489. [Google Scholar] [CrossRef] [Green Version]
- Malvezzi, A.; Higa, P.M.; Antonia, T.d.A.; Silva, G.M.; Gozzo, F.C.; Ferro, E.S.; Castro, L.M.; de Rezende, L.; Monteiro, G.; Demasi, M. The cysteine-rich protein thimet oligopeptidase as a model of the structural requirements for S-glutathiolation and oxidative oligomerization. PLoS ONE 2012, 7, e39408. [Google Scholar] [CrossRef] [Green Version]
- Dando, P.M.; Brown, M.A.; Barrett, A.J. Human thimet oligopeptidase. Biochem. J. 1993, 294, 451–457. [Google Scholar] [CrossRef]
- Acker, G.R.; Molineaux, C.; Orlowski, M. Synaptosomal membrane-bound form of endopeptidase-24.15 generates Leu-enkephalin from dynorphin1-8, alpha- and beta-neoendorphin, and Met-enkephalin from Met-enkephalin-Arg6-Gly7-Leu8. J. Neurochem. 1987, 48, 284–292. [Google Scholar] [CrossRef]
- Ferro, E.S.; Tambourgy, D.V.; Abreu, P.A.; Camargo, A.C.; Raw, I.; Ho, P.L. Characterization of an endooligopeptidase A-like protein in PC12 cells: Activity modulation by cAMP but not by basic fibroblast growth factor. J. Cell. Biochem. 1995, 57, 311–320. [Google Scholar] [CrossRef]
- Ferro, E.S.; Tambourgi, D.V.; Gobersztejn, F.; Gomes, M.D.; Sucupira, M.; Armelin, M.C.; Kipnis, T.L.; Camargo, A.C. Secretion of a neuropeptide-metabolizing enzyme similar to endopeptidase 22.19 by glioma C6 cells. Biochem. Biophys. Res. Commun. 1993, 191, 275–281. [Google Scholar] [CrossRef]
- Gomes, M.D.; Juliano, L.; Ferro, E.S.; Matsueda, R.; Camargo, A.C.M. Dynorphin-derived peptides reveal the presence of a critical cysteine for the activity of brain endo-oligopeptidase A. Biochem. Biophys. Res. Commun. 1993, 197. [Google Scholar] [CrossRef]
- Ferro, E.S.; Sucupira, M.; Marques, N.; Camargo, A.C.M.; Menna-Barreto, L. Circadian rhythm of the endopeptidase 22.19 (EC 3.4.22.19) in the rat brain. Chronobiol. Int. 1992, 9. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Hamassaki, D.E.; Camargo, A.C.; Britto, L.R. Endo-oligopeptidase A, a putative enkephalin-generating enzyme, in the vertebrate retina. J. Neurochem. 1991, 57, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Juliano, L.; Chagas, J.R.; Hirata, I.Y.; Carmona, E.; Sucupira, M.; Oliveira, E.S.; Oliveira, E.B.; Camargo, A.C. A selective assay for endooligopeptidase A based on the cleavage of fluorogenic substrate structurally related to enkephalin. Biochem. Biophys. Res. Commun. 1990, 173, 647–652. [Google Scholar] [CrossRef]
- Derewenda, U.; Tarricone, C.; Choi, W.C.; Cooper, D.R.; Lukasik, S.; Perrina, F.; Tripathy, A.; Kim, M.H.; Cafiso, D.S.; Musacchio, A.; et al. The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure 2007, 15, 1467–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Li, F.; Liang, Y.; Shen, Y.; Zhao, X.; Huang, Q.; Zhu, X. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 2003, 23, 1239–1250. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, M.A.; Guerreiro, J.R.; Charych, E.; Kamiya, A.; Barbosa, R.L.; Machado, M.F.; Campeiro, J.D.; Oliveira, V.; Sawa, A.; Camargo, A.C.; et al. Assessing the role of endooligopeptidase activity of Ndel1 (nuclear-distribution gene E homolog like-1) in neurite outgrowth. Mol. Cell. Neurosci. 2010, 44, 353–361. [Google Scholar] [CrossRef]
- Hayashi, M.A.; Portaro, F.C.; Bastos, M.F.; Guerreiro, J.R.; Oliveira, V.; Gorrao, S.S.; Tambourgi, D.V.; Sant’Anna, O.A.; Whiting, P.J.; Camargo, L.M.; et al. Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc. Natl. Acad. Sci. USA 2005, 102, 3828–3833. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, M.A.F.; Portaro, F.C.V.; Tambourgi, D.V.; Sucupira, M.; Yamane, T.; Fernandes, B.L.; Ferro, E.S.; Reboucas, N.A.; De Camargo, A.C.M. Erratum: Molecular and immunochemical evidences demonstrate that endooligopeptidase A is the predominant cytosolic oligopeptidase of rabbit brain (Biochemical and Biophysical Research Communications (2000) 269, 1 (7-13)). Biochem. Biophys. Res. Commun. 2000, 272. [Google Scholar]
- Hayashi, M.A.; Gomes, M.D.; Rebouças, N.A.; Fernandes, B.L.; Ferro, E.S.; de Camargo, A.C. Species specificity of thimet oligopeptidase (EC 3.4.24.15). Biol. Chem. Hoppe-Seyler 1996, 377, 283–291. [Google Scholar] [CrossRef]
- Dal Mas, C.; Nani, J.V.; Noto, C.; Yonamine, C.M.; da Cunha, G.R.; Mansur, R.B.; Ota, V.K.; Belangero, S.I.; Cordeiro, Q.; Kapczinski, F.; et al. Ndel1 oligopeptidase activity as a potential biomarker of early stages of schizophrenia. Schizophr. Res. 2019, 208, 202–208. [Google Scholar] [CrossRef]
- Paschoalin, T.; Carmona, A.K.; Oliveira, V.; Juliano, L.; Travassos, L.R. Characterization of thimet- and neurolysin-like activities in Escherichia coli M 3 A peptidases and description of a specific substrate. Arch. Biochem. Biophys. 2005, 441, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.C.; Graeff, F.G. Subcellular distribution and properties of the bradykinin inactivation system in rabbit brain homogenates. Biochem. Pharm. 1969, 18, 548–549. [Google Scholar] [CrossRef]
