Abstract
The aging process starts directly after birth and lasts for the entire lifespan; it manifests itself with a decline in an organism’s ability to adapt and is linked to the development of age-related diseases that eventually lead to premature death. This review aims to explore how microRNAs (miRNAs) are involved in skin functioning and aging. Recent evidence has suggested that miRNAs regulate all aspects of cutaneous biogenesis, functionality, and aging. It has been noted that some miRNAs were down-regulated in long-lived individuals, such as let-7, miR-17, and miR-34 (known as longevity-related miRNAs). They are conserved in humans and presumably promote lifespan prolongation; conversely, they are up-regulated in age-related diseases, like cancers. The analysis of the age-associated cutaneous miRNAs revealed the increased expression of miR-130, miR-138, and miR-181a/b in keratinocytes during replicative senescence. These miRNAs affected cell proliferation pathways via targeting the p63 and Sirtuin 1 mRNAs. Notably, miR-181a was also implicated in skin immunosenescence, represented by the Langerhans cells. Dermal fibroblasts also expressed increased the levels of the biomarkers of aging that affect telomere maintenance and all phases of the cellular life cycle, such as let-7, miR-23a-3p, 34a-5p, miR-125a, miR-181a-5p, and miR-221/222-3p. Among them, the miR-34 family, stimulated by ultraviolet B irradiation, deteriorates collagen in the extracellular matrix due to the activation of the matrix metalloproteinases and thereby potentiates wrinkle formation. In addition to the pro-aging effects of miRNAs, the plausible antiaging activity of miR-146a that antagonized the UVA-induced inhibition of proliferation and suppressed aging-related genes (e.g., p21WAF-1, p16, and p53) through targeting Smad4 has also been noticed. Nevertheless, the role of miRNAs in skin aging is still not fully elucidated and needs to be further discovered and explained.
1. Introduction
The aging process starts directly after birth and lasts through the entire lifespan. For the majority of the population, it becomes a pressing problem around age 40–60 when the middle-age period of life free of diseases is followed by the period associated with the beginning of age-related diseases (ARDs) that eventually leads to a shortening of life. There are numerous theories describing the causative effects that deteriorate health, and almost all of them are based on the mechanisms that affect homeostasis at the genetic, molecular, cellular, tissue, and organismal levels. Many theories suggest that the appearance of various aging phenotypes occurs because of the age-dependent accumulation of damages [1]. At the same time, some individuals demonstrate the delayed development of aging features or ARDs and have a significantly longer life span despite their socioeconomic status.
An increase in life expectancy around the world is positive, but it is a double-edged sword. On the one side, older people have more opportunities to live an active lifestyle, stay with their families, or continue to work and contribute to multiple areas of society. The extent of this possibility mainly depends on health. On the other side, an increased longevity is directly associated with a higher odds of developing ARDs. Therefore, this means that long-lived people will suffer from pain and disability for a much more extended period of time due to degenerative processes and ARDs (cardiovascular, diabetes, cancer, Parkinson’s, Alzheimer’s, and others). These morbidities drastically complicate a patient’s life and hasten mortality. It is worth noting that clinical manifestations of the abovementioned ARDs are preceded by long (10–20 years) asymptomatic periods of illness development [2]. If no changes are implemented, these trends will continue, and aging will become a significant social and economic burden [3]. The increased life spans have created a pressing need for a better understanding of the pathogenesis of the aging process and the identification of biomarkers and possible therapeutic targets for ARDs.
We know that human longevity is determined by multiple factors, including genetic and environmental influences. About a quarter of the changes in our genetic material over our life period are due to various mutations or epigenetic regulation [4]. The results of epidemiological studies show that centenarians usually demonstrate features of healthy aging, accompanied by the lack or the delayed onset of ARDs. The process of aging is associated with various physiological and pathological molecular mechanisms, including intricate patterns of changes in gene expression leading to changes in the performance of multiple cell types, tissues, and organs. The insulated genetic aberrations in aging-relevant pathways trigger segmental, tissue-selective aging phenotypes [5]. Nevertheless, for a majority of tissues, it remains unknown which age-related modifications play a prominent etiological role in the mechanism of aging, or which ones are just epiphenomena [6].
The most evident and visible symptoms of aging in humans are manifested by changing skin appearance due to continuous exposure to exogenous irritants. Skin is the first line of defense against abiotic and biotic environmental factors [7,8]. Despite an ongoing renewal process, the cutaneous regenerative capacity reduces with age. A decrease in regenerative potential is often linked to a decline in the elimination of senescent cells, and their gradual accumulation results in the physiological aging of the tissue itself [9]. Many questions remain as to how to achieve longevity and healthy aging [10]. Skin seems to be a perfect health sensor because an alteration in its appearance directly signals the ongoing pathological changes in the organism. Thus, research on dermal health and aging is essential for an overall understanding of the processes of aging.
Current the biology of aging includes studies that explore or combine the issues of genomic instability, telomere length, deterioration of mitochondrial function, cellular senescence, epigenetic modifications, proteostasis, exhaustion of stem cells, autophagy, altered intercellular communication, as well as the deteriorated sensing accuracy or trophic function [1,11,12].
A typical signature of aging is the down-regulation of the expression of proteins involved in the electron transport chain. It remains disputable whether this is a reason or an outcome of the aging process. The respiratory chain is controlled by the communication between mitochondria and the nucleus, and changes in the expression of mitochondrial genes depend on energetic demand and a reductive load [13]. A disruption or reductive overload of the respiratory chain leads to the overproduction and release of reactive oxygen species (ROS) or Ca2+ by mitochondria or elevated levels of AMP and ADP. Signals of the abovementioned mitochondrial dysfunction are sensed by the mammalian target of rapamycin (mTOR) or AMP-dependent protein kinase (AMPK) and calmodulin. Subsequently, these signals are transduced to PPARγ-coactivator 1α (PGC1A), which activates the switch in the nuclear mitochondrial biogenic program [14,15]. Dysfunction in these pathways has been described in association with age-related human pathologies and has been observed in aged tissues [13]. Nevertheless, AMPK/mTOR and PGC1A are not unique pathways involved in the regulation of mitochondrial and nuclear interactions in the functionality of the respiratory chain but also require the cellular redox state via Sirtuin 1 (SIRT1) and c-myc. It was found that the SIRT1 expression and nicotinamide adenine dinucleotide (NAD+) levels decrease throughout aging in murine skeletal muscle and lead to dysfunction in mitochondrial biogenesis activation via SIRT1 and c-myc in the AMPK/PGC1A-independent fashion [16].
The majority of genetics-based longevity studies are primarily focused on sirtuins SIRT1 and SIRT3 and calorie restriction. Sirtuins have been reported to contribute to the extension of lifespan due to the improvement of mitochondrial function during dietary restrictions [17,18]. Additionally, sirtuins SIRT1, SIRT6, and SIRT7 are involved in regulating the telomere length and integrity via deacetylase activity, which also facilitates chromatin condensation and histone modification [19,20]. The potential mechanism might include the TOR signaling pathway.
It is intriguing that within the last decade, it was discovered that microRNAs (miRNAs) were widely involved in the modulation of almost all processes in an organism, including aging. The potential role of miRNAs in aging biology is linked to the regulation of the expression of proteins maintaining the insulin-like growth factor (IGF)-1 and TOR signaling pathways [21]. Moreover, publications show that microRNAs play an essential role in regulating the balance between cellular proliferative capacity and replicative senescence [22]. The expression of various miRNAs was found to be directly associated with lifespan duration, ARDs, or their predisposition. Therefore, the expression of a subset of miRNAs also might be considered as one of the aging markers and as a predictor of chronological age, longevity and premature aging, the development of pathologies, or even a risk of mortality [23]. Unfortunately, there is a deficiency of robust information about miRNA functions in the processes of human aging and longevity, and many of these aspects still need to be discovered [24].
To date, there is a lack of information about the relation between miRNA involvement in the dermal and organismal aging process from the perspective of longevity and presumably skin disorders. Furthermore, there is currently a paucity of data that demonstrate possible links between miRNA expression and aging or age-related clinical outcomes [25]. In this review, we will focus on associations between miRNA expression and organismal and cutaneous aging.
2. Skin Structure and Biology
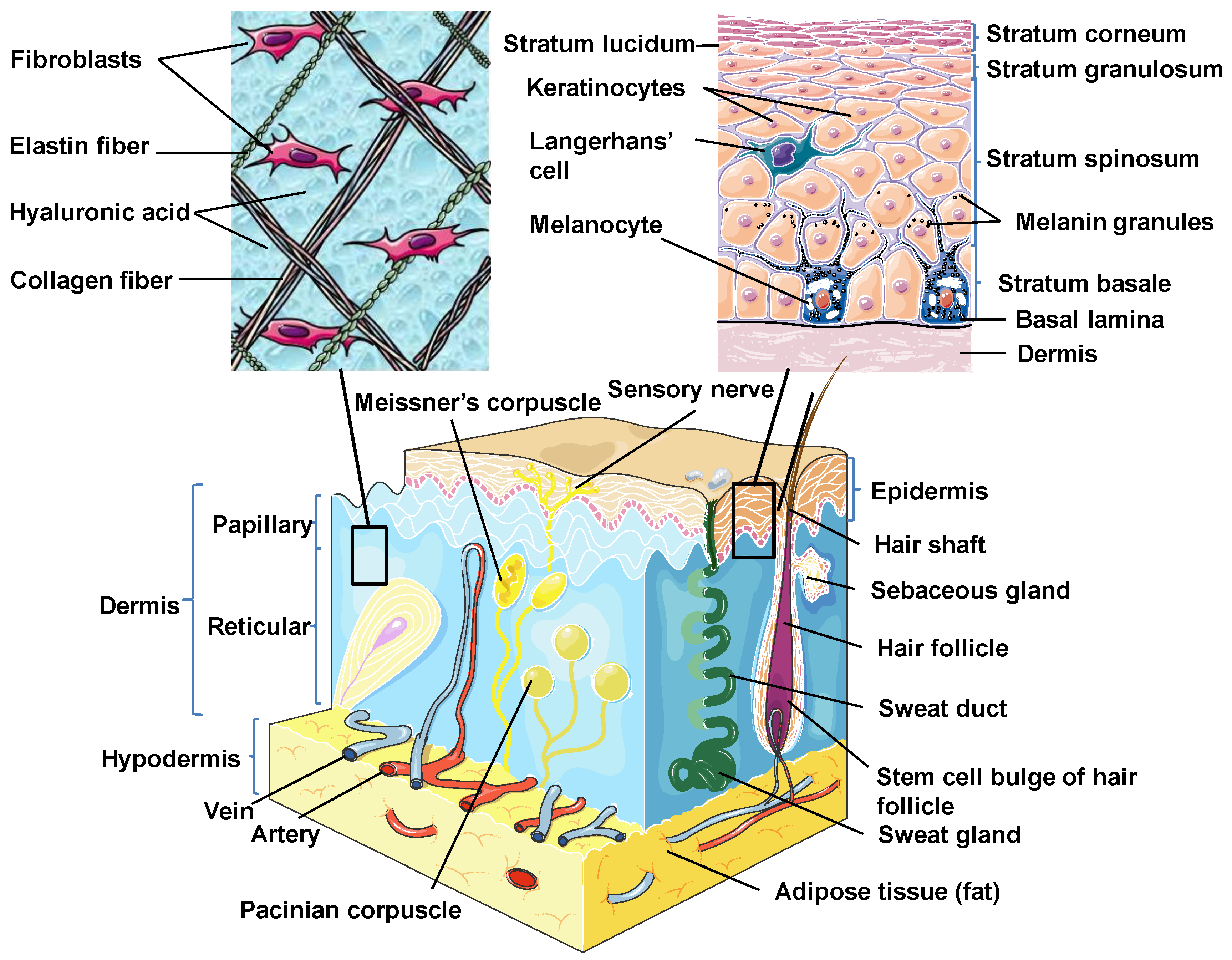
Skin is the outermost organ and the largest one in the human body. It occupies about 8% of the total body mass of an adult human, covering 1.8 m2 of surface area [26]. Apart from being a protective barrier, the skin is also responsible for maintaining homeostasis, including the prevention of the percutaneous loss of electrolytes, fluid, and proteins; it also controls immune activity and sensory perception as well as temperature regulation [27,28,29]. The skin consists of three separate but functionally interdependent layers: the epidermis, dermis, and hypodermis (Figure 1). Their cellular components maintain the mechanical defense, photoprotection, immunosurveillance, nutrient metabolism, repair, and rejuvenation [30,31].
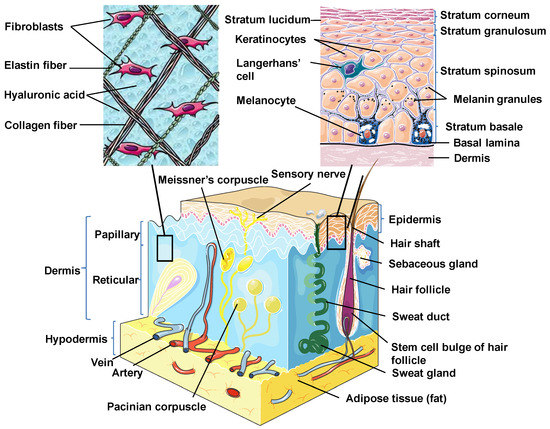
Figure 1.
Skin structure. Schematic diagram showing the structural components of the normal human skin. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (http://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
2.1. Epidermis
The epidermis is the outermost layer of the skin derived from the ectoderm and is tightly interlocked with dermal ridges. It mainly serves as a protective bulwark between the outside world and the internal environment of the body. Other epidermal functions are related to immunoprotection, thermoregulation, ultraviolet protection, resistance to trauma, and the maintenance of energy metabolism. The epidermis consists of keratinocytes, Langerhans cells, melanocytes, neuroendocrine (Merkel) cells, and inflammatory cells (Figure 1) [32]. Keratinocytes comprise over 90% of the cellular population of the epidermis and are connected by desmosomes and tight junctions. They produce and store an intracellular fibrous protein—keratin—that provides hardness and water-resistant properties to the skin and its appendages (hair and nails) [33]. Langerhans cells resemble macrophages in their functions and perform phagocytic activity. Melanocytes produce the pigment melanin that gives hair and skin its color and is also involved in UVR damage protection. Merkel cells function as a touch-perception receptor [32].
Depending on the location in the body, the skin is classified into two types: thin skin and thick skin. They are respectively made of four or five layers of epithelial cells in the epidermis. Thin skin consists of the following layers or strata (from the innermost layer to the outermost one): the stratum basale (the basal layer or stratum germinativum) which contains stem cells that attach the epidermis to the basal lamina; stratum spinosum (the spinous or prickle cell layer); stratum granulosum (the granular cell layer); stratum corneum (the cornified cell layer) (Figure 1). The fifth layer, located between the stratum corneum and the stratum granulosum, is known as the stratum lucidum (the clear cell layer), found predominantly in thick skin which covers the palms of the hands and the soles of the feet [31,33].
2.2. Dermis
The dermal layer delivers nutrients and supports the circulation of the skin [34]. It is separated from the epidermis by a structurally and chemically complex basement membrane zone presented by intertwining collagen fibers. The dermal-epidermal interface is tightly interlocked and stabilized by hemidesmosomes [33]. Structurally, the dermis is a connective tissue layer composed of an interconnected net of collagenous and elastin fibers embedded within the ground substance produced by fibroblasts; it is abundant with lymph and haemocirculation, nerves, and additional structural components (hair follicles and sweat glands). The majority of dermis cells are presented by fibroblasts that possess multifunctional activity but that are predominantly involved in the production of extracellular matrix (ECM) components [31]. Apart from fibroblasts, the dermal layer is composed of a broad number of endothelial and neural cells, supporting elements as well as mast cells embedded within a matrix of collagen and glycosaminoglycans. In the dermis, there are also dendritic and nondendritic monocyte/macrophages, myofibroblasts, along with dermal dendrocytes expressing factor XIIIa that are implicated in the hemostatic and inflammatory processes [35,36].
The dermis consists of two layers. The first one is a papillary layer, composed of loose areolar connective tissue with fibroblasts, collagen bundles, and thin elastic fibers that form a loose mesh and provide a mechanical anchorage and nutrients to the overlying epidermis. Additionally, it contains phagocytes, lymphatic capillaries, nerve fibers, and touch receptors known as the Meissner corpuscles [33]. The second layer is a reticular layer, which is a thicker layer located between the papillary dermis and the subcutaneous adipose tissue. It is made up of irregular dense connective tissue with thick collagen bundles and coarse elastic fibers and is supported by wide vascularization and the sensory and sympathetic nerve supply. Elastin fibers in this layer render the dermal elasticity and movements. While collagen fibers maintain the structural and tensile robustness, they are also responsible for the support of skin hydration by binding water. They are widespread in the papillary compartment and hypodermal layer [31].
