Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Research Object and Cultivation Conditions
2.3. Cell Treatment and Pellet Lysis
2.4. Proteomic Analysis
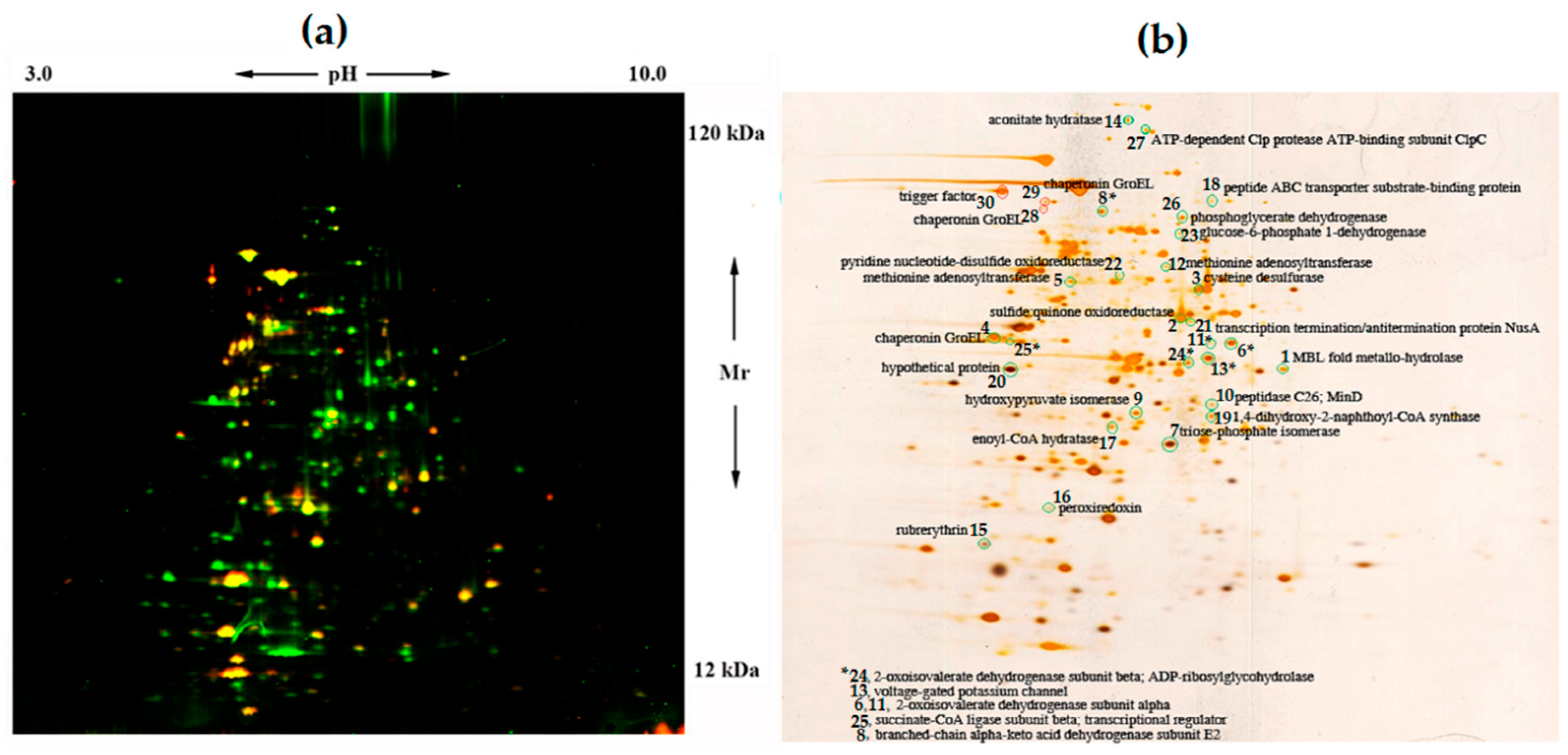
2.4.1. Two-Dimensional Difference Gel Electrophoresis (2D DIGE)
2.4.2. Trypsin Digestion and MALDI Mass Spectrometry
2.4.3. Protein Identification