- Greene, L.J.; Camargo, A.C.; Krieger, E.M.; Stewart, J.M.; Ferreira, S.H. Inhibition of the conversion of angiotensin I to II and potentiation of bradykinin by small peptides present in Bothrops jararaca venom. Circ. Res. 1972, 31 (Suppl. 2), 62–71. [Google Scholar]
- Camargo, A.C.; Shapanka, R.; Greene, L.J. Preparation, assay, and partial characterization of a neutral endopeptidase from rabbit brain. Biochemistry 1973, 12, 1838–1844. [Google Scholar] [CrossRef]
- Oliveira, E.B.; Martins, A.R.; Camargo, A.C. Isolation of brain endopeptidases: Influence of size and sequence of substrates structurally related to bradykinin. Biochemistry 1976, 15, 1967–1974. [Google Scholar] [CrossRef]
- Montiel, J.L.; Cornille, F.; Roques, B.P.; Noble, F. Nociceptin/orphanin FQ metabolism: Role of aminopeptidase and endopeptidase 24.15. J. Neurochem. 1997, 68, 354–361. [Google Scholar] [CrossRef]
- Oliveira, V.; Campos, M.; Melo, R.L.; Ferro, E.S.; Camargo, A.C.; Juliano, M.A.; Juliano, L. Substrate specificity characterization of recombinant metallo oligo-peptidases thimet oligopeptidase and neurolysin. Biochemistry 2001, 40, 4417–4425. [Google Scholar] [CrossRef]
- Berti, D.A.; Morano, C.; Russo, L.C.; Castro, L.M.; Cunha, F.M.; Zhang, X.; Sironi, J.; Klitzke, C.F.; Ferro, E.S.; Fricker, L.D. Analysis of intracellular substrates and products of thimet oligopeptidase in human embryonic kidney 293 cells. J. Biol. Chem. 2009, 284, 14105–14116. [Google Scholar] [CrossRef] [Green Version]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Camargo, A.C.; Gomes, M.D.; Reichl, A.P.; Ferro, E.S.; Jacchieri, S.; Hirata, I.Y.; Juliano, L. Structural features that make oligopeptides susceptible substrates for hydrolysis by recombinant thimet oligopeptidase. Biochem. J. 1997, 324, 517–522. [Google Scholar] [CrossRef]
- Turk, B.E.; Huang, L.L.; Piro, E.T.; Cantley, L.C. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 2001, 19, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.J.; Sampath, S.; Coll-Rodriguez, J.; Schmidt, J.; Ray, K.; Rodgers, D.W. Swapping the substrate specificities of the neuropeptidases neurolysin and thimet oligopeptidase. J. Biol. Chem. 2007, 282, 9722–9732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rioli, V.; Kato, A.; Portaro, F.C.; Cury, G.K.; te Kaat, K.; Vincent, B.; Checler, F.; Camargo, A.C.; Glucksman, M.J.; Roberts, J.L.; et al. Neuropeptide specificity and inhibition of recombinant isoforms of the endopeptidase 3.4.24.16 family: Comparison with the related recombinant endopeptidase 3.4.24.15. Biochem. Biophys. Res. Commun. 1998, 250, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Portaro, F.C.V.; Gomes, M.D.; Cabrera, A.; Fernandes, B.L.; Silva, C.L.; Ferro, E.S.; Juliano, L.; De Camargo, A.C.M. Thimet oligopeptidase and the stability of MHC class I epitopes in macrophage cytosol. Biochem. Biophys. Res. Commun. 1999, 255, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Fukami, S.; Watanabe, K.; Iwata, N.; Haraoka, J.; Lu, B.; Gerard, N.P.; Gerard, C.; Fraser, P.; Westaway, D.; St George-Hyslop, P.; et al. Abeta-degrading endopeptidase, neprilysin, in mouse brain: Synaptic and axonal localization inversely correlating with Abeta pathology. Neurosci. Res. 2002, 43, 39–56. [Google Scholar] [CrossRef]
- Matsas, R.; Kenny, A.J.; Turner, A.J. The metabolism of neuropeptides. The hydrolysis of peptides, including enkephalins, tachykinins and their analogues, by endopeptidase-24.11. Biochem. J. 1984, 223, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Healy, D.P.; Orlowski, M. Immunocytochemical localization of endopeptidase 24.15 in rat brain. Brain Res. 1992, 571, 121–128. [Google Scholar] [CrossRef]
- Massarelli, E.E.; Casatti, C.A.; Kato, A.; Camargo, A.C.; Bauer, J.A.; Glucksman, M.J.; Roberts, J.L.; Hirose, S.; Ferro, E.S. Differential subcellular distribution of neurolysin (EC 3.4.24.16) and thimet oligopeptidase (EC 3.4.24.15) in the rat brain. Brain Res. 1999, 851, 261–265. [Google Scholar] [CrossRef]
- Fontenele-Neto, J.D.; Massarelli, E.E.; Gurgel Garrido, P.A.; Beaudet, A.; Ferro, E.S. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J. Comp. Neurol. 2001, 438, 399–410. [Google Scholar] [CrossRef]
- Schumacher, T.N.; De Bruijn, M.L.; Vernie, L.N.; Kast, W.M.; Melief, C.J.; Neefjes, J.J.; Ploegh, H.L. Peptide selection by MHC class I molecules. Nature 1991, 350, 703–706. [Google Scholar] [CrossRef]
- Rock, K.L.; Rothstein, L.; Benacerraf, B. Analysis of the association of peptides of optimal length to class I molecules on the surface of cells. Proc. Natl. Acad. Sci. USA 1992, 89, 8918–8922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deres, K.; Schumacher, T.N.; Wiesmüller, K.H.; Stevanović, S.; Greiner, G.; Jung, G.; Ploegh, H.L. Preferred size of peptides that bind to H-2 Kb is sequence dependent. Eur. J. Immunol. 1992, 22, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Michalek, M.T.; Grant, E.P.; Gramm, C.; Goldberg, A.L.; Rock, K.L. A role for the ubiquitin-dependent proteolytic pathway in MHC class l-restricted antigen presentation. Nature 1993, 363, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Palacios, A.; Colston, M.; Lowrie, D. Mycobacterium leprae 65hsp antigen expressed from a retroviral vector in a macrophage cell line is presented to T cells in association with MHC class II in addition to MHC class I. Microb. Pathog. 1992, 12, 27–38. [Google Scholar] [CrossRef]