2.3. Hypodermis
The hypodermis (also called the subcutaneous layer of the skin or superficial fascia) is a layer directly below the dermis which transitions to the fascial layers. It consists of well-vascularized, loose, areolar connective tissue with a lesser collagen content and abundant adipose tissue that form a layer of variable thickness depending on its location in the body. The hypodermis facilitates cutaneous mobility. The adipose tissue serves as a storage of metabolic energy and provides thermal insulation, cushioning properties, and shock absorption for the integumentary system [31,33].
About 30,000 dermal cells die every minute. At the same time, cutaneous development and normal physiology is a highly adapted process that depends on the cooperation between genetic networks and various regulatory factors [37]. Despite daily exposure to environmental damage, the skin sustains continuous self-renewal to replace old or injured cells and repair the damaged tissue [38]. However, when pathogenic effects exceed the protective cutaneous capabilities or regenerative properties depleted with age, it potentiates cellular senescence and different dermatological diseases [39].
3. Molecular Aspects of Skin Development
Cutaneous formation begins within the first two weeks of development. The ectoderm eventually forms the epidermis and melanocytes as well as the nervous system. The latter one starts developing during the third week of fetal life, when the ectoderm creates the neural plate within it and initiates neural crest development. The mesoderm gives rise to blood vessels, muscles, bones, and fibroblasts, except for some subpopulations that are derived from the neural crest of ectoderm. Besides this, the endoderm is not involved in cutaneous formation [32]. It is believed that neural crest cells secrete the Wnt1 ligand, a signaling molecule that activates the transcription factor and cytoskeletal protein, β-catenin. The latter controls the epithelial differentiation and the functioning of stem cells and appendages. Fibroblast growth factors induce the neural fate, while bone morphogenetic proteins (BMPs) and WNT signaling regulate the epidermal fate [38,40].
3.1. The Development of Epidermis
The surface ectoderm stimulates the generation of a single layer of basal keratinocytes from the germinativum—a layer of cuboidal undifferentiated and mitotically active cells. Keratinocytes are famous for an abundant synthesis of intermediate filaments keratins. Germinativum expresses the gene p63, which is essential for epidermal differentiation. Experiments demonstrate mice lethality immediately following birth due to p63 deficiency [41]. In humans, TP63 mutation causes several autosomal dominant ectodermal dysplasias, which are characterized by different combinations of limb, ectodermal, and orofacial abnormalities as well as alopecia, suggesting the role of p63 in maintaining stem cell proliferation [42,43].
Numerous signaling pathways control the epidermis formation and stratification. For instance, the ligand Delta or Jagged binds the receptor Notch, which initiates transcription and epidermal differentiation. The inhibition of the Notch pathway results in the deficiency of the development of the cutaneous barrier [44]. The epidermis fails to stratify in cases with a low level of expression of the p63 transcription factor, which consequently deteriorates appendage formation [45]. The development of hair follicles and epidermal interfollicular lineages is regulated by the notch 1 and 2 signaling pathways [46]. The mitogen-activated protein kinases (MAPK) signal transduction pathway (also known as the Ras-Raf-MEK-ERK pathway) likewise regulates the epidermal proliferation and differentiation. The deletion of its key enzyme Mek1/2 (mitogen-activated protein dual kinases 1/2) results in cutaneous underdevelopment [47]. Conversely, the augmented activity of the epidermal growth factor (EGF) involved in the MAPK pathway potentiates proliferation and epidermal tumor growth [48].
Three distinct pools of stem cells are located in the interfollicular epidermis, the bulge, and the sebaceous gland supporting epidermal homeostasis. Skin stem cells undergo continuous self-renewal and differentiation into the required cell lineages to replenish cells such as keratinocytes, which die due to programmed termination or injury [28]. The renewal of the cutaneous barrier occurs via the spinous transition of basal cells in mature skin in parallel with changes in gene expression (e.g., the down-regulation of Keratin5/14 and up-regulation of Keratin1/10) controlled by p63 and the canonical Notch pathway [45].
3.2. The Development of Dermis
The dermis is formed from the mesodermal layer. The distinct boundary between the epidermis and the dermis appears at the eighth week of gestation. Fibroblasts represent the primary cell type that compose the dermis and demonstrate the ability to regulate the epithelial cell function. They also abundantly secrete collagens and other extracellular matrix molecules. In addition, fibroblasts show a high level of heterogeneity depending on the skin location. Thus, early embryonic dermal fibroblast progenitors can potentially differentiate into several cell types. For instance, the upper dermal fibroblast progenitor cells (PDGFRa, Blimp1, Dlk−, Irig1 markers) become the dermal papillae (a ball of fibroblasts that control hair keratinocytes) and the arrector pili muscle (the muscle attached to small hairs that causes goosebumps) [49,50]. In the upper dermis, there are papillary fibroblasts. The lower dermal fibroblast progenitor (PDGFRa, Blimp1−, Dlk1) generates the lower reticular fibroblasts and intradermal adipocytes/dermal white adipose tissues [40,50]. Reticular fibroblasts have a lower density and are biased for collagen I over collagen III production. Moreover, after an injury they differentiate into myofibroblasts, which promotes wound closure and presumably scarring [50]. The formation of the appendages is directly interrelated with the papillary dermis [40]. Skin development is not complete at birth because the final full barrier formation occurs afterwards.
4. Processes Involved in Skin Aging
Biological aging is an integral process. It consists of multiple interrelated processes based on internal biochemical reactions, genetic programs, and ongoing external influences. Aging affects all organs and systems, albeit at a different rate [51]. The skin, like all other organs, undergoes distinguishable changes due to the progressive deterioration of morphological and physiological functions with increasing age. Thus, the barrier function and mechanical protection are compromised as a result of a gradual diminishing in the replacement of cellular components. Therefore, wound healing and immune responses are delayed, which is also accompanied by disrupted thermoregulation along with depleted sebum and sweat production. Furthermore, the skin provides the first visible evidence of the aging process [52].
Cutaneous aging is a complex phenomenon involving two simultaneously occurring processes: intrinsic aging and extrinsic aging. Intrinsic aging, known as chronological or natural aging, is genetically determined, whereas extrinsic aging is caused by environmental factors, such as chronic sun exposure (the total sunlight spectrum contains 45% ultraviolet light), and is known as photoaging [53]. The main symptoms of dermal aging are represented by the gradual process of wrinkle development, combined with skin sagging and drooping. Naturally aged skin looks dry and has fine wrinkles, but is still smooth and light [54]. In contrast, extrinsically photo-aged skin has thick layers (a leathery aspect), coarse wrinkles, irregular pigmentation (“age-spots” which are actinic lentigines), capillary telangiectasia, elastosis, and precancerous lesions such as actinic keratosis and malignant tumors [55,56,57,58].
Both types of cutaneous aging reduce the proliferative capacity of fibroblasts, keratinocytes, and melanocytes [53]. The morphological manifestations are the result of a decreased extracellular matrix (ECM) synthesis in the dermis due to the increased expression of matrix-degrading enzymes and matrix metalloproteinases (MMPs), which are mainly secreted by epidermal keratinocytes and dermal fibroblasts [55]. MMPs lead to the degradation and accumulation of a nonfunctional matrix due to cross-links in collagen fibers (intrinsic aging) or partially degraded elastin fibers (extrinsic aging) in combination with an increased oxidative stress and inflammatory process [57].
Extrinsic and intrinsic types of dermal aging are based on the common pathogenic molecular pathways [59]. The interactive features of both types of cutaneous aging are the generation of reactive oxygen species (ROS), resulting in the degradation of the ECM by the overexpressed MMPs [60]. The accumulation of ROS predominantly leads to the activation of tyrosine kinases receptors (TKRs) via the inactivation of protein tyrosine phosphatases. Then, TKRs are phosphorylated, and their subsequent signaling pathways are activated. These include the three families of mitogen-activated protein kinases (MAPKs): p38MAPK, c-Jun N-terminal kinase (JNK), and ERK (extracellular signal-regulated kinases). The activation of these MAPKs is followed by the increased expression of transcription factor activator protein-1 (AP-1). It stimulates the expression of different metalloproteinases (MMP-1, MMP-3, and MMP-9) and prevents the appearance of procollagen-1 [57,60,61]. The enhanced activities of MMPs eventually lead to ECM degradation, which deteriorates the cutaneous structure and manifests as aged skin.
Although the fundamental mechanisms are still poorly understood, a growing body of evidence is in favor of multiple pathogenic pathways of skin aging. Chronological and photoaging skin types may occur in parallel or overlap, and their leading mechanism is intensified oxidative stress, which presumably has one of the most detrimental effects on cutaneous aging. At the same time, all the vital processes are simultaneously controlled by genetic and epigenetic mechanisms, including DNA methylation, histone modification, and noncoding RNA (ncRNA) regulation (long ncRNAs, microRNAs, and circular RNAs) [62]. Despite a lot of accumulated data, microRNAs remain one of the most puzzling parts of aging biology.
5. Biogenesis of microRNAs
To date, nearly 2700 human mature miRNAs have been identified [63]; they are presumed to target over 60% of protein-coding genes in humans [24]. Additionally, miRNAs are thought to modulate the expression of approximately one-third of all human genetic codes [22].
MicroRNAs are a class of small non-coding endogenous, evolutionarily conserved RNAs (~19–24 nucleotides in length) that generally function to modulate the levels/translation of messenger RNA (mRNA). Their primary function is related to the posttranscriptional regulation of gene expression [64,65]. miRNAs block the translation and perform the cleavage of mRNA targets, resulting in a reduction in protein levels. Additionally, it was shown that the binding of miRNA stimulates the initiation of mRNA decay factors and a subsequent potentiation of mRNA instability and degradation, resulting in a reduction in expression levels [25]. Furthermore, miRNAs were proven to participate in numerous pathobiological processes, such as cell proliferation, differentiation, apoptosis/necrosis, tissue degeneration, cancer, and age-related diseases [22].
The biogenesis of miRNAs occurs through a multi-step process that requires both nuclear and cytoplasmic phases. Typically, miRNAs are transcribed by RNA polymerase II and III as a long primary miRNA (pri-miRNA) with a cap and a poly-A tail in the nucleus. Pri-miRNA contains a terminal loop, which consists of two flanking unstructured single-stranded tails and a double-stranded stem of about 30 base pairs [34]. Pri-miRNAs get processed into short 60–70 nucleotide stem-loop structures called precursor miRNAs (pre-miRNAs) by the microprocessor complex, which consists of the RNase III enzyme Drosha and double-stranded RNA binding protein DGCR8 (Di George syndrome critical region 8 gene) [66,67,68]. The translocation of pre-miRNA into cytoplasm across the nuclear pore complex occurs with the aid of the Exportin5 (XPO5) complex, which belongs to the karyopherin β family bound to the Ras-related nuclear protein (Ran) GTPase [34,69]. In the cytoplasm, the hydrolysis of guanosine triphosphate (GTP) to guanosine diphosphate (GDP) results in pre-miRNA discharge [66]. The RNase III enzyme Dicer processes pre-miRNA and generates the miRNA duplex with phosphate at the 5′ end and a two-nucleotide overhang with a hydroxyl group at the 3′ end. The miRNA duplex gets loaded onto Argonaute (AGO) protein in the presence of the RNA-induced silencing complex (RISC). RISC consists of Dicer, its partner protein, which is also known as the transactivation response RNA binding protein (TRBP), and AGO. TRBP detects the guide and passenger strands in the miRNA duplex. Once it recognizes the strand with the less stable 5’ end, the protein loads the miRNA duplex in the correct orientation onto the AGO protein [70]. AGO unwinds the duplex and removes the passenger strand retaining the mature miRNA molecule, and only one strand remains to form an effector complex RISC. After being loaded into RISC, miRNA correctly positions itself alongside the target mRNA to direct the posttranscriptional repression [34], [66,68,71].
MicroRNAs interact with the 3′ untranslated region (UTR) of the target mRNA and, once bound, there appears to be two ways in which it can inactivate the mRNA, thus causing mRNA degradation and the inhibition of the target mRNA translation. The first mechanism is based on the complementary binding of miRNA to mRNA, with a subsequent deadenylation and degradation of the target mRNA. The second mechanism leads to the inhibition of translation, where the RISC complex prevents the ribosome subunit from binding [72].
6. MicroRNAs as Aging Biomarkers
Many miRNAs have been previously demonstrated to be differentially expressed in cells, tissues, whole blood [10,73,74,75], plasma [76,77,78], and serum [79,80,81] in relation to age.
The microRNA expression has been analyzed in the whole blood of 5221 adults, and 127 microRNAs that were differentially expressed depending on age and age-related diseases were identified. The targets of these miRNAs were associated with the main aging mechanisms, such as the regulation of transcription, translation, and immune responses. The authors suggested an epigenetic age prediction model based on the profiling of mRNA complementary to miRNA [82]. Kinser and Pincus (2020) reviewed the microRNA involvement in the modulation of aging, longevity, and age-related diseases within different species, including humans. They noticed that lin-4, let-7, miR-17, and miR-34 (known as longevity-related miRNAs) are conserved in humans and presumably promote lifespan prolongation [23].
Numerous experimental and computational data suggest that miRNAs regulate the expression of the majority of the known protein-coding genes at both the transcriptional and translational levels. In humans, microRNAs predominantly down-regulate their gene targets. The accurate prediction of gene targets and their involvement in the cellular regulations is necessary for understanding the physiological and pathological processes, pathogenesis of various diseases, and development of treatment strategies. Within the last decade, Dr. Xiaowei Wang and colleagues developed and improved an online database, miRDB (http://mirdb.org), for miRNA target prediction and functional annotations [83]. The miRDB database contains information about 7000 miRNAs and their 3.5 million predicted targets in five species: human, mouse, rat, dog, and chicken. Recently, the authors implemented an updated version of miRDB, the new MirTarget algorithm that allows analyzing custom miRNA data or gene target sequences (Tables S1–S10) for the transcriptome-wide prediction of gene targets or miRNA regulators for a flexible analysis of miRNA functions [83].
Zhang et al. (2015) profiled circulating miRNAs in serum from adults (40–70 years old) and identified significantly down-regulated age-dependent miRNAs, including miR-29b, miR-106b, miR-130b, miR-142-5p, and miR-340; and up-regulated miRNAs, including miR-92a, miR-222, and miR-375 [81]. Notably, miR-130b-5p was also involved in ribosome biogenesis and RNA processing and translation [10]. Interestingly, in long- versus short-lived individuals, there was a significant down-regulation of miR-340-3p, miR-374a-5p, and miR-376c in serum, which was negatively correlated with their lifespan [80]. miR-340-3p is predicted to have 316 targets (e.g., the RPTOR gene, a regulator of MTOR complex 1; the GNAI3 gene, G protein subunit alpha i3; the JAK gene, Janus kinase 3, etc.) outlined in the miRDB online database [63,83]. In addition, a longer lifespan was positively correlated with the up-regulation of miR-211-5p, miR-1225-3p, and miR-5095 [80]. Another large population-based study profiling the miRNAs in plasma from adults identified the age-dependent elevation of the following miRNAs: let-7a-5p, miR-30b-5p, miR-30c-5p (Table S2), miR-126-3p, miR-142-3p, and miR-210; meanwhile, the miR-93-5p (Table S7) expression level was decreased [77]. Similarly, there is an age-dependent up-regulation of miR-126-3p in the plasma of older individuals [78]. Noren Hooten et al. (2013) also found down-regulated miR-151a-5p, miR-181a-5p, and miR-1248 in the human serum of older individuals (a mean age of 64.6 years); these miRNAs were often found to be differentially expressed in the inflammatory pathways of chronic age-related pathologies [79,84].
Table 1 introduces the list of differentially expressed miRNAs with respect to their role in normal skin development.

Table 1.
The differentially expressed microRNAs (miRNAs) in the skin of long-lived individuals and their role in normal skin development.
The analysis of plasma circulating miRNA from a twin study conducted by the National Heart, Lung, and Blood Institute identified significantly up-regulated miR-3615 and miR-619 in younger deceased twins compared to their longer-lived co-twin. Additionally, the increased expression of miR-423-5p and miR-4305 was detected in less than 70% of short-lived twins [76]. In this study, changes in the remaining longevity-related miRNAs were not significant, potentially due to a small sample size.
The immune system deteriorates with age. There is significant evidence of miRNA participation in pathways regulating immunological function. For instance, the miR-24 identified in whole blood [10] and peripheral blood mononuclear cells [94] was shown to be involved in the activation of the immune response (Table 1). Furthermore, Brunner et al. (2012) discovered that the down-regulation of the expression of histone H2A family member X, which plays an essential role in DNA damage response, was initiated by the increased expression of miR-24 in T-cell lines [97].
In addition, miR-181a was reported to facilitate local immunological stability [98]. It was shown to be co-expressed with the CXCL16, RAB27A, and SPON2 genes responsible for immune response [82]. Cytotoxic T lymphocytes express RAB27A, which modulates the hexosaminidase and granzyme A granules secretion and immunological synapse activity [99]. Recent studies using senescence-accelerated mouse models discovered miR-301a-3p and miR-181a-1-3p to be differentially expressed in CD4+ T cells. Additionally, the up-regulated miR-301a was shown to facilitate the Th17 subset generation via the IL-6/23-STAT3 pathway, and miR-301a-3p was found to be significantly reduced in the CD4+ T cells of old mice. In contrast, miR-181a-1-3p was strongly elevated in the development of the age-related inflammatory phenotype, characterized by the enhanced levels of IL-6, IL-2, and TNF-α and the decreased levels of IL-10 in CD4+ T cells. Concomitantly, age-related alterations were found in the T cell differentiation-related pathways and their potential target genes, such as GNG12, MAPK8, PIK3CA, PIK3R1, PRKACB, PRKCA, and PRKCB regulated by miR-17-5p (Table S4), miR-144-3p, miR-451a, miR-301a-3p, and miR-582-5p [100]. Notably, numerous immune function-related genes (e.g., APOBEC3G, CCR7, CD28, CST7, CTSW, DPP4, ETS1, FAIM3, FLT3LG, ICOS, KIF13B, SLAMF7, TCF7, TGFBR3, TNFRSF17, TRAT1, etc.) were potential targets of miR-193b-3p. Similarly, miR-31-5p was predicted to target CD27, CST7, DPP4, ETS1, FASLG, F2R, FLT3LG, GIMAP5, GZMA, ICOS, INPP5D, KYNU, TCF7, TGFBR3, TRAT1, and many other immune function-related genes [82].
El Sharawy et al. (2012) validated eight miRNAs in the samples of 15 centenarians (a mean age of 96.4 years) and middle-aged individuals (a mean age of 45.9 years) that were proposed as longevity biomarkers. Among them, miR-106a, miR-126, miR-20a, miR-144*, and miR-18a were down-regulated, while miR-30d, miR-320d, and miR-320b were up-regulated in long-lived individuals [10]. Seven of these miRNAs were in line with the results of another study [101]. Storci et al. (2019) discovered longevity-related miRNAs in peripheral blood mononuclear cells and dermal fibroblasts from centenarians: down-regulated miR-21, miR-125a, miR-125b, miR-146a, and miR-155a (pro-inflammatory); up-regulated miR-335-5p, miR-532-5p, and miR-508-3p (anti-inflammatory). Simultaneously, the overexpressed pro-inflammatory and diminished levels of anti-inflammatory miRNAs were often linked to age-related diseases [74].
Due to the differential expression of microRNAs in long-lived individuals compared to younger ones, it was suggested that changes in the appearance of these miRNAs during physiological aging might inhibit carcinogenesis [21] and multiple pathologies associated with age [102]. For example, miR-17 (Table S1), miR-18a, miR18b, miR-20a (Table S5), and miR-106a (Table S8) in inflammation-associated conditions remained poorly expressed in centenarians [103,104], while they were up-regulated in cancers [21]. Conversely, the up-regulated miRNAs related to longevity were reported to be down-regulated in cancers; these miRNAs included miR-320d in colon cancer stem cells [105], miR-339-5p in breast cancer tissues [106], and miR-30d in chronic lymphoblastic leukemia [107] (Table 2). In comparison to healthy individuals, some age-related neurodegenerative diseases such as Alzheimer’s disease and spinocerebellar ataxia type 1 were associated with the increased expression of miR-101, miR-130, and miR-144 in the cerebellum and cortex [108]. The up-regulation of miR-130a was also found in the brain tissues of rats with cerebral infarction and was correlated with subsequent elevated neuronal apoptosis, accompanied by the liberation of inflammatory mediators and the inhibition of the PTEN/PI3K/Akt signaling pathway and anti-apoptotic genes [109].

Table 2.
The association of miRNAs differentially expressed in long-lived individuals with different pathological conditions.
The inverse expression of microRNAs in long-lived individuals and age-related diseases may signify the pathogenetic mechanisms of the latter and could potentially be used for diagnostic preventive purposes and treatment strategies. However, there is still incomplete information on the involvement of miRNA in various mechanisms of organismal aging and the skin in particular.
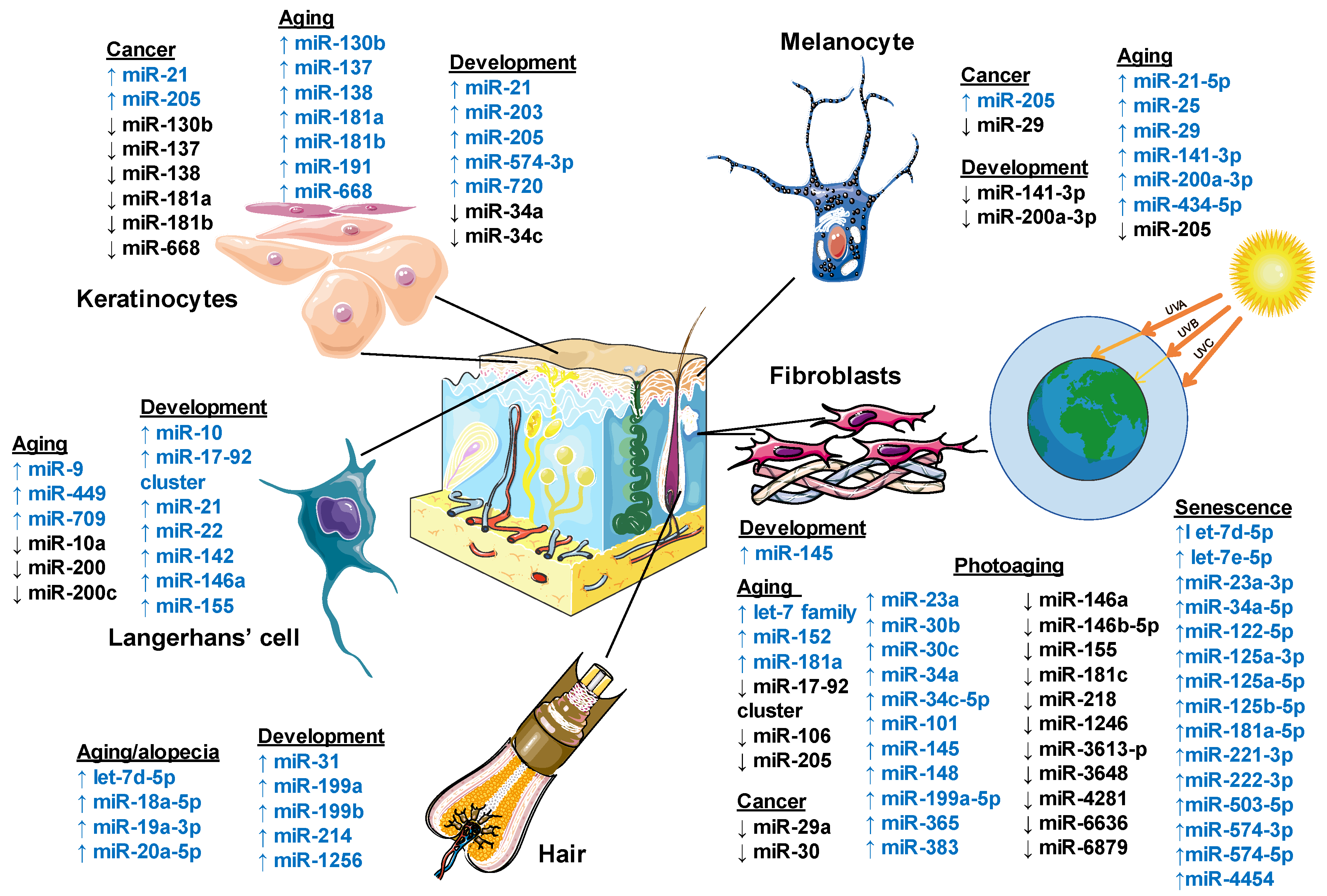
Figure 2 summarizes the miRNAs involved in normal development and skin pathologies.
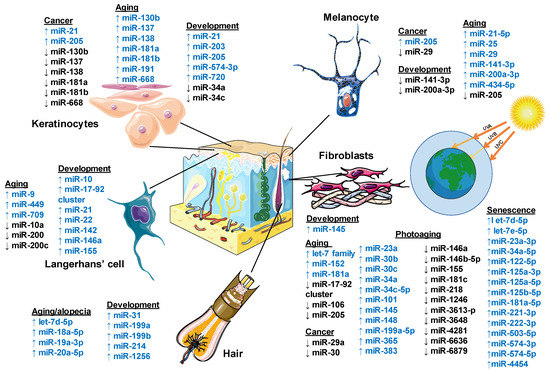
Figure 2.
Role of microRNA regulation in human skin. Schematic diagram showing the involvement of miRNA in the cutaneous components’ development, aging processes, and cancer. MicroRNAs are shown as up-regulated in blue color and down-regulated in black color. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License. (http://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Table 2 introduces the list of age-related differentially expressed miRNAs with respect to their involvement in various pathological conditions.
7. Epigenetic Regulation in Cutaneous Cellular Senescence
Cellular senescence is associated with the halting of mitotic cell divisions and the loss of proliferative potential, along with preserved viability or metabolic activity [126,127]. The senescent stage is represented by deviations in the protein expression levels and activity and, in particular, by changes in the p53/p21 and the pRB/p16 tumor suppressor pathways [128]. Indeed, tumor suppressor p53, also known as the Guardian of the Genome, down-regulates the expression of genes required for telomere maintenance, DNA repair, and centromere structure; its expression is enhanced by the progressive passaging of human fibroblasts [129]. One of the leading reasons for replicative senescence is telomere attrition, which works as a mitotic clock [130]. MicroRNAs likely play a role in cellular aging due to their involvement in proliferation-associated gene silencing. To date, about 30 proteins have been discovered to be implicated in cutaneous aging. Furthermore, most of the miRNAs associated with these proteins in the aging skin were reported to be down-regulated. For instance, filaggrin that undertakes the modulation of the cutaneous barrier function, hydration, and pH, and presumably the buffering capacity (via its breakdown products), was decreased 2.7-fold in aged skin [131]. Epigenetic mechanisms, including DNA methylation, histone modification, chromatin remodeling, and non-coding RNA control, also guide cellular senescence [132].
Within recent decades, robust evidence has supported an essential role of chromatin changes, including DNA methylation and post-translational histone modifications, in the regulation of gene expression. The latter depends on chromatin accessibility: more “relaxed” chromatin makes a gene more accessible for transcription. DNA and histone modifications establish an epigenetic “code” that controls gene function by favoring or limiting the transcriptional activity of genomic domains. DNA can be chemically modified by the methylation of cytosine nucleotides in a CpG context. DNA methylation is established and maintained by methylating enzymes, DNA methyltransferases (DNMTs). The hypermethylated DNA segment has a decreased chance of being transcribed into mRNA, and therefore genes located in this region typically have a low expression level. In contrast, hypomethylated DNA regions allow active transcription. Simultaneously, DNA methylation changes DNA–histone interactions, where unmethylated DNA is typically associated with acetylated histones, whereas methylated DNA is associated with deacetylated histones [133].
Epigenetic regulation plays a critical role in controlling the expression of various genetic elements. For example, DNA methylation and histone modifications are responsible for X chromosome inactivation and the repression of repetitive elements [134], such as transposable elements, telomeres, and centromeric DNA [4]. In turn, condensed chromatin limits recombination and decreases the risk of mutation, therefore facilitating genomic stability. Apart from the control of gene expression, epigenetic modifications control recombination, repair, stem cell development, and various processes of cellular differentiation. The impairment of epigenetic regulation is associated with aging and aging-related diseases, such as cancer [135]. For instance, extracellular matrix molecules such as ECM1 maintain the differentiation and maturation of the epidermis, keratinocyte adhesion and signaling, neovascularization, and the dermis functioning via ECM formation. The increased expression of ECM1 is associated with enhanced invasiveness and metastasis in different cancers. Recent studies demonstrated that DNA methylation and hydroxymethylation regulate the tumor-suppressive role of ECM1, ATF5, and EOMES in human hepatocellular carcinoma [136]. Other studies found significant differences in the DNA methylation levels in skin samples from young and old as well as sun-exposed and protected body sites; differences in DNA methylation were found at particular gene loci [137], correlating with the level of damage in the skin [29]. Site-specific changes in DNA methylation in response to aging or skin damage can be used for predicting the aging phenotype or the risk of development of specific dermal diseases.
The epigenetic regulation of the majority of biological processes depends on DNA methyltransferase 1 (DNMT1). DNMT1 maintains CpG methylation, and its decline in activity with age leads to a decrease in global methylation or epigenome changes (drift) in aging cells [138]. A recent study demonstrated that the DNMT1 expression was decreased in passage-aged fibroblasts, and its silencing in young fibroblasts induced the senescence phenotype. However, the microarray analyses showed that the down-regulation of miR-217 in old fibroblasts stimulated a partial reversion of the senescence phenotype. The authors also demonstrated that miR-217 promoted senescence by suppressing the DNMT1-mediated methylation of p16 and pRb by targeting the DNMT1 3′-untranslated region (3′-UTR) in skin tissues and aged fibroblasts [139]. A more detailed description of the role of methylation and DNMT1 in aging is beyond the scope of this review.
8. The Role of microRNAs in the Dermal Aging
The significance of miRNAs in the regulation of skin development and homeostasis has started to unfold within the last few years. Numerous evidence-based studies have shed light on some novel miRNAs that regulate gene expression in stem cell biology and developmental biology. To date, we have accumulated a significant amount of knowledge about the miRNAs’ involvement in specific steps of the organismal development, physiology, and pathogenesis of certain conditions. Nevertheless, the role of miRNAs in regulating skin development, maturation, functioning, and aging is not yet fully characterized; therefore, multiple studies are in progress to elucidate the related mechanisms. In this section, we will concentrate on the mechanisms of molecular regulation of the aging process maintained by miRNAs and their main skin targets (i.e., keratinocytes, dermal fibroblasts, melanocytes, etc.) (Figure 2).
8.1. MicroRNAs Influence Keratinocyte Aging
One of the first studies focused on senescence-associated skin cells revealed a set of miRNAs involved in the regulation of epidermal normal human keratinocytes (NHKs). Among 126 senescence-associated miRNAs specified in senescent NHKs, 9 miRNAs (7%) were down-regulated and 117 miRNAs (93%) were up-regulated. Shin et al. (2011) found significantly up-regulated miR-137 and miR-668 during the replicative senescence of three independent NHK cultures and organismal aging. The authors reported a strong direct correlation between the above-mentioned miRNAs and an elevation in senescence-associated β-galactosidase activity, along with an elevation in p16INK4A and p53 senescence markers of the ARF/p53 and p16INK4A/RB pathways, respectively [140]. In parallel, they considered senescence as a potent tumor-suppressive pathway based on the fact that miR-137 and miR-668 were down-regulated in many human head and neck squamous cell carcinoma cell lines. Apart from the inhibition of cancer cell growth, miR-137 is involved in the induction of the cell cycle G1 arrest by targeting CDK6 and Cdc42. miR-137 likely works through the suppression of either the Cdc42-PAK1-MLC or ERK signaling pathway and the proliferation/invasion of colorectal cancer cells by mimicking the effects of Cdc42 knockdown [122,123].