- Silva, C.L.; Portaro, F.C.; Bonato, V.L.; de Camargo, A.C.; Ferro, E.S. Thimet oligopeptidase (EC 3.4.24.15), a novel protein on the route of MHC class I antigen presentation. Biochem. Biophys. Res. Commun. 1999, 255, 591–595. [Google Scholar] [CrossRef]
- York, I.A.; Mo, A.X.; Lemerise, K.; Zeng, W.; Shen, Y.; Abraham, C.R.; Saric, T.; Goldberg, A.L.; Rock, K.L. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity 2003, 18, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.I.; Pabon, A.; Swanson, T.A.; Glucksman, M.J. Regulation of cell-surface major histocompatibility complex class I expression by the endopeptidase EC3.4.24.15 (thimet oligopeptidase). Biochem. J. 2003, 375, 111–120. [Google Scholar] [CrossRef]
- Kessler, J.H.; Khan, S.; Seifert, U.; Le Gall, S.; Chow, K.M.; Paschen, A.; Bres-Vloemans, S.A.; de Ru, A.; van Montfoort, N.; Franken, K.L.; et al. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat. Immunol. 2011, 12, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Guinan, A.F.; Rochfort, K.D.; Fitzpatrick, P.A.; Walsh, T.G.; Pierotti, A.R.; Phelan, S.; Murphy, R.P.; Cummins, P.M. Shear stress is a positive regulator of thimet oligopeptidase (EC3.4.24.15) in vascular endothelial cells: Consequences for MHC1 levels. Cardiovasc. Res. 2013, 99, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Gewehr, M.C.F.; Teixeira, A.A.S.; Santos, B.A.C.; Biondo, L.A.; Gozzo, F.C.; Cordibello, A.M.; Eichler, R.A.S.; Reckziegel, P.; Da Silva, R.N.O.; Dos Santos, N.B.; et al. The Relevance of Thimet Oligopeptidase in the Regulation of Energy Metabolism and Diet-Induced Obesity. Biomolecules 2020, 10, 321. [Google Scholar] [CrossRef] [Green Version]
- Santos, N.B.D.; Franco, R.D.; Camarini, R.; Munhoz, C.D.; Eichler, R.A.S.; Gewehr, M.C.F.; Reckziegel, P.; Llanos, R.P.; Dale, C.S.; Silva, V.; et al. Thimet Oligopeptidase (EC 3.4.24.15) Key Functions Suggested by Knockout Mice Phenotype Characterization. Biomolecules 2019, 9, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Procaccini, C.; De Rosa, V.; Pucino, V.; Formisano, L.; Matarese, G. Animal models of Multiple Sclerosis. Eur. J. Pharm. 2015, 759, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Castelli, L.; Goverman, J.M. MHC class I-restricted myelin epitopes are cross-presented by Tip-DCs that promote determinant spreading to CD8⁺ T cells. Nat. Immunol. 2013, 14, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Eason, D.D.; Cannon, J.P.; Haire, R.N.; Rast, J.P.; Ostrov, D.A.; Litman, G.W. Mechanisms of antigen receptor Evolution. In Proceedings of Seminars in Immunology; Elsevier Ltd.: Oxford, UK, 2004; Volume 16, pp. 215–226. [Google Scholar]
- Danchin, E.; Vitiello, V.; Vienne, A.; Richard, O.; Gouret, P.; McDermott, M.F.; Pontarotti, P. The major histocompatibility complex origin. Immunol. Rev. 2004, 198, 216–232. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; McKinney, E.C.; Flajnik, M.F.; Ishibashi, T. The evolutionary origin of the major histocompatibility complex: Polymorphism of class II alpha chain genes in the cartilaginous fish. Eur. J. Immunol. 1993, 23, 2160–2165. [Google Scholar] [CrossRef]
- Hedrick, P.W. Evolutionary Genetics of the Major Histocompatibility Complex. Am. Nat. 1994, 143, 945–964. [Google Scholar] [CrossRef]
- Hughes, A.L.; Nei, M. Evolutionary relationships of the classes of major histocompatibility complex genes. Immunogenetics 1993, 37, 337–346. [Google Scholar] [CrossRef]
- Gille, C.; Goede, A.; Schloetelburg, C.; Preissner, R.; Kloetzel, P.M.; Gobel, U.B.; Frommel, C. A comprehensive view on proteasomal sequences: Implications for the evolution of the proteasome. J. Mol. Biol. 2003, 326, 1437–1448. [Google Scholar] [CrossRef]
- Robinson, C.V.; Sali, A.; Baumeister, W. The molecular sociology of the cell. Nature 2007, 450, 973–982. [Google Scholar] [CrossRef]
- Ferro, E.S.; Hyslop, S.; Camargo, A.C. Intracellullar peptides as putative natural regulators of protein interactions. J. Neurochem. 2004, 91, 769–777. [Google Scholar] [CrossRef]
- Fricker, L.D. Proteasome Inhibitor Drugs. Annu. Rev. Pharm. Toxicol. 2020, 60, 457–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippton, H.; Lin, B.; Gumusel, B.; Witriol, N.; Wasserman, A.; Knight, M. Hemopressin, a hemoglobin fragment, dilates the rat systemic vascular bed through release of nitric oxide. Peptides 2006, 27, 2284–2288. [Google Scholar] [CrossRef] [PubMed]