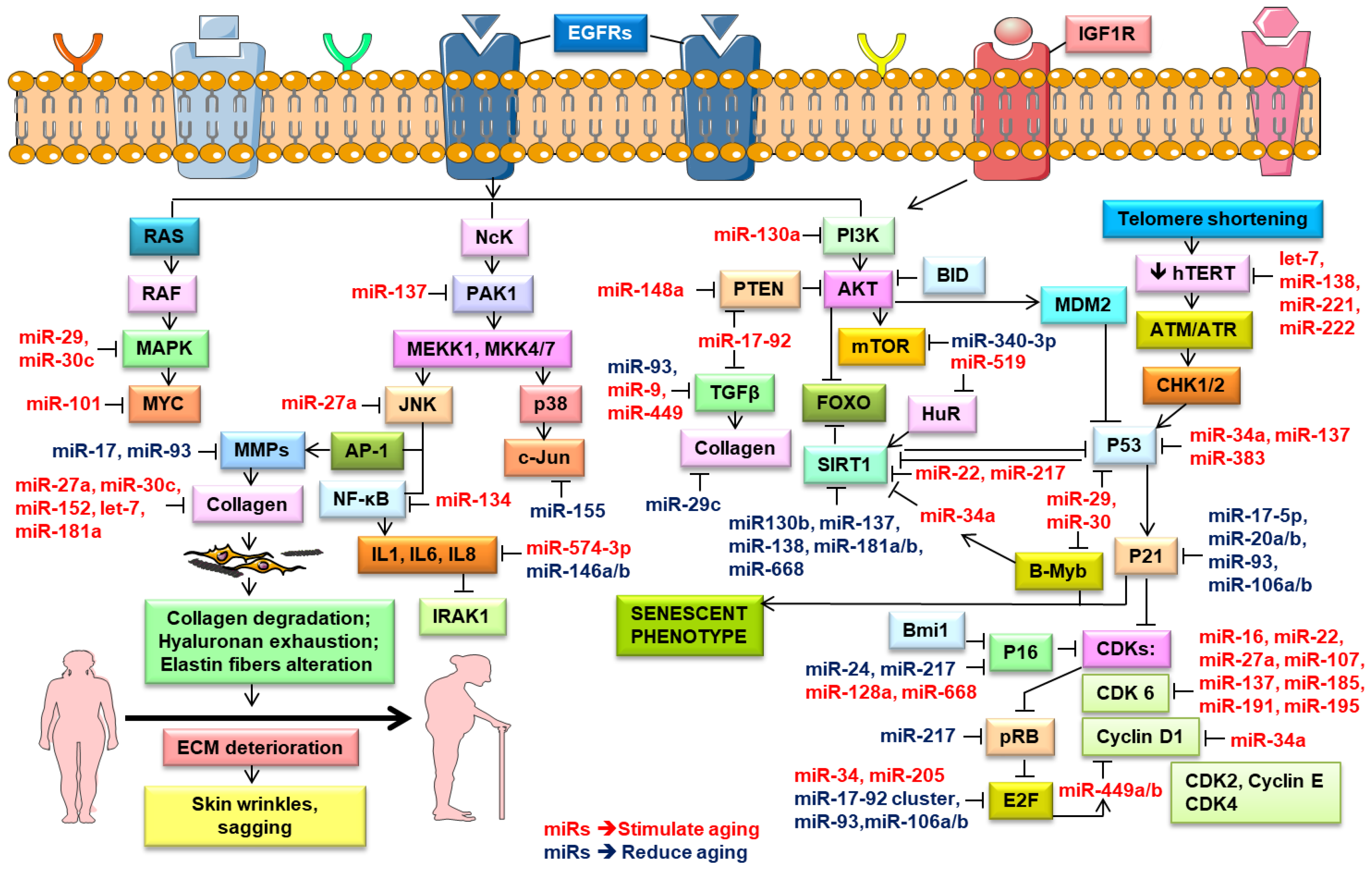
Rivetti et al. (2012) identified up-regulated miR-138, miR-181a, miR-181b, and miR-130b in keratinocytes during replicative senescence [125]. They pointed out that these four miRNAs modulated the cell proliferation pathways by targeting p63 and Sirtuin 1 (SIRT1) mRNAs (Figure 3). SIRT1, an NAD-dependent histone deacetylase, modulated cellular differentiation, metabolism, immune and stress responses, and replicative senescence, and reduced p53-mediated apoptosis and FOXO-induced apoptosis [141]. SIRT1 extended the lifespan in mammals [142], while its knockdown induced cellular senescence [125]. Aging is associated with a decrease in the SIRT1 levels and their protective effects, leading to numerous age-related diseases [17]. Down-regulated miR-138, miR-181a, miR-181b, and miR-130b as well as miR-137 and miR-668 were found in different cancers and were considered to be tumor-suppressor miRNAs [124,125,143]. Concomitantly, many research groups noted the role of miR-130b in targeting p63 mRNA. The P63 gene was a transcription factor of the p53 family that played a vital part in the epithelial development. Based on in vitro and in vivo studies, p63 was found to counteract cellular senescence and aging [9]. Besides, p63 directly inhibited the expression of the senescence-inducing miRNAs (e.g., miR-130b, miRNA-138, miRNA-181a, and miR-181b) [125]. Moreover, the p63-mediated regulation of miRNA activity was demonstrated in tumor and metastasis suppression through the transcriptional regulation of Dicer and miR-130b [88].
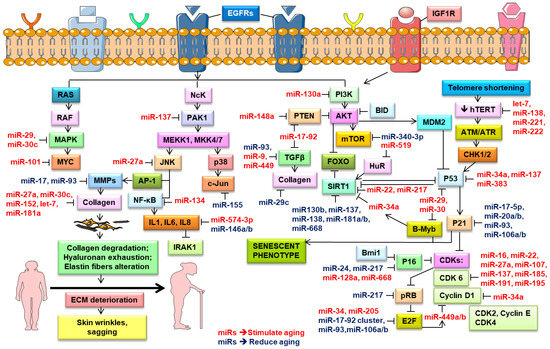
Figure 3.
Role of microRNA in the regulation of the key molecular signaling pathways in the skin aging process. Schematic diagram shows the miRNAs that regulate cellular signaling mechanisms and potentially stimulate (depicted in red color) and reduce (represented in blue color) aging. Pointed arrow defines activation, while blunted – inhibition. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (http://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Mancini et al. (2014) has broadly reviewed the role of senescence-associated miR-191 during keratinocyte senescence. MiR-191 is capable of blocking the G1–S phase transition through the control of cyclin-dependent protein kinase 6 (CDK6) gene expression. This blockage is manifested as a cell cycle arrest and a quiescent state that contributes to the development of senescence processes [9]. In parallel, miR-191 down-regulates CDK6 and SATB1 (special adenine- and thymine-rich binding protein 1) mRNA, accompanied by an increase in senescence-associated markers [144]. SATB1 acts as a docking site for chromatin remodeling enzymes; it also recruits corepressors (HDACs) or coactivators (HATs) directly to promoters [145]. miR-191 targets CDK9, NOTCH2, and RPS6KA3; it also represses the proliferation of primary human fibroblasts. Besides this, it plays a role in multiple cancer types, including gastric, colorectal, breast, thyroid, and hepatocellular carcinoma [146].
In addition to keratinocytes, the epidermis contains other cell types, such as melanocytes, Langerhans cells (LCs), Merkel cells, and stem cells [32] (Figure 1).
8.2. The Role of microRNAs in the Age-Dependent Regulation of Immunomodulatory Skin Cells
Langerhans cells (LCs) are immature skin-residential dendritic cells (DCs) that recognize and present antigens. LCs, like all peripheral DCs, can traffic peripherally acquired antigens to regional lymph nodes, where they present antigens to naive and memory T cells and initiate adaptive immune responses or induce tolerance. These immune skin cells reside either in the epidermis or in the suprabasal layer of the stratified squamous epithelium of the upper part of the digestive system mucosae [147,148]. The epidermis is an epithelial layer that is composed primarily of keratinocytes, albeit LCs constitute 3–8% of the entire epidermal cellular population [149]. LCs are also involved in the pathogenesis of multiple cutaneous illnesses, including autoimmune diseases, allergy contact dermatitis, and cancer [148,149]. Considerable progress has been made in the characterization of various developmental processes in LCs [147], whereas the mechanisms involved in LC senescence and aging are still not clear.
The differential regulation of miRNAs in LCs with age contributes to immunosenescence by increasing the susceptibility to skin viral and fungal infections as well as to cutaneous cancer development. Several miRNAs are linked to LC development (miR-22 and miR-142 via the interferon regulatory factor 8 (IRF8), maturation, differentiation (miR-21, miR-34a, miR-99b, miR-223, and miR-511) and the immune function (miR-10, miR-21, miR-142-3p, miR-146a, and miR-155) [150]. Aging LCs are associated with the up-regulated expression of miR-709, miR-449, and miR-9 alongside with down-regulated miR-200c and miR-10a. miR-449 and miR-9 target the components of TGF-β signaling, such as TGFβ1, TGFβRI, TGFβRII, RUNX3, and C/EBP (CCAAT/enhancer-binding protein). Hence, the overexpressed miR-449 and miR-9 in aging LCs may down-regulate the TGF-β signaling pathway and also block the development of Langerhans cells [151]. LC homeostasis and development is also mediated by colony stimulating factor 1 (CSF1)/CSFR and IRF8 [147,152]. In mice, the overexpression of miR-709 and miR-449 in LCs down-regulates the expression of IRF8 and CSFR, causing a deficiency in the development of LCs [151]. Another study found that the transcription factor Gfi1 (growth factor independence 1) maintains LC homeostasis and is a potential target of miR-200 [153]. Interestingly, the down-regulation of miR-200 in aging LCs may directly up-regulate Gfi1, with a subsequent blockage of the LCs’ activity [151].
Besides this, the homeostasis and maturation of LCs are also dependent on the RANKL/RANK system, which is a putative target for miR-9 or miR-20 [151]. Hence, RANKL maintains lymphoid tissue formation and bone homeostasis. RANKL is also an apoptosis regulator gene, a binding partner of osteoprotegerin (OPG), a ligand for the receptor RANK that controls cell proliferation through cyclins D1, ID2, and ID4 [154,155]. The overexpression of miR-9 diminishes cellular migration and survival by repressing the expression of the proto-oncogene Cbl, which enhances the amount of pro-apoptotic protein Bim [156,157]. The miR-17-92 cluster encoding six miRNAs—miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1—is also highly expressed in skin-specific dendritic cells—i.e., the Langerhans cells [89]. Nevertheless, the miR-17-92 cluster has an unclear role in the process of survival, maturation, antigen presentation, and migration ability in these cells, presumably due to the functional redundancy [147,158]. Thereby, age-regulated miRNAs have the potential to deteriorate LC development, maturation, and function by targeting various signaling pathways.
8.3. The Age-Dependent microRNA Regulation of Various Processes in Melanocytes
Melanocytes are another network of epidermal cells that produce the pigment protecting from ultraviolet radiation [159]. Apart from the skin, these melanin-producing cells can be found in the hair follicles, eyes, inner ear, bones, heart, and brain. Melanins (eumelanin, pheomelanin, and neuromelanin) are pigment molecules that are most often synthesized by melanocytes (melanogenesis). In the skin, it subsequently distributes itself in the epidermal keratinocytes. Melanin absorbs light in the skin and hair, thus leading to photoreceptor shielding, thermoregulation, and photoprotection. Furthermore, it is a potent cation chelator and is able to act as a free radical sink, reducing the risk of folate depletion and dermal degradation, leading to premature aging and decreasing the odds of the development of malignant melanoma [160]. After the age of 30, melanocytes decrease in number by 8% to 20% per decade. The reduction in melanocyte numbers is smaller in sun-exposed areas. However, these cells are irregularly spaced and functionally compromised, producing an abnormal pigmentation upon sun exposure [37].
Interestingly, miRNAs and long non-coding RNAs were reported to play an essential role in melanogenesis-regulating molecules in melanocytes. The α-melanocyte-stimulating hormone (α-MSH), one of the leading factors in melanogenesis, was found to down-regulate the expression levels of miR-141-3p and miR-200a-3p. Conversely, the suppressed melanogenesis and tyrosinase activity were induced by the overexpression of miR-141-3p and miR-200a-3p in B16-4A5 cells, which is a melanoma-derived cell line. Furthermore, both miR-141-3p and miR-200a-3p directly targeted the microphthalmia-associated transcription factor (MITF), a key regulator of color genes coding for tyrosinase and hyaluronidase [66]. Tyrosinase, a melanocytic membrane-bound glycoprotein, is critical for melanin biosynthesis in the skin and hair. Hyaluronidase degrades subcutaneous hyaluronan, a central anti-aging component of the extracellular matrix, facilitating a mechanism for skin wrinkle formation [161]. MITF controls survival, proliferation, pigmentation, invasion, and oxygen stress in melanocytes by regulating the downstream gene expression. The up-regulation of miR-141-3p and miR-200a-3p decrease cutaneous pigmentation by targeting MITF [162]. There is also evidence that melanin production is controlled by miR-25, which suppresses MITF, whereas miR-21-5p regulates MITF indirectly via Sox-5 and miR-434-5p via tyrosinase and hyaluronidase targeting [161,163,164]. Besides this, the abatement of growth factors (e.g., SCF and α-MSH) may contribute to premature melanocyte cell senescence and the consequent clearance by the immune system [165]. Moreover, the inhibition of melanogenesis deteriorates melanocyte protective functions and might potentiate premature aging mechanisms. Indeed, senescent melanocytes induce paracrine telomere damage and senescence in the surrounding keratinocytes. This spontaneously affects the proliferative potential of keratinocytes, resulting in age-related epidermal atrophy [166].
Recent studies report that miRNAs might induce a senescence phenotype in melanocytes and inhibit melanoma tumor progression. The results of miRNA expression profiling analyses showed a significantly down-regulated miR-205 in melanoma compared with nevi; the miR-205 expression was correlated inversely with melanoma growth. The examination of miRNA target databases predicted E2F1 and E2F5 as plausible targets of miR-205 [167]. E2F1 is a member of the E2F family and regulates the G1/S transition phase in the cell cycle, which is tightly controlled by the retinoblastoma protein (Rb) (Figure 3). E2F1 is able to induce or inhibit apoptosis [168]. Furthermore, E2F1 potentiates cellular survival through the AKT pathway, which mediates cell growth, cell survival, and the inhibition of apoptosis [169]. Its increased expression prompts cells towards oncogenic transformation [170]. Another E2F family member, E2F5, similar to E2F1, is an oncogenic cell cycle regulator that is also found to be amplified in numerous tumors, including melanoma [171]. miR-205 in melanoma cell lines inversely correlates with the expression levels of E2F1 and E2F5. The increased expression of miR-205 reduces E2F-regulated AKT phosphorylation and suppresses the proliferative capacity of melanoma cells. The overexpression of miR-205 initiates a caspase cascade to induce apoptosis by suppressing the phosphorylation of caspase-9 and Bcl-2-associated death promoter (BAD), and concomitantly increasing the cleavage of caspase-3 and Poly (ADP-ribose) polymerase (PARP) with a subsequent release of cytochrome c. A continuous up-regulation of miR-205 suppresses melanoma cell proliferation, colony formation, and tumor cell growth in vivo and induces a senescence phenotype. The up-regulation of trimethyl-histone H3 (Lys-9), another marker of senescence at the chromatin level, is accompanied by the elevated expression of p16INK4A [45]. In turn, the up-regulation of p16INK4a activates Rb by suppressing its phosphorylation through the inhibition of CDK4/6 kinase activities and thus induces a senescence program. Altogether, these data indicate a tumor suppressor role of miR-205 in melanoma [167]. Another recent study suggests that the miR-29 family also prevents melanoma progression downstream of MAPK signaling by repressing the transcription factor MAFG (MAF bZIP transcription factor G) [172]; more studies are needed to substantiate these findings.
8.4. The Role of microRNA Regulation in the Aging of Dermal Fibroblasts
The dermal layer comprises large amounts of extracellular matrix (ECM), composed of proteins and specialized carbohydrates. Fibroblasts represent most of the dermal cells. In contrast, Schwann cells and nerve endings, endothelial cells organized in vessels, pericytes, mast cells, tissue macrophages, and other cells of the immune system are a minority. The dermis consists of the papillary (upper) and reticular (lower) layers. The papillary layer starts at the epidermal basement membrane and is more densely populated by fibroblasts than the reticular dermis, which contains thicker collagen fibers. However, the ECM becomes less organized due to the altered ECM protein turnover or/and post-translational modifications accumulated with age [159].
Fibroblasts produce and regenerate components of ECM; participate in wound healing, etc.; and, like other cells, deteriorate in functional capabilities with age. To date, we have accumulated a significant amount of evidence that miRNAs can also mediate the functions of fibroblasts. MiRNAs are linked to the reduced expression of transmembrane receptors (e.g., integrins) and components of the ECM (e.g., collagens, elastins) in senescent skin fibroblasts. Mancini et al. (2014) found that miR-152 and miR-181a induce senescence in proliferating human dermal fibroblasts. Indeed, miR-152 can significantly decrease dermal fibroblast adhesion through the down-regulation of integrin alpha 5 (ITGA5) by direct targeting [9]. Integrin-α5 participates in cell-surface-mediated signaling through the fibronectin receptor by enhancing cell adhesion and migration. Additionally, ITGA5 is one key functional miR-205 target in the re-epithelialization process in the epidermis. Similar to the effect of TGF-β1, the epidermal down-regulation of miR-205 promotes ITGA5 keratinocyte migration, whereas an increased miR-205 level is associated with delayed wound healing and chronic wound development [173]. Interestingly, the expression of collagen XVI (COL16A1) in senescent fibroblasts is down-regulated and directly targeted by the up-regulated miR-181a. COL16A is found in fibroblasts, keratinocytes, smooth muscle cells, and amnion; however, it is a minor component of the cutaneous ECM [174]. Collagen XVI provides the mechanical anchoring by connecting ECM proteins to cells; it is expressed in the dermal-epidermal junction zone of the papillary dermis [175]. It appears that both miR-152 and miR-181a may have an intricate role in the dermal ECM remodeling of aged skin [9]. Recently, more microRNAs as biomarkers of aging were identified in human dermal senescent fibroblasts that affected telomere maintenance and all phases of a cellular life cycle. Among the identified biomarkers were 15 up-regulated miRNAs, such as let-7d-5p (Table S3), let-7e-5p, miR-23a-3p, 34a-5p, miR-122-5p, miR-125a-3p, miR-125a-5p, miR-125b-5p, miR-181a-5p, miR-221-3p, miR-222-3p (Table S10), miR-503-5p, miR-574-3p (Table S1), miR-574-5p, and miR-4454; in fact, miR-221/222 were presented in both replicative and oxidative stress-induced senescence [85].
Martinez et al. (2011) demonstrated the up-regulation of the miR-29 and miR-30 microRNA families during induced and replicative fibroblast senescence, along with the activation of the Rb pathway. The binding of Rb-E2F complexes to the Myb-related protein B (B-Myb) promoter repressed B-Myb transcription and its expression, which could result in senescence. They found that the overexpressed miR-29a and miR-30 stimulated senescence directly suppressing the expression of the B-Myb transcription factor [176]. B-Myb is involved in cell cycle progression and cellular senescence by possessing both activator and repressor activities. The cyclin A/cyclin-dependent kinase 2 phosphorylated B-Myb during the S-phase of a cell cycle and was subsequently degraded during late G2 in a ubiquitin-dependent manner [177]. B-Myb was present in all mitotically cycling cells, thus activating the cell division cycle 2, cyclin D1 and insulin-like growth factor-binding protein 5 genes [178]. Interestingly, miR-29c was reduced in mice with isoproterenol-induced myocardial fibrogenesis. However, the increased expression of the anti-fibrotic factor miR-29c down-regulated its target genes, such as Collagen Type I Alpha 1 Chain (COL1A1), COL1A2, COL3A1, COL5A1, fibrillin-1, and TGFβ1, thus exerting protective effects against fibrosis and myocardial injury [179]. Conversely, the overexpression of B-Myb and factors mediating its activation strongly correlated with the down-regulated miR-29a and miR-30 and cancer progression, respectively [177]. miR-29a [180,181] and the entire miRNA-30 family (i.e., miR-30a (p = 0.024), miR-30b (p = 0.021), and miR-30c (p = 0.009)) [182] were found to be down-regulated in acute myeloid leukemia (AML).
In turn, the senescence process in dermal fibroblasts was also associated with down-regulated miRNAs, such as the miR-17-92 cluster and miR-106 [112]. The miR-17-92 cluster transcriptionally regulated genes involved in cell cycle control and tumor development, such as BCL2L11 (BIM), p63, p57, p27, and p21. It potentiated the proliferation, and thereby was believed to counteract senescence [9]. Transcription factors E2F1 and E2F3 activated the miR-17-92 cluster transcription [183], while p53 repressed it [184]. Hence, an age-dependent decrease in E2F1 and E2F3 led to a reduced expression of the miR-17-92 cluster, affecting the P53, ERB, and MAPK signaling pathways (Figure 3), and consequently accelerated apoptosis and inhibited growth [179]. Furthermore, the inhibition of miR-17-92 and miR-106b-25 in a c-MYC-dependent manner led to an increase in the BIM, PTEN, and P21 levels [185].
The data described here show that the expression of senescence-associated miRNAs affects the functioning of various genes in cutaneous cells and might promote or counteract cellular senescence. Moreover, many of these genes are expressed in other body tissues (e.g., p63, SIRT1) and are directly linked to organismal aging [22].
Table 3 summarizes the information about miRNAs that are differentially expressed in long-lived individuals and lists the top predicted targets of these miRNAs. A more detailed list of these targeted genes is presented in Table S1. All the possible targets of differentially expressed miRNAs are listed in Table S1–S10.

Table 3.
MicroRNA expression in the skin of long-lived individuals with a prediction of functional miRNA targets.
ADAM28, ADAM metallopeptidase domain 28; BMP2, bone morphogenetic protein 2; BNIP3L, BCL2 interacting protein 3 like; CASP3, caspase 3; CCND2, cyclin D2; CNR1, cannabinoid receptor 1 (CB1); CNRIP1 cannabinoid receptor interacting protein 1; COL1A2, collagen type I alpha 2 chain; CSF1, colony stimulating factor 1; E2F3, E2F transcription factor 3; EGFR, epidermal growth factor receptor; HA, hyaluronic acid; HAS2, hyaluronan synthase 2; IL6, interleukin 6; INF-α/β/ɣ, interferon-alpha/beta/gamma; JAK1, Janus kinase 1; KRT5, keratin 5; KRT10, keratin 10; LCs, Langerhans cells; MITF, melanocyte inducing transcription factor; MMP3, matrix metallopeptidase 3; NOTCH2NLA, notch 2 N-terminal like A; PAK5, p21 (RAC1) activated kinase 5; PIP3, phosphatidylinositol-3,4,5-trisphosphate; pRB, retinoblastoma protein; PTEN, phosphatase and tensin homolog; RXRA, retinoid X receptor alpha; SIRT5, sirtuin 5; SIRT7, sirtuin 7; SMAD1, SMAD family member 1; SMAD4, SMAD family member 4; SPTBN1, spectrin beta, non-erythrocytic 1; TF, transcription factors; TGF-β, transforming growth factor-beta; USP45, ubiquitin specific peptidase 45.
Currently, various types of clinical trials directed towards studying aging and age-related pathologies are running worldwide. Table 4 summarizes ongoing clinical trials in which the prominent common feature is a microRNAs assessment in different tissues, including skin, and their role in the pathogenesis of aging-related processes; more detailed information about these clinical trials are published at https://clinicaltrials.gov/.

Table 4.
Ongoing clinical trials associated with aging and age-related conditions which use miRNAs as a target or a marker.
9. The Role of miRNAs in Dermal Photoaging
The continuous exogenous effect of solar ultraviolet (UV) exposure is one of the primary reasons for premature skin aging and is called photoaging. Daily, humans are typically exposed to the UV wavelengths of sunlight, which are primarily composed of UVA (90–95%) and UVB (5–10%). These wavelengths of light are absorbed by multiple cellular biomolecules, including genomic DNA [188]. The short-wave radiations (UVB under 300 nm and UVC) are the most dangerous. They can damage DNA because their wavelength coincides with the absorption spectra of DNA, RNA, and proteins [189]. DNA damage caused by UV exposure induces abundant bulky lesions, such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4 PPs) [190]. CPDs formation occurs preferentially at thymine-containing dipyrimidine sites and likely also at methyl CpG-associated dipyrimidine sites, which include the TCG sequence [191]. It can lead directly or indirectly to DNA strand breaks that typically manifest in the form of mutations, neoplastic transformation, and premature cutaneous aging [192].
Recently, Zhang et al. (2020) demonstrated the UVB-induced up-regulation of miR-27a in keratinocytes [187]. They discovered that, following UVB irradiation, miR-27a facilitated the removal of CPDs and reduced cell apoptosis. Furthermore, miR-27a (Table S6) decreased the expression of TARDBP (target genes transactive response DNA-binding protein) and APAF-1 (apoptotic protease activating factor-1) [187]. The overexpression of miR-1246 directly up-regulates UVB-induced apoptosis through the suppression of the Rhotekin 2 (RTKN2) expression [193]. An increase in the miR-26a in HaCaT cells promotes UVB-induced apoptosis by the histone methyltransferase enhancer zeste homolog 2 (EZH2), which it turn depends on the MYC expression level [187,194,195]. UVB irradiation also targets the 3′-UTR of EZH2 by the up-regulation of miR-101 and induces senescence in human fibroblasts. Nevertheless, both the overexpressed miR-101 and the down-regulated EZH2 may independently induce senescence in the absence of UVB exposure [196]. The UVB-stimulated up-regulation of miR-141 expression inversely correlates with PTEN (Phosphatase and tensin homologue deleted on chromosome 10) expression levels in HaCaT cells. In contrast, anti-miR-141 increased the PTEN expression and reduced apoptosis in the UVB-treated cells [197].
It was lately found that UVA- and UVB-exposure of keratinocytes in vitro induced hypermethylation and histone modifications, which silenced tumor suppressor genes (Cip1/p21/p16INK4a) relevant for photo-carcinogenesis. The DNA hypermethylation of the CDKN2A/B locus in dermal fibroblasts was also observed in gerontology donors [198]. A global genome analysis of DNA methylation patterns obtained from the sun-protected and sun-exposed skin areas of young and old individuals indicates that chronic insolation (i.e., extrinsic aging) results in global DNA hypomethylation, whereas the intrinsic aging of sun-protected skin may cause the widespread hypermethylation of CpG islands [6,199].
UV radiation types A and B are able to penetrate deep into the skin, harmfully affecting the epidermal and dermal layers. Despite the superficial location, epidermal keratinocytes are less sensitive to UV radiation than dermal fibroblasts [9]. Notably, UVA up-regulates c-Jun expression; the transcription of c-Jun is autoregulated by its product Jun, and c-Jun affects the collagen gene activity in human skin fibroblasts. MiR-155 is considered a direct modulator of c-Jun expression. UVA exposure down-regulates miR-155, leading to the up-regulation of c-Jun [200].
To date, several studies implicate miRNAs in premature cellular senescence caused by UVB irradiation. For example, the UVB-exposed cells have increased the expression of miR-34c-5p that targets the 3′-UTR of E2F3. Like other E2F family members, E2F3 participates in cell cycle progression, proliferation, and development. Additionally, it protects dermal fibroblasts from the UVB-induced premature senescence, because it regulates senescence-related genes (e.g., p53 and p21WAF-1), and is also bound specifically to the retinoblastoma protein pRB in a cell-cycle dependent manner [201]. Recently, Blackstone et al. (2020) found a significant overexpression of the miR-34 family and a simultaneous increase in the Dicer1 expression along with the decreased fibroblast proliferation and reduced diameter of fibrils of dermal collagen caused by chronic UVB exposure in vivo. The disintegrated collagen structure entirely deteriorates the deposition of the extracellular matrix and alters the skin biomechanics, leading to skin weakness, stiffness, and reduced elasticity and pliability [202].
Differential miRNA expression profiles have been identified in photoaged primary human fibroblasts irradiated with long-wave UVA light [186]. They linked five up-regulated miRNAs (miR-30b, miR-30c, miR-148a (Table S9), miR-199a-5p, and miR-365) and seven down-regulated ones (miR-146a, miR-146b-5p, miR-181c, miR-218, miR-1246, miR-3613-3p, and miR-4281) in the photoaging process in dermal fibroblasts. The authors suggested that the predicted miRNA targets were associated with pathways in cancers, including the TGF-β pathway. They found that miR-146a targeted Smad4, TRAF6, IRAK1, and FADD; and they also noticed that the UVA-induced overexpression of miR-146a could moderately decrease the expression of Smad4 in fibroblasts via base pairing with Smad4 3′-UTR [186]. Curiously, another group of authors found that the overexpression of miR-146a antagonized the UVA-induced inhibition of proliferation and suppressed the up-regulation of aging-related genes (e.g., p21WAF-1, p16, and p53) through targeting Smad4 [203]. In another study, Bhaumik et al. (2009) showed the up-regulation of miRNAs 146a and 146b in response to the IL-1 receptor signaling as a part of a negative feedback loop that restrained the excessive senescence-associated secretory phenotype (SASP) activity. In particular, the ectopic expression of miR-146a/b in skin fibroblasts suppressed IL-6 and IL-8 secretion and down-regulated IRAK1 (Interleukin-1 receptor-associated kinase 1), a crucial component of the IL-1 receptor signal transduction pathway [102]. Even so, aging biology needs more evidence-based studies to support the role of miR-146 as an anti-aging photoprotective miRNA (Figure 3).
In the recent study, using the microarray analysis, Srivastava et al. (2018) revealed the differentiated expression of miRNAs between the sun-protected and sun-exposed skin of young and aged groups [204]. In comparison with the sun-protected skin, in the sun-exposed skin the expression of miR-34a, miR-145, and miR-383 was increased, while that of miR-663b, miR-3648, and miR-6879 was decreased. The authors also found that miR-34a, miR-134, and miR-383 were differentially expressed in all three age groups studied. They explained the up-regulated levels of miR-34a and miR-383 in both chronological and photoaging. It was proposed that these miRNAs targeted the p53-gene network via a G1 phase arrest of the cell cycle and the suppression of cell-cycle regulator cyclin, such as D1 and Cdk4, resulting in senescence. During chronological and photoaging, the up-regulation of miR-134 was linked to the activation of NF-κB signaling alongside pro-inflammatory cytokines (e.g., IL-1β, TNF-α, and IL-8) and the inhibition of a longevity gene SIRT1 [204].
Several studies reported the involvement of miRNAs in the modulation of autophagic pathways associated with aging. Indeed, the development of an aging phenotype was reciprocally related to the reduced autophagic activity [62]. Zhang et al. (2016) found that the PUVA (psoralen and ultraviolet A) and UVB-induced premature senescence was linked to the suppression of the autophagy signaling pathway via the overexpression of miR-23a. In this study, the authors elucidated that miR-23a inhibited the AMBRA1 gene, [205] which is implicated in the formation of the autophagosome core complex at an early step of autophagy regulated by rapamycin complex 1 (mTORC1) [206]. Besides, in the model of the UVB stress-induced premature senescence (UVB-SIPS) of human fibroblasts, it was found that the overexpression of miR-23a-5p reduced cellular proliferation and accelerated a cell cycle arrest. At the same time, in UVB-SIPS fibroblasts it had an opposite effect on the photoprotective circulatory RNA_100797, which acted as a sponge of miR-23a-5p [207].
The progress made in recent years demonstrates beneficial results in the rejuvenation strategies on the models of SISP in fibroblasts. Thus, UVB-aged human dermal fibroblasts treated with exosomes from the induced pluripotent stem cells (iPSCs) showed the reduced cellular alterations and expression of matrix-degrading enzymes (MMP-1/3) that was directly involved in ECM degradation [208]. Interestingly, exosomes contain extracellular miRNAs, functional messenger RNAs, and some proteins generated in the host cells [209]. In the photoaged fibroblasts, the iPSCs-derived exosomes also restored the expression of collagen type I; they potentially might be used for future rejuvenation practices for skin aging treatment [208,210].
10. Conclusions
This review illustrates the role of microRNAs in skin development and functioning and describes the pathogenetic mechanisms of cutaneous aging. Numerous microRNAs are inversely expressed within different age groups, suggesting their role in the regulation of the age-related signaling pathways. Evidence of differentially regulated miRNAs during the chronological and photoaging of the skin was also presented. Although some studies have proposed miRNA senescence-regulated mechanisms in specific cutaneous cells, there is still controversy in the published results. The process of the regulation of senescence and aging in the skin by miRNAs is still not fully elucidated, which delays the development of antiaging therapies.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/15/5281/s1.
Author Contributions
Conceptualization, M.G.; writing, reviewing, and editing, M.G., V.C., I.K. and O.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding from MITACS and NSERC grants.