- Dale, C.S.; Pagano Rde, L.; Rioli, V.; Hyslop, S.; Giorgi, R.; Ferro, E.S. Antinociceptive action of hemopressin in experimental hyperalgesia. Peptides 2005, 26, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Heimann, A.S.; Gomes, I.; Dale, C.S.; Pagano, R.L.; Gupta, A.; de Souza, L.L.; Luchessi, A.D.; Castro, L.M.; Giorgi, R.; Rioli, V.; et al. Hemopressin is an inverse agonist of CB1 cannabinoid receptors. Proc. Natl. Acad. Sci. USA 2007, 104, 20588–20593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanova, E.V.; Lee, J.E.; Kelleher, N.L.; Sweedler, J.V.; Gulley, J.M. Comparative peptidomics analysis of neural adaptations in rats repeatedly exposed to amphetamine. J. Neurochem. 2012, 123, 276–287. [Google Scholar] [CrossRef]
- Lee, J.E.; Zamdborg, L.; Southey, B.R.; Atkins, N., Jr.; Mitchell, J.W.; Li, M.; Gillette, M.U.; Kelleher, N.L.; Sweedler, J.V. Quantitative peptidomics for discovery of circadian-related peptides from the rat suprachiasmatic nucleus. J. Proteome Res. 2013, 12, 585–593. [Google Scholar] [CrossRef]
- Southey, B.R.; Lee, J.E.; Zamdborg, L.; Atkins, N., Jr.; Mitchell, J.W.; Li, M.; Gillette, M.U.; Kelleher, N.L.; Sweedler, J.V. Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal. Chem. 2014, 86, 443–452. [Google Scholar] [CrossRef]
- Fricker, L.D. Analysis of mouse brain peptides using mass spectrometry-based peptidomics: Implications for novel functions ranging from non-classical neuropeptides to microproteins. Mol. Biosyst. 2010, 6, 1355–1365. [Google Scholar] [CrossRef]
- Berezniuk, I.; Sironi, J.J.; Wardman, J.; Pasek, R.C.; Berbari, N.F.; Yoder, B.K.; Fricker, L.D. Quantitative peptidomics of Purkinje cell degeneration mice. PLoS ONE 2013, 8, e60981. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef]
- Fesenko, I.A.; Arapidi, G.P.; Skripnikov, A.Y.; Alexeev, D.G.; Kostryukova, E.S.; Manolov, A.I.; Altukhov, I.A.; Khazigaleeva, R.A.; Seredina, A.V.; Kovalchuk, S.I.; et al. Specific pools of endogenous peptides are present in gametophore, protonema, and protoplast cells of the moss Physcomitrella patens. BMC Plant Biol. 2015, 15, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fesenko, I.; Khazigaleeva, R.; Govorun, V.; Ivanov, V. Analysis of Endogenous Peptide Pools of Physcomitrella patens Moss. Methods Mol. Biol. 2018, 1719, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Filippova, A.; Lyapina, I.; Kirov, I.; Zgoda, V.; Belogurov, A.; Kudriaeva, A.; Ivanov, V.; Fesenko, I. Salicylic acid influences the protease activity and posttranslation modifications of the secreted peptides in the moss Physcomitrella patens. J. Pept. Sci. 2019, 25, e3138. [Google Scholar] [CrossRef] [PubMed]
- Gelman, J.S.; Sironi, J.; Castro, L.M.; Ferro, E.S.; Fricker, L.D. Peptidomic analysis of human cell lines. J. Proteome Res. 2011, 10, 1583–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, X.; Wang, F.; You, L.; Xu, P.; Cao, Y.; Chen, L.; Wen, J.; Guo, X.; Cui, X.; et al. Identification of intracellular peptides associated with thermogenesis in human brown adipocytes. J. Cell. Physiol. 2019, 234, 7104–7114. [Google Scholar] [CrossRef] [PubMed]
- Cafe-Mendes, C.C.; Ferro, E.S.; Torrao, A.S.; Crunfli, F.; Rioli, V.; Schmitt, A.; Falkai, P.; Britto, L.R.; Turck, C.W.; Martins-de-Souza, D. Peptidomic analysis of the anterior temporal lobe and corpus callosum from schizophrenia patients. J. Proteom. 2017, 151, 97–105. [Google Scholar] [CrossRef]
- Gelman, J.S.; Dasgupta, S.; Berezniuk, I.; Fricker, L.D. Analysis of peptides secreted from cultured mouse brain tissue. Biochim. Biophys. Acta 2013, 1834, 2408–2417. [Google Scholar] [CrossRef] [Green Version]
- Gelman, J.S.; Sironi, J.; Castro, L.M.; Ferro, E.S.; Fricker, L.D. Hemopressins and other hemoglobin-derived peptides in mouse brain: Comparison between brain, blood, and heart peptidome and regulation in Cpefat/fat mice. J. Neurochem. 2010, 113, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Gelman, J.S.; Fricker, L.D. Hemopressin and other bioactive peptides from cytosolic proteins: Are these non-classical neuropeptides? AAPS J. 2010, 12, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, C.M.M.; Correa, C.N.; Iwai, L.K.; Ferro, E.S.; Castro, L.M. Characterization of Intracellular Peptides from Zebrafish (Danio rerio) Brain. Zebrafish 2019, 16, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Heimann, A.S.; Favarato, M.H.; Gozzo, F.C.; Rioli, V.; Carreno, F.R.; Eberlin, M.N.; Ferro, E.S.; Krege, J.H.; Krieger, J.E. ACE gene titration in mice uncovers a new mechanism for ACE on the control of body weight. Physiol. Genom. 2005, 20, 173–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, M.F.; Cunha, F.M.; Berti, D.A.; Heimann, A.S.; Klitzke, C.F.; Rioli, V.; Oliveira, V.; Ferro, E.S. Substrate phosphorylation affects degradation and interaction to endopeptidase 24.15, neurolysin, and angiotensin-converting enzyme. Biochem. Biophys. Res. Commun. 2006, 339, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.C.