Acknowledgments
We acknowledge the use of Servier Medical Art Commons Attribution 3.0 Unported License (http://smart.servier.com) to generate Figure 1, Figure 2, and Figure 3.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferrucci, L.; Gonzalez-Freire, M.; Fabbri, E.; Simonsick, E.; Tanaka, T.; Moore, Z.; Salimi, S.; Sierra, F.; De Cabo, R. Measuring biological aging in humans: A quest. Aging Cell 2019, 19, e13080. [Google Scholar] [CrossRef]
- Umansky, S.R. Aging and aging-associated diseases: A microRNA-based endocrine regulation hypothesis. Aging 2018, 10, 2557–2569. [Google Scholar] [CrossRef]
- Matjusaitis, M.; Chin, G.; Sarnoski, E.A.; Stolzing, A. Biomarkers to identify and isolate senescent cells. Ageing Res. Rev. 2016, 29, 1–12. [Google Scholar] [CrossRef]
- Sidler, C.; Kovalchuk, O.; Kovalchuk, I. Epigenetics of aging and age-related disorders. In Pharmacoepigenetics; Academic Press: New York, NY, USA, 2019; pp. 871–883. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Tigges, J.; Krutmann, J.; Fritsche, E.; Haendeler, J.; Schaal, H.; Fischer, J.W.; Kalfalah, F.; Reinke, H.; Reifenberger, G.; Stühler, K.; et al. The hallmarks of fibroblast ageing. Mech. Ageing Dev. 2014, 138, 26–44. [Google Scholar] [CrossRef]
- Choi, M.; Lee, C. Immortalization of primary keratinocytes and its application to skin research. Biomol. Ther. 2015, 23, 391–399. [Google Scholar] [CrossRef]
- Lämmermann, I.; Terlecki-Zaniewicz, L.; Weinmüllner, R.; Schosserer, M.; Dellago, H.; Branco, A.D.D.M.; Autheried, D.; Sevcnikar, B.; Kleissl, L.; Berlin, I.; et al. Blocking negative effects of senescence in human skin fibroblasts with a plant extract. NPJ Aging Mech. Dis. 2018, 4, 1–10. [Google Scholar] [CrossRef]
- Mancini, M.; Lena, A.M.; Saintigny, G.; Mahé, C.; Daniele, N.D.; Melino, G.; Candi, E. MicroRNAs in human skin ageing. Ageing Res. Rev. 2014, 17, 9–15. [Google Scholar] [CrossRef]
- ElSharawy, A.; Keller, A.; Flachsbart, F.; Wendschlag, A.; Jacobs, G.; Kefer, N.; Brefort, T.; Leidinger, P.; Backes, C.; Meese, E.; et al. Genome-wide miRNA signatures of human longevity. Aging Cell 2012, 11, 607–616. [Google Scholar] [CrossRef]
- Caravia, X.M.; Roiz-Valle, D.; Morán-Álvarez, A.; Lopez-Otin, C. Functional relevance of miRNAs in premature ageing. Mech. Ageing Dev. 2017, 168, 10–19. [Google Scholar] [CrossRef]
- Wong, S.Q.; Kumar, A.V.; Mills, J.; Lapierre, L.R. Autophagy in aging and longevity. Qual. Life Res. 2019, 139, 277–290. [Google Scholar] [CrossRef]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The retrograde response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Jager, S.; Handschin, C.; St.-Pierre, J.; Spiegelman, B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1a. Proc. Natl. Acad. Sci. USA 2007, 104, 12017–12022. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Rodgers, J.T.; Arlow, D.H.; Vazquez, F.; Mootha, V.K.; Puigserver, P. mTOR controls mitochondrial oxidative function through a YY1–PGC-1α transcriptional complex. Nature 2007, 450, 736–740. [Google Scholar] [CrossRef]
- Kalfalah, F.; Sobek, S.; Bornholz, B.; Götz-Rösch, C.; Tigges, J.; Fritsche, E.; Krutmann, J.; Köhrer, K.; Deenen, R.; Ohse, S.; et al. Inadequate mito-biogenesis in primary dermal fibroblasts from old humans is associated with impairment of PGC1A-independent stimulation. Exp. Gerontol. 2014, 56, 59–68. [Google Scholar] [CrossRef]
- Korman, B. Evolving insights into the cellular and molecular pathogenesis of fibrosis in systemic sclerosis. Transl. Res. 2019, 209, 77–89. [Google Scholar] [CrossRef]
- Kishimoto, S.; Uno, M.; Nishida, E. Molecular mechanisms regulating lifespan and environmental stress responses. Inflamm. Regen. 2018, 38, 22. [Google Scholar] [CrossRef]
- Rossi, M.; Gorospe, M. Noncoding RNAs controlling telomere homeostasis in senescence and aging. Trends Mol. Med. 2020, 26, 422–433. [Google Scholar] [CrossRef]
- Amano, H.; Sahin, E. Telomeres and sirtuins: At the end we meet again. Mol. Cell Oncol. 2019, 6, e1632613. [Google Scholar] [CrossRef]
- Grillari, J.; Hackl, M.; Grillari-Voglauer, R. miR-17–92 cluster: Ups and downs in cancer and aging. Biogerontology 2010, 11, 501–506. [Google Scholar] [CrossRef]
- Balistreri, C.R.; Candore, G.; Scapagnini, G.; Caruso, C. Aging and Antiaging Strategies. In Textbook of Aging Skin, 2nd ed.; Farage, M.A., Miller, K.W., Maibach, H.I., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1817–1827. [Google Scholar] [CrossRef]
- Kinser, H.E.; Pincus, Z. MicroRNAs as modulators of longevity and the aging process. Qual. Life Res. 2019, 139, 291–308. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA–disease associations. Nucleic Acids Res. 2018, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Rajeswari, R.; Venugopal, J.; Sundarrajan, S.; Sridhar, R.; Shayanti, M.; Ramakrishna, S. Polysaccharide nanofibrous scaffolds as a model for in vitro skin tissue regeneration. J. Mater. Sci. Mater. Electron. 2012, 23, 1511–1519. [Google Scholar] [CrossRef]
- Cavinato, M.; Koziel, R.; Romani, N.; Weinmüllner, R.; Jenewein, B.; Hermann, M.; Dubrac, S.; Ratzinger, G.; Grillari, J.; Schmuth, M.; et al. UVB-Induced Senescence of Human Dermal Fibroblasts Involves Impairment of Proteasome and Enhanced Autophagic Activity. J. Gerontol. 2016, 72, 632–639. [Google Scholar] [CrossRef][Green Version]
- Horsburgh, S.; Fullard, N.; Roger, M.; Degnan, A.; Todryk, S.; Przyborski, S.; O’Reilly, S. MicroRNAs in the skin: Role in development, homoeostasis and regeneration. Clin. Sci. 2017, 131, 1923–1940. [Google Scholar] [CrossRef]
- Farage, M.A.; Miller, K.W.; Maibach, H.I. (Eds.) Degenerative changes in aging skin. In Textbook of Aging Skin, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 15–30. [Google Scholar] [CrossRef]
- Bíró, T.; Tóth, B.I.; Haskó, G.; Paus, R.; Pacher, P. The endocannabinoid system of the skin in health and disease: Novel perspectives and therapeutic opportunities. Trends Pharmacol. Sci. 2009, 30, 411–420. [Google Scholar] [CrossRef]
- Elder, D.E.; Elenitsas, R.; Rosenbach, M.; Murphy, G.F.; Rubin, A.I.; Xu, X. Lever’s Histopathology of the Skin, 11th ed.; Elder, D.E., Rosenbach, M., Murphy, G.F., Adam, I., Rubin, X.X., Eds.; Wolters Kluwer: Philadelphia, PA, USA, 2015. [Google Scholar]
- Kang, S. Fitzpatrick’s Dermatology, 9th ed.; Kang, S., Amagai, M., Anna, L., Bruckner, A.H.E., Margolis, D.J., McMichael, A.J., Orringer, J.S., Eds.; McGraw-Hill Education: New York, NY, USA, 2019. [Google Scholar]
- Kierszenbaum, L.A.; Tres, L. Histology and Cell Biology: An Introduction to Pathology E-Book, 4th ed.; Elsevier Saunders: Philadelphia, PA, USA, 2016. [Google Scholar]
- Singhvi, G.; Manchanda, P.; Rapalli, V.K.; Dubey, S.K.; Gupta, G.; Dua, K. MicroRNAs as biological regulators in skin disorders. Biomed. Pharmacother. 2018, 108, 996–1004. [Google Scholar] [CrossRef]
- Pagliari, C.; Sotto, M.N. Correlation of factor XIIIa + dermal dendrocytes with paracoccidioidomycosis skin lesions. Med. Mycol. 2002, 40, 407–410. [Google Scholar] [CrossRef]
- Włodarczyk, M.; Sobolewska, A.; Lesiak, A.; Narbutt, J. The role of factor XIII-A in the development of inflammatory skin lesions. Open Life Sci. 2014, 9, 869–873. [Google Scholar] [CrossRef][Green Version]
- Fore, J. A review of skin and the effects of aging on skin structure and function. Ostomy Wound Manag. 2006, 52, 24–35. [Google Scholar]
- Pal, D.; Sen, C.K. MicroRNAs in skin development, function and disorders. In MicroRNAs in Diseases and Disorders: Emerging Therapeutic Targets, 1st ed.; Peplow, P.V., Martinez, B., Calin, G.A., Esquela-Kerscher, A., Eds.; Croydon: Royal Society of Chemistry: Cambridge, UK, 2019; Volume 6, pp. 275–292. [Google Scholar] [CrossRef]
- Huang, H.-C.; Chang, T.-M.; Chang, Y.-J.; Wen, H.-Y. UVB Irradiation regulates ERK1/2- and p53-dependent thrombomodulin expression in human keratinocytes. PLoS ONE 2013, 8, e67632. [Google Scholar] [CrossRef]
- Thulabandu, V.; Chen, D.; Atit, R.P. Dermal fibroblast in cutaneous development and healing. Wiley Interdiscip. Rev. Dev. Biol. 2017, 7, e307. [Google Scholar] [CrossRef]
- Mills, A.A.; Zheng, B.; Wang, X.-J.; Vogel, H.; Roop, D.R.; Bradley, A.; Vogel, H. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef]
- Senoo, M.; Pinto, F.; Crum, C.P.; McKeon, F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 2007, 129, 523–536. [Google Scholar] [CrossRef]
- Duchatelet, S.; Russo, C.; Osterburg, C.; Mallet, S.; Bole-Feysot, C.; Nitschké, P.; Richard, M.-A.; Dötsch, V.; Missero, C.; Nassif, A.; et al. A TP63 mutation causes prominent alopecia with mild ectodermal dysplasia. J. Investig. Dermatol. 2019, 140, 1103–1106. [Google Scholar] [CrossRef]
- Pan, Y.; Lin, M.-H.; Tian, X.; Cheng, H.-T.; Gridley, T.; Shen, J.; Kopan, R. γ-secretase functions through notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell 2004, 7, 731–743. [Google Scholar] [CrossRef]
- Blanpain, C.; Fuchs, E. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009, 10, 207–217. [Google Scholar] [CrossRef]
- Yamamoto, N.; Tanigaki, K.; Han, H.; Hiai, H.; Honjo, T. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 2003, 13, 333–338. [Google Scholar] [CrossRef]
- Scholl, F.A.; Dumesic, P.A.; Barragan, D.I.; Harada, K.; Bissonauth, V.; Charron, J.; Khavari, P.A. Mek1/2 MAPK kinases are essential for mammalian development, homeostasis, and raf-induced hyperplasia. Dev. Cell 2007, 12, 615–629. [Google Scholar] [CrossRef]
- Janes, S.M.; Watt, F.M. New roles for integrins in squamous-cell carcinoma. Nat. Rev. Cancer 2006, 6, 175–183. [Google Scholar] [CrossRef]
- Fujiwara, H.; Ferreira, M.; Donati, G.; Marciano, D.K.; Linton, J.M.; Sato, Y.; Hartner, A.; Sekiguchi, K.; Reichardt, L.F.; Watt, F.M. The Basement Membrane of Hair Follicle Stem Cells Is a Muscle Cell Niche. Cell 2011, 144, 577–589. [Google Scholar] [CrossRef]
- Driskell, R.R.; Lichtenberger, B.; Hoste, E.; Kretzschmar, K.; Simons, B.D.; Charalambous, M.; Ferrón, S.R.; Herault, Y.; Pavlovic, G.; Ferguson-Smith, A.C.; et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 2013, 504, 277–281. [Google Scholar] [CrossRef]
- Berman, A.E.; Leontieva, O.V.; Natarajan, V.; McCubrey, J.A.; Demidenko, Z.N.; Nikiforov, M.A. Recent progress in genetics of aging, senescence and longevity: Focusing on cancer-related genes. Oncotarget 2012, 3, 1522–1532. [Google Scholar] [CrossRef]
- Makrantonaki, E.; Zouboulis, C.C. Characteristics and Pathomechanisms of Endogenously Aged Skin. Dermatology 2007, 214, 352–360. [Google Scholar] [CrossRef]
- Dudonné, S.; Coutière, P.; Woillez, M.; Mérillon, J.-M.; Vitrac, X. DNA macroarray study of skin aging-related genes expression modulation by antioxidant plant extracts on a replicative senescence model of human dermal fibroblasts. Phytother. Res. 2010, 25, 686–693. [Google Scholar] [CrossRef]
- Tobin, D. Introduction to skin aging. J. Tissue Viability 2017, 26, 37–46. [Google Scholar] [CrossRef]
- Kang, S.M.; Han, S.; Oh, J.-H.; Lee, Y.M.; Park, C.-H.; Shin, C.-Y.; Lee, D.; Chung, J.H. A synthetic peptide blocking TRPV1 activation inhibits UV-induced skin responses. J. Dermatol. Sci. 2017, 88, 126–133. [Google Scholar] [CrossRef][Green Version]
- Berneburg, M.; Plettenberg, H.; Medve-Konig, K.; Pfahlberg, A.; Gers-Barlag, H.; Gefeller, O.; Krutmann, J.; Medve-K, K. Induction of the photoaging-associated mitochondrial common deletion in vivo in normal human skin. J. Investig. Dermatol. 2004, 122, 1277–1283. [Google Scholar] [CrossRef]
- Toutfaire, M.; Bauwens, E.; Debacq-Chainiaux, F. The impact of cellular senescence in skin ageing: A notion of mosaic and therapeutic strategies. Biochem. Pharmacol. 2017, 142, 1–12. [Google Scholar] [CrossRef]
- Gkogkolou, P.; Böhm, M. Advanced glycation end products: Key players in skin aging? Derm.-Endocrinol. 2012, 4, 259–270. [Google Scholar] [CrossRef]
- Wlaschek, M.; Tantcheva-Poór, I.; Naderi, L.; Ma, W.; Schneider, L.A.; Razi-Wolf, Z.; Schüller, J.; Scharffetter-Kochanek, K. Solar UV irradiation and dermal photoaging. J. Photochem. Photobiol. B Biol. 2001, 63, 41–51. [Google Scholar] [CrossRef]
- Rittié, L.; Fisher, G.J. Natural and sun-induced aging of human skin. Cold Spring Harb. Perspect. Med. 2015, 5, a015370. [Google Scholar] [CrossRef]
- Batista, L.F.Z.; Kaina, B.; Meneghini, R.; Menck, C.F.M. How DNA lesions are turned into powerful killing structures: Insights from UV-induced apoptosis. Mutat. Res. Mutat. Res. 2009, 681, 197–208. [Google Scholar] [CrossRef]
- Lee, L.-Y.; Liu, S.-X. Pathogenesis of photoaging in human dermal fibroblasts. Int. J. Dermatol. Venereol. 2020, 3, 37–42. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; A Grässer, F.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2019, 48, D101–D110. [Google Scholar] [CrossRef]
- Abdallah, F.; Pichon, C. MicroRNAs in skin biology: Biogenesis, regulations and functions in homeostasis and diseases. Immunome Res. 2019, 15, 1–19. [Google Scholar] [CrossRef][Green Version]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef]
- Sunderland, N.; Skroblin, P.; Barwari, T.; Huntley, R.P.; Lu, R.; Joshi, A.; Lovering, R.; Mayr, M. MicroRNA biomarkers and platelet reactivity. Circ. Res. 2017, 120, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Fuchs, E. MicroRNA-mediated control in the skin. Cell Death Differ. 2009, 17, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Laffont, B.; Corduan, A.; Plé, H.; Duchez, A.-C.; Cloutier, N.; Boilard, E.; Provost, P. Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles. Blood 2013, 122, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Teruel-Montoya, R.; Rosendaal, F.R.; Martinez, C. MicroRNAs in hemostasis. J. Thromb. Haemost. 2014, 13, 170–181. [Google Scholar] [CrossRef]
- Soliman, A.M.; Das, S.; Ghafar, N.A.; Teoh, S.L. Role of MicroRNA in Proliferation Phase of Wound Healing. Front. Genet. 2018, 9, 38. [Google Scholar] [CrossRef]
- Meder, B.; Backes, C.; Haas, J.; Leidinger, P.; Stähler, C.; Großmann, T.; Vogel, B.; Frese, K.; Giannitsis, E.; Katus, H.A.; et al. Influence of the confounding factors age and sex on microrna profiles from peripheral blood. Clin. Chem. 2014, 60, 1200–1208. [Google Scholar] [CrossRef]
- Storci, G.; De Carolis, S.; Papi, A.; Bacalini, M.G.; Gensous, N.; Marasco, E.; Tesei, A.; Fabbri, F.; Arienti, C.; Zanoni, M.; et al. Genomic stability, anti-inflammatory phenotype, and up-regulation of the RNAseH2 in cells from centenarians. Cell Death Differ. 2019, 26, 1845–1858. [Google Scholar] [CrossRef]
- Majidinia, M.; Karimian, A.; Alemi, F.; Yousefi, B.; Safa, A.; Alami, F. Targeting miRNAs by polyphenols: Novel therapeutic strategy for aging. Biochem. Pharmacol. 2020, 173, 113688. [Google Scholar] [CrossRef]
- Wu, S.; Kim, T.-K.; Wu, X.; Scherler, K.; Baxter, D.; Wang, K.; Krasnow, R.E.; Reed, T.; Dai, J. Circulating microRNAs and life expectancy among identical twins. Ann. Hum. Genet. 2016, 80, 247–256. [Google Scholar] [CrossRef]
- Ameling, S.; Kacprowski, T.; Chilukoti, R.K.; Malsch, C.; Liebscher, V.; Suhre, K.; Pietzner, M.; Friedrich, N.; Homuth, G.; Hammer, E.; et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med. Genom. 2015, 8, 61. [Google Scholar] [CrossRef]
- Olivieri, F.; Bonafè, M.; Spazzafumo, L.; Gobbi, M.; Prattichizzo, F.; Recchioni, R.; Marcheselli, F.; La Sala, L.; Galeazzi, R.; Rippo, M.R.; et al. Age- and glycemia-related miR-126-3p levels in plasma and endothelial cells. Aging 2014, 6, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Hooten, N.N.; Fitzpatrick, M.; Wood, W.H.; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microRNA levels in serum. Aging 2013, 5, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Smith-Vikos, T.; Liu, Z.; Parsons, C.; Gorospe, M.; Ferrucci, L.; Gill, T.M.; Slack, F.J. A serum miRNA profile of human longevity: Findings from the Baltimore Longitudinal Study of Aging (BLSA). Aging 2016, 8, 2971–2987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, H.; Zhang, C.; Jing, Y.; Wang, C.; Liu, C.; Zhang, R.; Wang, J.; Zhang, J.; Zen, K.; et al. Investigation of MicroRNA expression in human serum during the aging process. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 70, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Huan, T.; Chen, G.; Liu, C.; Bhattacharya, A.; Rong, J.; Chen, B.H.; Seshadri, S.; Tanriverdi, K.; Freedman, J.E.; Larson, M.G.; et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 2018, 17, e12687. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef]
- Hooten, N.N.; Martin-Montalvo, A.; Dluzen, D.F.; Zhang, Y.; Bernier, M.; Zonderman, A.B.; Becker, K.G.; Gorospe, M.; De Cabo, R.; Evans, M.K. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 2016, 15, 572–581. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Vartholomatos, G.; Tzavaras, T.; Hatziapostolou, M.; Fackelmayer, F.O.; Sandaltzopoulos, R.; Polytarchou, C.; Kolettas, E. Senescence-associated microRNAs target cell cycle regulatory genes in normal human lung fibroblasts. Exp. Gerontol. 2017, 96, 110–122. [Google Scholar] [CrossRef]
- Antonini, D.; Russo, M.T.; De Rosa, L.; Gorrese, M.; Del Vecchio, L.; Missero, C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J. Investig. Dermatol. 2010, 130, 1249–1257. [Google Scholar] [CrossRef]
- Chikh, A.; Matin, R.N.H.; Senatore, V.; Hufbauer, M.; Lavery, D.; Raimondi, C.; Ostano, P.; Mello-Grand, M.; Ghimenti, C.; Bahta, A.; et al. iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J. 2011, 30, 4261–4273. [Google Scholar] [CrossRef]
- Schneider, M.R. MicroRNAs as novel players in skin development, homeostasis and disease. Br. J. Dermatol. 2011, 166, 22–28. [Google Scholar] [CrossRef]
- Kuo, G.; Wu, C.-Y.; Yang, H.-Y. MiR-17-92 cluster and immunity. J. Formos. Med. Assoc. 2019, 118, 2–6. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Li, N.; Teng, W.; Wang, M.; Zhang, Y.; Xiao, Z. TGF-β1 promotes scar fibroblasts proliferation and transdifferentiation via up-regulating MicroRNA-21. Sci. Rep. 2016, 6, 32231. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Yang, D.; Xiao, Z.; Chen, X. MicroRNA-21 affects proliferation and apoptosis by regulating expression of PTEN in human keloid fibroblasts. Plast. Reconstr. Surg. 2014, 134, 561e–573e. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.I.; Mardaryev, A.N.; Lewis, C.J.; Sharov, A.A.; Botchkareva, N. MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J. Cell Sci. 2011, 124, 3399–3404. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Simões, A.; Chen, Z.; Zhao, Y.; Wu, X.; Dai, Y.; DiPietro, L.A.; Zhou, X. Overexpression of the oral mucosa-specific microRNA-31 promotes skin wound closure. Int. J. Mol. Sci. 2019, 20, 3679. [Google Scholar] [CrossRef] [PubMed]
- Hooten, N.N.; Abdelmohsen, K.; Gorospe, M.; Ejiogu, N.; Zonderman, A.B.; Evans, M.K. microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE 2010, 5, e10724. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Yang, J.; Xu, L.; Zhang, C. Cell-specific effects of miR-221/222 in vessels: Molecular mechanism and therapeutic application. J. Mol. Cell. Cardiol. 2012, 52, 245–255. [Google Scholar] [CrossRef]
- Brunner, S.; Herndler-Brandstetter, D.; Arnold, C.R.; Wiegers, G.J.; Villunger, A.; Hackl, M.; Grillari, J.; Moreno-Villanueva, M.; Bürkle, A.; Grubeck-Loebenstein, B. Upregulation of miR-24 is associated with a decreased DNA damage response upon etoposide treatment in highly differentiated CD8+ T cells sensitizing them to apoptotic cell death. Aging Cell 2012, 11, 579–587. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Fan, H.; Zhao, X.; Liu, D.; Hu, Y.; Kidd, A.R.; Bao, J.; Hou, Y. MicroRNA-181a Regulates Local immune balance by inhibiting proliferation and immunosuppressive properties of mesenchymal stem cells. Stem Cells 2012, 30, 1756–1770. [Google Scholar] [CrossRef]
- Stinchcombe, J.C.; Barral, D.C.; Mules, E.H.; Booth, S.; Hume, A.N.; Machesky, L.M.; Seabra, M.C.; Griffiths, G. Rab27a is required for regulated secretion in Cytotoxic T lymphocytes. J. Cell Biol. 2001, 152, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, J.; Xiao, L.; Wang, Y.; Liu, G.; Li, J.; Liang, F. Identification of differentially expressed miRNAs in the response of spleen CD4+ T cells to electroacupuncture in senescence-accelerated mice. Cell Biophys. 2020, 78, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Wu, M.-H.; Liu, F.; Li, Y.; Li, N.; Li, G.-Y.; Shen, S.-R. Differential miRNA expression and their target genes between NGX6-positive and negative colon cancer cells. Mol. Cell. Biochem. 2010, 345, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Orjalo, A.V.; Rodier, F.; Lithgow, G.J.; Campisi, J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging 2009, 1, 402–411. [Google Scholar] [CrossRef]
- Balia, C.; Giordano, M.; Scalise, V.; Neri, T.; Fontanini, G.; Basolo, F.; Celi, A.; Pedrinelli, R. miR-19a and miR-20a and Tissue Factor Expression in Activated Human Peripheral Blood Mononuclear Cells. Thrombosis 2017, 2017, 1076397. [Google Scholar] [CrossRef]
- Wei, X.-J.; Li, X.-W.; Lu, J.-L.; Long, Z.-X.; Liang, J.-Q.; Wei, S.-B.; Lu, C.-X.; Lu, W.-Z. MiR-20a regulates fibroblast-like synoviocyte proliferation and apoptosis in rheumatoid arthritis. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3886–3893. [Google Scholar]
- Zhang, H.; Li, W.; Nan, F.; Ren, F.; Wang, H.; Xu, Y.; Zhang, F. MicroRNA expression profile of colon cancer stem-like cells in HT29 adenocarcinoma cell line. Biochem. Biophys. Res. Commun. 2011, 404, 273–278. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, Q.; Wang, C.-Q.; Wang, X.-N.; Wang, Y.; Zhao, J.-J.; Mao, S.-S.; Zhang, G.-H.; Zhang, N.; Xu, X.-C. MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer 2010, 10, 542. [Google Scholar] [CrossRef]
- Márton, S.; Garcia, M.R.; Robello, C.; Persson, H.; Trajtenberg, F.; Pritsch, O.; Rovira, C.; Naya, H.; Dighiero, G.; Cayota, A. small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia 2007, 22, 330–338. [Google Scholar] [CrossRef]
- Persengiev, S.; Kondova, I.; Otting, N.; Koeppen, A.H.; Bontrop, R.E. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol. Aging 2011, 32, 2316. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, J.; Hu, L.; Kong, L.; Wang, T.; Di, M.; Li, C.; Gui, S. miR-130a alleviates neuronal apoptosis and changes in expression of Bcl-2/Bax and caspase-3 in cerebral infarction rats through PTEN/PI3K/Akt signaling pathway. Exp. Ther. Med. 2020, 19, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Zheng, M.; Hayashi, M.; Lee, J.-D.; Yoshino, O.; Lin, S.; Han, J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J. Clin Investig. 2008, 118, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Stratz, C.; Nührenberg, T.G.; Binder, H.; Valina, C.M.; Trenk, D.; Hochholzer, W.; Neumann, F.; Fiebich, B.L. Micro-array profiling exhibits remarkable intra-individual stability of human platelet micro-RNA. Thromb. Haemost. 2012, 107, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Hackl, M.; Brunner, S.; Fortschegger, K.; Schreiner, C.; Micutkova, L.; Mück, C.; Laschober, G.T.; Lepperdinger, G.; Sampson, N.; Berger, P.; et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 2010, 9, 291–296. [Google Scholar] [CrossRef]
- Banerjee, J.; Chan, Y.C.; Sen, C.K. MicroRNAs in skin and wound healing. Physiol. Genom. 2010, 43, 543–556. [Google Scholar] [CrossRef]
- Bouvy, C.; Wannez, A.; George, F.; Graux, C.; Chatelain, C.; Dogné, J.-M. Circulating MicroRNAs as biomarkers in diffuse large B-cell lymphoma: A pilot prospective longitudinal clinical study. Biomark. Cancer 2018, 10, 1179299X18781095. [Google Scholar] [CrossRef]
- Husni, A.S.; McCurdy, C.R.; Radwan, M.M.; Ahmed, S.A.; Slade, D.; Ross, S.A.; ElSohly, M.A.; Cutler, S.J. Evaluation of phytocannabinoids from high potency cannabis sativa using in vitro bioassays to determine structure-activity relationships for cannabinoid receptor 1 and cannabinoid receptor 2. Med. Chem. Res. 2014, 23, 4295–4300. [Google Scholar] [CrossRef]
- Rendon, A.; Schäkel, K. Psoriasis pathogenesis and treatment. Int. J. Mol. Sci. 2019, 6, 1475. [Google Scholar] [CrossRef]
- Harmanci, D.; Erkan, E.P.; Kocak, A.; Akdogan, G.G. Role of the microRNA-29 family in fibrotic skin diseases. Biomed. Rep. 2017, 6, 599–604. [Google Scholar] [CrossRef]
- Henderson, J.; Distler, J.; O’Reilly, S. The role of epigenetic modifications in systemic sclerosis: A druggable target. Trends Mol. Med. 2019, 25, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Altorok, N.; Almeshal, N.; Wang, Y.; Kahaleh, B. Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatol 2014, 54, 1759–1770. [Google Scholar] [CrossRef]
- Suárez, Y.; Fernández-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gorski, D.H. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 2008, 111, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lang, N.; Qiu, M.; Xu, F.; Li, Q.; Tang, Q.; Chen, J.; Chen, X.; Zhang, S.; Liu, Z.; et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int. J. Cancer 2011, 128, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Kozaki, K.-I.; Imoto, I.; Mogi, S.; Omura, K.; Inazawa, J. Exploration of tumor-suppressive MicroRNAs silenced by DNA Hypermethylation in oral cancer. Cancer Res. 2008, 68, 2094–2105. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, L.; Wang, A.; Yu, J.; Shi, F.; Zhou, X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009, 286, 217–222. [Google Scholar] [CrossRef]
- Cervo, P.R.D.V.; Lena, A.M.; Nicoloso, M.; Rossi, S.; Mancini, M.; Zhou, H.; Saintigny, G.; Dellambra, E.; Odorisio, T.; Mahé, C.; et al. p63-microRNA feedback in keratinocyte senescence. Proc. Natl. Acad. Sci. USA 2012, 109, 1133–1138. [Google Scholar] [CrossRef]
- Hayflick, L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Kang, M.-J.; Xu, J.; Selimyan, R.; Yoon, J.-H.; Martindale, J.L.; De, S.; Wood, W.H.; Becker, K.G.; et al. SAL-RNAs: Senescence-associated long non-coding RNAs. Aging Cell 2013, 12, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Toufektchan, E.; Toledo, F. The Guardian of the genome revisited: p53 downregulates genes required for telomere maintenance, DNA repair, and centromere structure. Cancers 2018, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Ning, M.S.; Andl, T. Control by a hair’s breadth: The role of microRNAs in the skin. Cell Mol. Life Sci. 2012, 70, 1149–1169. [Google Scholar] [CrossRef] [PubMed]
- Rinnerthaler, M.; Duschl, J.; Steinbacher, P.; Salzmann, M.; Bischof, J.; Schuller, M.; Wimmer, H.; Peer, T.; Bauer, J.W.; Richter, K. Age-related changes in the composition of the cornified envelope in human skin. Exp. Dermatol. 2013, 22, 329–335. [Google Scholar] [CrossRef]
- Simboeck, E.; Ribeiro, J.D.; Teichmann, S.; Di Croce, L. Epigenetics and senescence: Learning from the INK4-ARF locus. Biochem. Pharmacol. 2011, 82, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, O.; Kovalchuk, I. Genome Stability: From Virus to Human Application, 1st ed.; Kovalchuk, I., Kovalchuk, O., Eds.; Elsevier Inc./Academic Press: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Ming, X.; Zhang, Z.; Zou, Z.; Lv, C.; Dong, Q.; He, Q.; Yi, Y.; Li, Y.; Wang, H.; Zhu, B. Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res. 2020, 1–17. [Google Scholar] [CrossRef]
- Hashimoto, H.; Vertino, P.M.; Cheng, X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2010, 2, 657–669. [Google Scholar] [CrossRef]
- Gao, F.; Xia, Y.; Wang, J.-W.; Lin, Z.-L.; Ou, Y.; Liu, X.; Liu, W.; Zhou, B.; Luo, H.; Zhou, B.; et al. Integrated analyses of DNA methylation and hydroxymethylation reveal tumor suppressive roles of ECM1, ATF5, and EOMES in human hepatocellular carcinoma. Genome Biol. 2014, 15, 533. [Google Scholar] [CrossRef]
- Grönniger, E.; Weber, B.; Heil, O.; Peters, N.; Stäb, F.; Wenck, H.; Korn, B.; Winnefeld, M.; Lyko, F. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010, 6, e1000971. [Google Scholar] [CrossRef]
- De Paoli-Iseppi, R.; Deagle, B.E.; McMahon, C.R.; Hindell, M.A.; Dickinson, J.L.; Jarman, S.N. Measuring animal age with DNA methylation: From humans to wild animals. Front. Genet. 2017, 8, 106. [Google Scholar] [CrossRef]
- Wang, B.; Du, R.; Xiao, X.; Deng, Z.-L.; Jian, D.; Xie, H.-F.; Li, J. Microrna-217 modulates human skin fibroblast senescence by directly targeting DNA methyltransferase 1. Oncotarget 2017, 8, 33475–33486. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-H.; Pucar, A.; Kim, R.H.; Bae, S.D.; Chen, W.; Kang, M.K.; Park, N.-H. Identification of senescence-inducing microRNAs in normal human keratinocytes. Int. J. Oncol. 2011, 39, 1205–1211. [Google Scholar] [CrossRef][Green Version]
- Glass, D.; Viñuela, A.; Davies, M.N.; Ramasamy, A.; Parts, L.; Knowles, D.A.; Brown, A.A.; Hedman, A.K.; Small, K.S.; Buil, A.; et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013, 14, R75. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Shah, P.P.; Nativio, R.; Berger, S.L. Epigenetic mechanisms of longevity and aging. Cell 2016, 166, 822–839. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Cheng, Z.; Zhang, J.; Li, R.; Zhao, P.; Fu, Z.; You, Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008, 1236, 185–193. [Google Scholar] [CrossRef]
- Lena, A.; Mancini, M.; Cervo, P.R.D.V.; Saintigny, G.; Mahé, C.; Melino, G.; Candi, E. MicroRNA-191 triggers keratinocytes senescence by SATB1 and CDK6 downregulation. Biochem. Biophys. Res. Commun. 2012, 423, 509–514. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Purbey, P.K.; Sinha, C.K.; Notani, D.; Limaye, A.; Jayani, R.S.; Galande, S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell 2006, 22, 231–243. [Google Scholar] [CrossRef]