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S. Inhibition of thimet oligopeptidase by siRNA alters specific intracellular peptides and potentiates isoproterenol signal transduction. FEBS Lett. 2012, 586, 3287–3292. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Fishman, M.A.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Fricker, L.D. Effect of Protein Denaturation and Enzyme Inhibitors on Proteasomal-Mediated Production of Peptides in Human Embryonic Kidney Cells. Biomolecules 2019, 9, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Fishman, M.; Mahallati, H.; Castro, L.; Tashima, A.; Ferro, E.; Fricker, L. Reduced Levels of Proteasome Products in a Mouse Striatal Cell Model of Huntington’s Disease. PLoS ONE 2015, 10, e0145333. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, S.; Castro, L.M.; Dulman, R.; Yang, C.; Schmidt, M.; Ferro, E.S.; Fricker, L.D. Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS ONE 2014, 9, e103604. [Google Scholar] [CrossRef]
- Gelman, J.S.; Sironi, J.; Berezniuk, I.; Dasgupta, S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S.; Fricker, L.D. Alterations of the Intracellular Peptidome in Response to the Proteasome Inhibitor Bortezomib. PLoS ONE 2013, 8, e53263. [Google Scholar] [CrossRef] [Green Version]
- Fricker, L.D.; Gelman, J.S.; Castro, L.M.; Gozzo, F.C.; Ferro, E.S. Peptidomic analysis of HEK293T cells: Effect of the proteasome inhibitor epoxomicin on intracellular peptides. J. Proteome Res. 2012, 11, 1981–1990. [Google Scholar] [CrossRef] [Green Version]
- Berti, D.A.; Russo, L.C.; Castro, L.M.; Cruz, L.; Gozzo, F.C.; Heimann, J.C.; Lima, F.B.; Oliveira, A.C.; Andreotti, S.; Prada, P.O.; et al. Identification of intracellular peptides in rat adipose tissue: Insights into insulin resistance. Proteomics 2012, 12, 2668–2681. [Google Scholar] [CrossRef] [PubMed]
- Soughayer, J.S.; Wang, Y.; Li, H.; Cheung, S.-H.; Rossi, F.M.; Stanbridge, E.J.; Sims, C.E.; Allbritton, N.L. Characterization of TAT-mediated transport of detachable kinase substrates. Biochemistry 2004, 43, 8528–8540. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, C.B.; Russo, L.C.; Castro, L.M.; Forti, F.L.; do Monte, E.R.; Rioli, V.; Gozzo, F.C.; Colquhoun, A.; Ferro, E.S. A novel intracellular peptide derived from g1/s cyclin d2 induces cell death. J. Biol. Chem. 2014, 289, 16711–16726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monte, E.R.; Rossato, C.; Llanos, R.P.; Russo, L.C.; de Castro, L.M.; Gozzo, F.C.; de Araujo, C.B.; Peron, J.P.; Sant’Anna, O.A.; Ferro, E.S.; et al. Interferon-gamma activity is potentiated by an intracellular peptide derived from the human 19S ATPase regulatory subunit 4 of the proteasome. J. Proteom. 2017, 151, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.C.; Araujo, C.B.; Iwai, L.K.; Ferro, E.S.; Forti, F.L. A Cyclin D2-derived peptide acts on specific cell cycle phases by activating ERK1/2 to cause the death of breast cancer cells. J. Proteom. 2017, 151, 24–32. [Google Scholar] [CrossRef]
- Cunha, F.M.; Berti, D.A.; Ferreira, Z.S.; Klitzke, C.F.; Markus, R.P.; Ferro, E.S. Intracellular peptides as natural regulators of cell signaling. J. Biol. Chem. 2008, 283, 24448–24459. [Google Scholar] [CrossRef] [Green Version]
- Cotter, E.J.; von Offenberg Sweeney, N.; Coen, P.M.; Birney, Y.A.; Glucksman, M.J.; Cahill, P.A.; Cummins, P.M. Regulation of endopeptidases EC3.4.24.15 and EC3.4.24.16 in vascular endothelial cells by cyclic strain: Role of Gi protein signaling. Arter. Thromb Vasc. Biol. 2004, 24, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Lasdun, A.; Reznik, S.; Molineaux, C.J.; Orlowski, M. Inhibition of endopeptidase 24.15 slows the in vivo degradation of luteinizing hormone-releasing hormone. J. Pharmacol. Exp. Ther. 1989, 251, 439–447. [Google Scholar]
- Orlowski, M.; Reznik, S.; Ayala, J.; Pierotti, A.R. Endopeptidase 24.15 from rat testes. Isolation of the enzyme and its specificity toward synthetic and natural peptides, including enkephalin-containing peptides. Biochem. J. 1989, 261, 951–958. [Google Scholar] [CrossRef] [Green Version]
- Garrido, P.A.; Vandenbulcke, F.; Ramjaun, A.R.; Vincent, B.; Checler, F.; Ferro, E.; Beaudet, A. Confocal microscopy reveals thimet oligopeptidase (EC 3.4.24.15) and neurolysin (EC 3.4.24.16) in the classical secretory pathway. DNA Cell Biol. 1999, 18, 323–331. [Google Scholar] [CrossRef]
- Ferro, E.S.; Tullai, J.W.; Glucksman, M.J.; Roberts, J.L. Secretion of metalloendopeptidase 24.15 (EC 3.4.24.15). DNA Cell Biol. 1999, 18, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Carreno, F.R.; Goni, C.N.; Castro, L.M.; Ferro, E.S. 14-3-3 epsilon modulates the stimulated secretion of endopeptidase 24.15. J. Neurochem. 2005, 93, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Crack, P.J.; Wu, T.J.; Cummins, P.M.; Ferro, E.S.; Tullai, J.W.; Glucksman, M.J.; Roberts, J.L. The association of metalloendopeptidase EC 3.4.24.15 at the extracellular surface of the AtT-20 cell plasma membrane. Brain Res. 1999, 835. [Google Scholar] [CrossRef]