- Polioudakis, D.; Abell, N.S.; Iyer, V.R. MiR-191 regulates primary human fibroblast proliferation and directly targets multiple oncogenes. PLoS ONE 2015, 10, e0126535. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, J.; Yu, F.-S.; Zhou, L.; Mi, Q.-S. TGF-β1-induced transcription factor networks in Langerhans cell development and maintenance. Allergy 2016, 71, 758–764. [Google Scholar] [CrossRef]
- Sumpter, T.L.; Balmert, S.C.; Kaplan, D.H. Cutaneous immune responses mediated by dendritic cells and mast cells. JCI Insight 2019, 4, e123947. [Google Scholar] [CrossRef]
- Zegarska, B.; Pietkun, K.; Giemza-Kucharska, P.; Zegarski, T.; Nowacki, M.S.; Romańska-Gocka, K. Changes of Langerhans cells during skin ageing. Adv. Dermatol. Allergol. 2017, 34, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wu, L. The development and function of dendritic cell populations and their regulation by miRNAs. Protein Cell 2017, 8, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-P.; Qi, R.-Q.; Chen, W.-B.; Shi, Y.; Cui, Z.; Gao, X.-H.; Chen, H.-D.; Zhou, L.; Mi, Q.-S. Aging affects epidermal Langerhans cell development and function and alters their miRNA gene expression profile. Aging 2012, 4, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Tacke, F.; Angeli, V.; Bogunovic, M.; Loubeau, M.; Dai, X.-M.; Stanley, E.R.; Randolph, G.J.; Merad, M. Langerhans cells arise from monocytes in vivo. Nat. Immunol. 2006, 7, 265–273. [Google Scholar] [CrossRef]
- Rathinam, C.; Geffers, R.; Yücel, R.; Buer, J.; Welte, K.; Möröy, T.; Klein, C. The transcriptional repressor gfi1 controls STAT3-dependent dendritic cell development and function. Immunity 2005, 22, 717–728. [Google Scholar] [CrossRef]
- Schramek, D.; Sigl, V.; Penninger, J.M. RANKL and RANK in sex hormone-induced breast cancer and breast cancer metastasis. Trends Endocrinol. Metab. 2011, 22, 188–194. [Google Scholar] [CrossRef]
- Mueller, C.G.; Hess, E. Emerging Functions of RANKL in Lymphoid Tissues. Front. Immunol. 2012, 3, 261. [Google Scholar] [CrossRef]
- Wang, S.; Tang, C.; Zhang, Q.; Chen, W. Reduced miR-9 and miR-181a expression down-regulates Bim concentration and promote osteoclasts survival. Int. J. Clin. Exp. Pathol. 2014, 7, 2209–2218. [Google Scholar]
- Lozano, C.; Duroux, I.; Firat, H.; Schordan, E.; Apparailly, F. MicroRNAs: Key regulators to understand osteoclast differentiation? Front. Immunol. 2019, 10, 375. [Google Scholar] [CrossRef]
- Zhou, L.; Qi, R.-Q.; Liu, M.; Xu, Y.-P.; Li, G.; Weiland, M.; Kaplan, D.H.; Mi, Q.-S. microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun. 2013, 15, 57–61. [Google Scholar] [CrossRef]
- Gruber, F.; Kremslehner, C.; Eckhart, L.; Tschachler, E. Cell aging and cellular senescence in skin aging—Recent advances in fibroblast and keratinocyte biology. Exp. Gerontol. 2020, 130, 110780. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-L.; Chen, J.S.; Chang, D.C.; Lin, S.-L. Mir-434-5p mediates skin whitening and lightening. Clin. Cosmet. Investig. Dermatol. 2008, 1, 19. [Google Scholar] [CrossRef][Green Version]
- Itoh, T.; Fukatani, K.; Nakashima, A.; Suzuki, K. MicroRNA-141-3p and microRNA-200a-3p regulate α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; He, J.; Jia, X.; Jiang, J.; Bai, R.; Yu, X.; Lv, L.; Fan, R.; He, X.; Geng, J.; et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in alpaca (Lama pacos) skin melanocytes. Domest. Anim. Endocrinol. 2010, 38, 200–209. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Fan, R.; Chen, T.; Dong, C. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int. J. Mol. Sci. 2016, 17, 959. [Google Scholar] [CrossRef]
- Bellei, B.; Picardo, M. Premature cell senescence in human skin: Dual face in chronic acquired pigmentary disorders. Ageing Res. Rev. 2019, 57, 100981. [Google Scholar] [CrossRef]
- Victorelli, S.; Lagnado, A.; Halim, J.; Moore, W.; Talbot, D.; Barrett, K.; Chapman, J.; Birch, J.; Ogrodnik, M.; Meves, A.; et al. Senescent human melanocytes drive skin ageing via paracrine telomere dysfunction. EMBO J. 2019, 38, e101982. [Google Scholar] [CrossRef]
- Dar, A.A.; Majid, S.; De Semir, D.; Nosrati, M.; Bezrookove, V.; Kashani-Sabet, M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J. Biol. Chem. 2011, 286, 16606–16614. [Google Scholar] [CrossRef]
- Jiang, Y.; Saavedra, H.I.; Holloway, M.P.; Leone, G.; Altura, R.A. Aberrant regulation of survivin by the RB/E2F family of proteins. J. Biol. Chem. 2004, 279, 40511–40520. [Google Scholar] [CrossRef]
- Thomas, G.V.; Horvath, S.; Smith, B.L.; Crosby, K.; Lebel, L.A.; Schrage, M.; Said, J.; De Kernion, J.; Reiter, R.E.; Sawyers, C.L. Antibody-based profiling of the phosphoinositide 3-kinase pathway in clinical prostate cancer. Clin. Cancer Res. 2004, 10, 8351–8356. [Google Scholar] [CrossRef]
- Pierce, A.M.; Schneider-Broussard, R.; Gimenez-Conti, I.B.; Russell, J.L.; Conti, C.J.; Johnson, D.G. E2F1 Has both oncogenic and tumor-suppressive properties in a transgenic model. Mol. Cell. Biol. 1999, 19, 6408–6414. [Google Scholar] [CrossRef]
- Kothandaraman, N.; Bajic, V.B.; Pang, B.; Chan, Y.H.; Keow, P.B.; Razvi, K.; Salto-Tellez, M.; Choolani, M. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer 2010, 10, 64. [Google Scholar] [CrossRef]
- Vera, O.; Bok, I.; Jasani, N.; Nakamura, K.; Xu, X.; Mecozzi, N.; Karreth, F.A. MAPK-induced miR-29 restrains melanoma progression by targeting MAFG. bioRxiv 2020, 01, 922153. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, N.; Long, S.; Ge, L.; Wang, A.; Sun, H.; Ran, X.; Zou, Z.; Wang, J.; Su, Y. Downregulation of miR-205 in migrating epithelial tongue facilitates skin wound re-epithelialization by derepressing ITGA5. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 1443–1452. [Google Scholar] [CrossRef]
- Ratzinger, S.; Grassel, S.; Dowejko, A.; Reichert, T.E.; Bauer, R. Induction of type XVI collagen expression facilitates proliferation of oral cancer cells. Matrix Biol. 2011, 30, 118–125. [Google Scholar] [CrossRef]
- Kassner, A.; Hansen, U.; Miosge, N.; Reinhardt, D.P.; Aigner, T.; Bruckner-Tuderman, L.; Bruchner, P.; Grässel, S. Discrete integration of collagen XVI into tissue-specific collagen fibrils or beaded microfibrils. Matrix Biol. 2003, 22, 131–143. [Google Scholar] [CrossRef]
- Martinez, I.; Cazalla, D.; Almstead, L.L.; Steitz, J.A.; DiMaio, D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc. Natl. Acad. Sci. USA 2010, 108, 522–527. [Google Scholar] [CrossRef]
- Werwein, E.; Cibis, H.; Hess, D.; Klempnauer, K.-H. Activation of the oncogenic transcription factor B-Myb via multisite phosphorylation and prolylcis/transisomerization. Nucleic Acids Res. 2018, 47, 103–121. [Google Scholar] [CrossRef]
- Tanno, B.; Negroni, A.; Vitali, R.; Pirozzoli, M.C.; Cesi, V.; Mancini, C.; Calabretta, B.; Raschellà, G. Expression of insulin-like growth factor-binding protein 5 in neuroblastoma cells is regulated at the transcriptional level by c-Myb and B-Myb via direct and indirect mechanisms. J. Biol. Chem. 2002, 277, 23172–23180. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, H.; Gao, J.; Liu, Y.; Li, J.; Wang, J. Bioactive Ingredients in Chinese Herbal Medicines That Target Non-coding RNAs: Promising New Choices for Disease Treatment. Front. Pharmacol. 2019, 10, 515. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, X.-S.; Yang, G.-H.; Zhai, P.-F.; Xiao, Z.; Xia, L.-Y.; Chen, L.-R.; Wang, Y.; Wang, X.-Z.; Bi, L.-X.; et al. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol. Biol. Rep. 2011, 39, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Oorschot, A.A.D.-V.; Kuipers, J.E.; Arentsen-Peters, S.; Schotte, D.; De Haas, V.; Trka, J.; Baruchel, A.; Reinhardt, D.; Pieters, R.; Zwaan, C.M.; et al. Differentially expressed miRNAs in cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Pediatr. Blood Cancer 2011, 58, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Fuster, O.; Llop, M.; Dolz, S.; Garcia, P.; Such, E.; Ibáñez, M.; Luna, I.; Gómez, I.; López, M.; Cervera, J.; et al. Adverse prognostic value of MYBL2 overexpression and association with microRNA-30 family in acute myeloid leukemia patients. Leuk. Res. 2013, 37, 1690–1696. [Google Scholar] [CrossRef]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; De Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.-G.; Negrini, M.; et al. E2F1-regulated MicroRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.-L.; Xue, G.; Mei, Q.; Wang, Y.-Z.; Ding, F.-X.; Liu, M.-F.; Lu, M.-H.; Tang, Y.; Yu, H.-Y.; Sun, S.-H. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009, 28, 2719–2732. [Google Scholar] [CrossRef]
- Li, S.-G.; Shi, Q.-W.; Yuan, L.-Y.; Qin, L.-P.; Wang, Y.; Miao, Y.-Q.; Chen, Z.; Ling, C.-Q.; Qin, W.-X. C-Myc-dependent repression of two oncogenic miRNA clusters contributes to triptolide-induced cell death in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2018, 37, 51. [Google Scholar] [CrossRef]
- Li, W.; Zhou, B.-R.; Hua, L.-J.; Guo, Z.; Luo, D. Differential miRNA profile on photoaged primary human fibroblasts irradiated with ultraviolet A. Tumor Biol. 2013, 34, 3491–3500. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Yang, S.; Guo, Z. MiRNA-27a decreases ultraviolet B irradiation-induced cell damage. J. Cell. Biochem. 2019, 121, 1032–1038. [Google Scholar] [CrossRef]
- Marionnet, C.; Pierrard, C.; Lejeune, F.; Sok, J.; Thomas, M.; Bernerd, F. Different oxidative stress response in keratinocytes and fibroblasts of reconstructed skin exposed to non extreme daily-ultraviolet radiation. PLoS ONE 2010, 5, e12059. [Google Scholar] [CrossRef]
- Gebhard, D.; Matt, K.; Burger, K.; Bergemann, J. Shortwave UV-induced damage as part of the solar damage spectrum is not a major contributor to mitochondrial dysfunction. J. Biochem. Mol. Toxicol. 2014, 28, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Spandau, D.F.; Travers, J.B. impact of age and insulin-like growth factor-1 on dna damage responses in UV-irradiated human skin. Molecules 2017, 22, 356. [Google Scholar] [CrossRef] [PubMed]
- Ikehata, H. Mechanistic considerations on the wavelength-dependent variations of UVR genotoxicity and mutagenesis in skin: The discrimination of UVA-signature from UV-signature mutation. Photochem. Photobiol. Sci. 2018, 17, 1861–1871. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; LeDuc, C.; Verbeke, A.; Toussaint, O. UV, stress and aging. Derm.-Endocrinol. 2012, 4, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, Y.-F.; Xu, R.-H.; Lu, H.; Hu, C.; Qian, H. miR-1246 releases RTKN2-dependent resistance to UVB-induced apoptosis in HaCaT cells. Mol. Cell. Biochem. 2014, 394, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, B.; Liu, W.; Xu, Y.; Wu, D.; Yin, Z.; Permatasari, F.; Luo, D. MiR-23a regulates DNA damage repair and apoptosis in UVB-irradiated HaCaT cells. J. Dermatol. Sci. 2013, 69, 68–76. [Google Scholar] [CrossRef]
- Zhang, T.; Qian, H.; Hu, C.; Lu, H.; Li, J.-B.; Wu, Y.-F.; Li, W. MiR-26a mediates ultraviolet B-induced apoptosis by targeting histone methyltransferase EZH2 depending on Myc expression. Cell. Physiol. Biochem. 2017, 43, 1188–1197. [Google Scholar] [CrossRef]
- Greussing, R.; Hackl, M.; Charoentong, P.; Pauck, A.; Monteforte, R.; Cavinato, M.; Hofer, E.; Scheideler, M.; Neuhaus, M.; Micutkova, L.; et al. Identification of microRNA-mRNA functional interactions in UVB-induced senescence of human diploid fibroblasts. BMC Genom. 2013, 14, 224. [Google Scholar] [CrossRef]
- Li, W.; Di, W.; Hua, L.; Zhou, B.; Guo, Z.; Luo, D. UVB suppresses PTEN expression by upregulating miR-141 in HaCaT cells. J. Biomed. Res. 2011, 25, 135–140. [Google Scholar] [CrossRef]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; O Bauerschlag, D.; Jöckel, K.-H.; Erbel, D.R.; Mühleisen, T.W.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef]
- Sidler, C.; Kovalchuk, O.; Kovalchuk, I. Epigenetic regulation of cellular senescence and aging. Front. Genet. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Liu, P.; Yang, Z.; Li, L.; Su, H.; Lu, N.; Peng, Z. MiR-155 negatively regulates c-Jun expression at the post-transcriptional level in human dermal fibroblastsin vitro: Implications in UVA Irradiation-induced photoaging. Cell. Physiol. Biochem. 2012, 29, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.-R.; Guo, X.-F.; Zhang, J.-A.; Xu, Y.; Li, W.; Wu, D.; Yin, Z.-Q.; Permatasari, F.; Luo, D. Elevated miR-34c-5p mediates dermal fibroblast senescence by ultraviolet irradiation. Int. J. Biol. Sci. 2013, 9, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Blackstone, B.N.; Wilgus, T.A.; Roy, S.; Wulff, B.C.; Powell, H.M. Skin biomechanics and miRNA expression following chronic UVB irradiation. Adv. Wound Care 2020, 9, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Younis, S.; Komine, M.; Tomic-Canic, M.; Blumenberg, M. Skinomics: A new toolbox to understand skin aging. In Textbook of Aging Skin, 2nd ed.; Farage, M.A., Miller, K.W., Maibach, H.I., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1361–1379. [Google Scholar] [CrossRef]
- Srivastava, A.; Karlsson, M.; Marionnet, C.; Bernerd, F.; Gueniche, A.; Rawadi, C.E.L.; Ståhle, M.; Sonkoly, E.; Breton, L.; Pivarcsi, A. Identification of chronological and photoageing-associated microRNAs in human skin. Sci. Rep. 2018, 8, 12990. [Google Scholar] [CrossRef]
- Zhang, J.-A.; Zhou, B.-R.; Xu, Y.; Chen, X.; Liu, J.; Gozali, M.; Wu, D.; Yin, Z.-Q.; Luo, D. MiR-23a-depressed autophagy is a participant in PUVA- and UVB-induced premature senescence. Oncotarget 2016, 7, 37420–37435. [Google Scholar] [CrossRef]
- Cianfanelli, V.; De Zio, D.; Di Bartolomeo, S.; Nazio, F.; Strappazzon, F.; Cecconi, F. Ambra1 at a glance. J. Cell Sci. 2015, 128, 2003–2008. [Google Scholar] [CrossRef]
- Si, C.; Wang, J.; Ma, W.; Hua, H.; Zhang, M.; Qian, W.; Zhou, B.-R.; Luo, D. Circular RNA expression profile in human fibroblast premature senescence after repeated ultraviolet B irradiations revealed by microarray. J. Cell. Physiol. 2019, 234, 18156–18168. [Google Scholar] [CrossRef]
- Oh, M.; Lee, J.; Kim, Y.J.; Rhee, W.J.; Park, J.H. Exosomes derived from human induced pluripotent stem cells ameliorate the aging of skin fibroblasts. Int. J. Mol. Sci. 2018, 19, 1715. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Ullah, M.; Ng, N.N.; Concepcion, W.; Thakor, A.S. Emerging role of stem cell-derived extracellular microRNAs in age-associated human diseases and in different therapies of longevity. Ageing Res. Rev. 2020, 57, 100979. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).