- Oliveira, V.; Garrido, P.A.G.; Rodrigues, C.C.; Colquhoun, A.; Castro, L.M.; Almeida, P.C.; Shida, C.S.; Juliano, M.A.; Juliano, L.; Camargo, A.C.M.; et al. Calcium modulates endopeptidase 24.15 (EC 3.4.24.15) membrane association, secondary structure and substrate specificity. FEBS J. 2005, 272. [Google Scholar] [CrossRef] [PubMed]
- Jeske, N.A.; Berg, K.A.; Cousins, J.C.; Ferro, E.S.; Clarke, W.P.; Glucksman, M.J.; Roberts, J.L. Modulation of bradykinin signaling by EP24.15 and EP24.16 in cultured trigeminal ganglia. J. Neurochem. 2006, 97. [Google Scholar] [CrossRef]
- Jeske, N.A.; Glucksman, M.J.; Roberts, J.L. Metalloendopeptidase EC3.4.24.15 is constitutively released from the exofacial leaflet of lipid rafts in GT1-7 cells. J. Neurochem. 2004, 90, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Lew, R.A.; Tomoda, F.; Evans, R.G.; Lakat, L.; Boublik, J.H.; Pipolo, L.A.; Smith, A.I. Synthetic inhibitors of endopeptidase EC 3.4.24.15: Potency and stability in vitro and in vivo. Br. J. Pharm. 1996, 118, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.J.; Pierotti, A.R.; Jakubowski, M.; Sheward, W.J.; Glucksman, M.J.; Smith, A.I.; King, J.C.; Fink, G.; Roberts, J.L. Endopeptidase EC 3.4.24.15 presence in the rat median eminence and hypophysial portal blood and its modulation of the luteinizing hormone surge. J. Neuroendocrinol. 1997, 9, 813–822. [Google Scholar] [CrossRef]
- Wang, B.; Yang, H.; Liu, Y.C.; Jelinek, T.; Zhang, L.; Ruoslahti, E.; Fu, H. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 1999, 38, 12499–12504. [Google Scholar] [CrossRef]
- Masters, S.C.; Pederson, K.J.; Zhang, L.; Barbieri, J.T.; Fu, H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 1999, 38, 5216–5221. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.V.; Poulsen, E.G.; Rebula, C.A.; Hartmann-Petersen, R. Protein quality control in the nucleus. Biomolecules 2014, 4, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Wietrzynski, W.; Lee, C.W.; Schaffer, M.; Beck, F.; Schuller, J.M.; Salomé, P.A.; Plitzko, J.M.; Baumeister, W.; Engel, B.D. Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl. Acad. Sci. USA 2020, 117, 1069–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.I.; Tetaz, T.; Roberts, J.L.; Glucksman, M.; Clarke, I.J.; Lew, R.A. The role of EC 3.4.24.15 in the post-secretory regulation of peptide signals. Biochimie 1994, 76, 288–294. [Google Scholar] [CrossRef]
- Lew, R.A.; Tetaz, T.J.; Glucksman, M.J.; Roberts, J.L.; Smith, A.I. Evidence for a two-step mechanism of gonadotropin-releasing hormone metabolism by prolyl endopeptidase and metalloendopeptidase EC 3.4.24.15 in ovine hypothalamic extracts. J. Biol. Chem. 1994, 269, 12626–12632. [Google Scholar]
- Kest, B.; Orlowski, M.; Molineaux, C.J.; Bodnar, R.J. Antinociceptive properties of inhibitors of endopeptidase 24.15. Int. J. Neurosci. 1991, 56, 141–149. [Google Scholar] [CrossRef]
- Smith, A.I.; Lew, R.A.; Shrimpton, C.N.; Evans, R.G.; Abbenante, G. A novel stable inhibitor of endopeptidases EC 3.4.24.15 and 3.4.24.16 potentiates bradykinin-induced hypotension. Hypertension 2000, 35, 626–630. [Google Scholar] [CrossRef]
- Zeisel, A.; Hochgerner, H.; Lönnerberg, P.; Johnsson, A.; Memic, F.; van der Zwan, J.; Häring, M.; Braun, E.; Borm, L.E.; La Manno, G.; et al. Molecular Architecture of the Mouse Nervous System. Cell 2018, 174, 999–1014.e1022. [Google Scholar] [CrossRef] [Green Version]
- Kandel, E.R.; Schwartz, J.H.; Jessell, T.M.; Siegelbaum, S.A.; Hudspeth, A.J. Principles of Neural Science, 5th ed.; McGraw-Hill Education: New York, NY, USA, 2012. [Google Scholar]
- Karna, S.R.; Kongara, K.; Singh, P.M.; Chambers, P.; Lopez-Villalobos, N. Evaluation of analgesic interaction between morphine, dexmedetomidine and maropitant using hot-plate and tail-flick tests in rats. Vet. Anaesth. Analg. 2019, 46, 476–482. [Google Scholar] [CrossRef]
- Turner, A.J.; Matsas, R.; Kenny, A.J. Are there neuropeptide-specific peptidases? Biochem. Pharmacol. 1985, 34, 1347–1356. [Google Scholar] [CrossRef]
- Molineaux, C.J.; Ayala, J.M. An inhibitor of endopeptidase-24.15 blocks the degradation of intraventricularly administered dynorphins. J. Neurochem. 1990, 55, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Lasdun, A.; Orlowski, M. Inhibition of endopeptidase 24.15 greatly increases the release of luteinizing hormone and follicle stimulating hormone in response to luteinizing hormone/releasing hormone. J. Pharmacol. Exp. Ther. 1990, 253, 1265–1271. [Google Scholar] [PubMed]
- Pierotti, A.R.; Lasdun, A.; Ayala, J.M.; Roberts, J.L.; Molineaux, C.J. Endopeptidase-24.15 in rat hypothalamic/pituitary/gonadal axis. Mol. Cell. Endocrinol. 1991, 76, 95–103. [Google Scholar] [CrossRef]
- Tullai, J.W.; Cummins, P.M.; Pabon, A.; Roberts, J.L.; Lopingco, M.C.; Shrimpton, C.N.; Smith, A.I.; Martignetti, J.A.; Ferro, E.S.; Glucksman, M.J. The neuropeptide processing enzyme EC 3.4.24.15 is modulated by protein kinase A phosphorylation. J. Biol. Chem. 2000, 275, 36514–36522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, C.; Lebrethon, M.C.; Vandersmissen, E.; Gerard, A.; Purnelle, G.; Lemaitre, M.; Wilk, S.; Bourguignon, J.P. Early prepubertal ontogeny of pulsatile gonadotropin-releasing hormone (GnRH) secretion: I. Inhibitory autofeedback control through prolyl endopeptidase degradation of GnRH. Endocrinology 1999, 140, 4609–4615. [Google Scholar] [CrossRef]
- Peitl, V.; Štefanović, M.; Karlović, D. Depressive symptoms in schizophrenia and dopamine and serotonin gene polymorphisms. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 77, 209–215. [Google Scholar] [CrossRef]
- Sundstrom, J.M.; Hernandez, C.; Weber, S.R.; Zhao, Y.; Dunklebarger, M.; Tiberti, N.; Laremore, T.; Simo-Servat, O.; Garcia-Ramirez, M.; Barber, A.J.; et al. Proteomic Analysis of Early Diabetic Retinopathy Reveals Mediators of Neurodegenerative Brain Diseases. Invest. Ophthalmol. Vis. Sci. 2018, 59, 2264–2274. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.-Y.; Schaefer, C.; Krauss, R.M.; et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef]
- Klarin, D.; Damrauer, S.M.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; DuVall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef]
- Puppala, S.; Li, C.; Glenn, J.P.; Saxena, R.; Gawrieh, S.; Quinn, A.; Palarczyk, J.; Dick, E.J., Jr.; Nathanielsz, P.W.; Cox, L.A. Primate fetal hepatic responses to maternal obesity: Epigenetic signalling pathways and lipid accumulation. J. Physiol. 2018, 596, 5823–5837. [Google Scholar] [CrossRef]
- Cavalcanti, D.M.L.P.; Castro, L.M.; Rosa Neto, J.C.; Seelaender, M.; Neves, R.X.; Oliveira, V.; Forti, F.L.; Iwai, L.K.; Gozzo, F.C.; Todiras, M.; et al. Neurolysin knockout mice generation and initial phenotype characterization. J. Biol. Chem. 2014, 289, 15426–15440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, L.; Shuai, L.; Zhu, M.; Liu, P.; Xie, Z.F.; Jiang, S.; Jiang, H.W.; Li, J.; Zhao, Y.; Li, J.Y.; et al. The Landscape of Histone Modifications in a High-Fat Diet-Induced Obese (DIO) Mouse Model. Mol. Cell. Proteom. 2017, 16, 1324–1334. [Google Scholar] [CrossRef] [Green Version]
- Atshaves, B.P.; Storey, S.M.; McIntosh, A.L.; Petrescu, A.D.; Lyuksyutova, O.I.; Greenberg, A.S.; Schroeder, F. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J. Biol. Chem. 2001, 276, 25324–25335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.S. Peptides and Peptidomimetics as Potential Antiobesity Agents: Overview of Current Status. Front. Nutr. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Bravo-San Pedro, J.M.; Sica, V.; Martins, I.; Pol, J.; Loos, F.; Maiuri, M.C.; Durand, S.; Bossut, N.; Aprahamian, F.; Anagnostopoulos, G.; et al. Acyl-CoA-Binding Protein Is a Lipogenic Factor that Triggers Food Intake and Obesity. Cell Metab. 2019, 30, 754–767.e759. [Google Scholar] [CrossRef] [Green Version]
- Pai, J.; Yoon, T.; Kim, N.D.; Lee, I.-S.; Yu, J.; Shin, I. High-throughput profiling of peptide-RNA interactions using peptide microarrays. J. Am. Chem. Soc. 2012, 134, 19287–19296. [Google Scholar] [CrossRef]
- Meckelein, B.; de Silva, H.A.; Roses, A.D.; Rao, P.N.; Pettenati, M.J.; Xu, P.T.; Hodge, R.; Glucksman, M.J.; Abraham, C.R. Human endopeptidase (THOP1) is localized on chromosome 19 within the linkage region for the late-onset alzheimer disease AD2 locus. Genomics 1996, 31, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.P.; Prange, C.; Lennon, G. Human endopeptidase 24.15 (THOP1) is localized on chromosome 19p13.3 and is excluded from the linkage region for late-onset Alzheimer disease. Genomics 1998, 53, 239–240. [Google Scholar] [CrossRef] [Green Version]
- Pollio, G.; Hoozemans, J.J.; Andersen, C.A.; Roncarati, R.; Rosi, M.C.; van Haastert, E.S.; Seredenina, T.; Diamanti, D.; Gotta, S.; Fiorentini, A.; et al. Increased expression of the oligopeptidase THOP1 is a neuroprotective response to Abeta toxicity. Neurobiol. Dis. 2008, 31, 145–158. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, H.; Yang, C.; Xu, K.; Cai, Y.; Wang, Z.; Zhao, Z.; Shao, T.; Li, Y. Transcriptomic Analyses for Identification and Prioritization of Genes Associated With Alzheimer’s Disease in Humans. Front. Bioeng. Biotechnol. 2020, 8, 31. [Google Scholar] [CrossRef]
- Qi, L.; Li, S.H.; Si, L.B.; Lu, M.; Tian, H. Expression of THOP1 and its relationship to prognosis in non-small cell lung cancer. PLoS ONE 2014, 9, e106665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhao, H. Prognosis related miRNAs, DNA methylation, and epigenetic interactions in lung adenocarcinoma. Neoplasma 2019, 66, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, S.; Hishida, M.; Inokawa, Y.; Takano, N.; Kanda, M.; Nishikawa, Y.; Fujii, T.; Koike, M.; Sugimoto, H.; Kodera, Y. Expression analysis of THOP1 in background liver, a prognostic predictive factor in hepatocellular carcinoma, extracted by multiarray analysis. Ann. Surg. Oncol. 2014, 21, S443–S450. [Google Scholar] [CrossRef] [PubMed]
- Shchetynsky, K.; Diaz-Gallo, L.M.; Folkersen, L.; Hensvold, A.H.; Catrina, A.I.; Berg, L.; Klareskog, L.; Padyukov, L. Discovery of new candidate genes for rheumatoid arthritis through integration of genetic association data with expression pathway analysis. Arthritis Res. 2017, 19, 19. [Google Scholar] [CrossRef] [Green Version]
- Benjamim, C.F.; Ferreira, S.H.; Cunha, F.d.Q. Role of Nitric Oxide in the Failure of Neutrophil Migration in Sepsis. J. Infect. Dis. 2000, 182, 214–223. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Orlowski, M.; Michaud, C.; Molineaux, C.J. Substrate-related potent inhibitors of brain metalloendopeptidase. Biochemistry 1988, 27, 597–602. [Google Scholar] [CrossRef]
- Cardozo, C.; Orlowski, M. Evidence that enzymatic conversion of N-[1(R,S)-carboxy-3-phenylpropyl]-Ala-Ala-Phe-p-aminobenzoate, a specific inhibitor of endopeptidase 24.15, to N-[1(R,S)-carboxy-3-phenylpropyl]-Ala-Ala is necessary for inhibition of angiotensin converting enzyme. Peptides 1993, 14, 1259–1262. [Google Scholar] [CrossRef]
- Lew, R.A.; Boulos, E.; Stewart, K.M.; Perlmutter, P.; Harte, M.F.; Bond, S.; Aguilar, M.I.; Smith, A.I. Bradykinin analogues with beta-amino acid substitutions reveal subtle differences in substrate specificity between the endopeptidases EC 3.4.24.15 and EC 3.4.24.16. J. Pept. Sci. 2000, 6, 440–445. [Google Scholar] [CrossRef]
- Jirácek, J.; Yiotakis, A.; Vincent, B.; Lecoq, A.; Nicolaou, A.; Checler, F.; Dive, V. Development of highly potent and selective phosphinic peptide inhibitors of zinc endopeptidase 24-15 using combinatorial chemistry. J. Biol. Chem. 1995, 270, 21701–21706. [Google Scholar] [CrossRef] [Green Version]
- Hines, C.S.; Ray, K.; Schmidt, J.J.; Xiong, F.; Feenstra, R.W.; Pras-Raves, M.; de Moes, J.P.; Lange, J.H.; Melikishvili, M.; Fried, M.G.; et al. Allosteric inhibition of the neuropeptidase neurolysin. J. Biol. Chem. 2014, 289, 35605–35619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirali, S.; Botham, A.; Voisin, V.; Xu, C.; St-Germain, J.; Sharon, D.; Hoff, F.W.; Qiu, Y.; Hurren, R.; Gronda, M.; et al. The mitochondrial peptidase, neurolysin, regulates respiratory chain supercomplex formation and is necessary for AML viability. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabasi, A.L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, I.A.; Luck, K.; Spirohn, K.; Wang, Y.; Pollis, C.; Schlabach, S.; Bian, W.; Kim, D.K.; Kishore, N.; Hao, T.; et al. Network-based prediction of protein interactions. Nat. Commun. 2019, 10, 1240. [Google Scholar] [CrossRef] [Green Version]
- The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar] [CrossRef]



| mRNA | microRNA | THOP1−/− Effect on Expression |
|---|---|---|
| ACE1 | nd | higher (ST and HC) |
| NEP | nd | higher (ST); lower (female retroperitoneal adipose tissue) |
| Proteasome beta5 subunit | nd | lower (ST and PFC); lower (female retroperitoneal adipose tissue) |
| DPP4 | nd | lower (female retroperitoneal adipose tissue) |
| POP | nd | lower (female retroperitoneal adipose tissue) |
| IDE | nd | higher (male retroperitoneal adipose tissue) |
| 5HT receptor | nd | higher (HC) |
| Dopamine DR2D receptor | nd | lower (ST and HC) |
| Adrenergic beta 1 receptor | nd | higher (male retroperitoneal adipose tissue) |
| Adrenergic beta 3 receptor | nd | higher (male retroperitoneal adipose tissue) |
| Adrenergic beta 3 receptor | nd | lower (female retroperitoneal adipose tissue) |
| CD36 | nd | lower (male liver tissue; male/female inguinal adipose tissue) |
| LPL | nd | lower (male/female inguinal adipose tissue) |
| CD206 | nd | lower (male/female inguinal adipose tissue) |
| CD11C | nd | lower (male inguinal adipose tissue) |
| CD11C | nd | higher (female inguinal adipose tissue) |
| F4/80 | nd | lower (male/female inguinal adipose tissue) |
| PPAR-γ | nd | higher (male inguinal adipose tissue of THOP1−/− fed HD) |
| FAS | nd | higher (male inguinal adipose tissue of THOP1−/− fed HD) |
| miR-212 | higher (male liver tissue) | |
| miR-127 | higher (male liver tissue) | |
| miR-143 | higher (male retroperitoneal adipose tissue) | |
| miR-222 | unaltered THOP1−/− fed HD (male retroperitoneal adipose tissue) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferro, E.S.; Gewehr, M.C.F.; Navon, A. Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions. Biomolecules 2020, 10, 1229. https://doi.org/10.3390/biom10091229
Ferro ES, Gewehr MCF, Navon A. Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions. Biomolecules. 2020; 10(9):1229. https://doi.org/10.3390/biom10091229
Chicago/Turabian StyleFerro, Emer S., Mayara C. F. Gewehr, and Ami Navon. 2020. "Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions" Biomolecules 10, no. 9: 1229. https://doi.org/10.3390/biom10091229