2.5. Protein Characterization and Pathway Mapping
2.6. Analytical Techniques
2.7. Statistical Methods
3. Results and Discussion
3.1. Toxic Effect of Sulfide Concentrate on Bacterial Growth and Oxidative Activity
3.2. Metal Accumulation by Cells
3.3. Proteome Reorganization in Response to the Pyrite-Arsenopyrite Concentrate
3.3.1. Stress Response
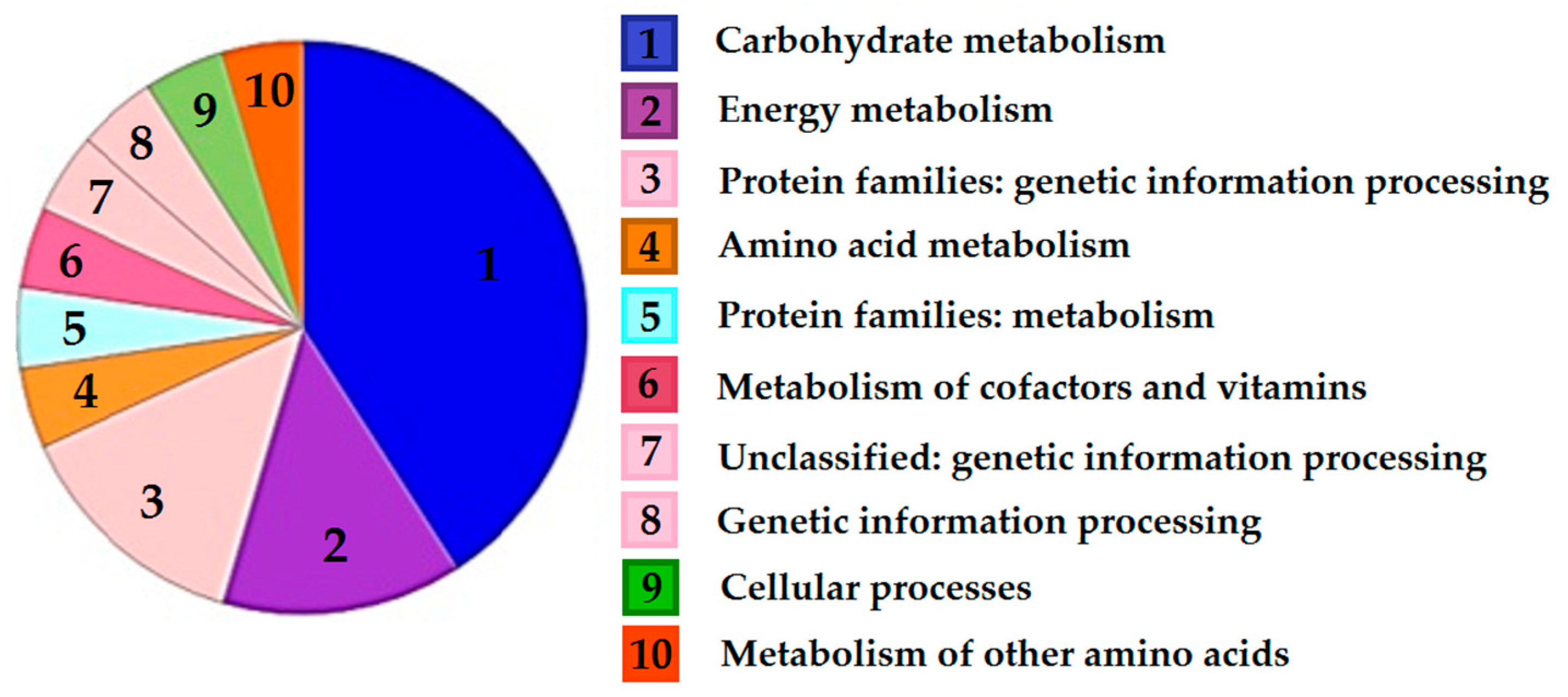
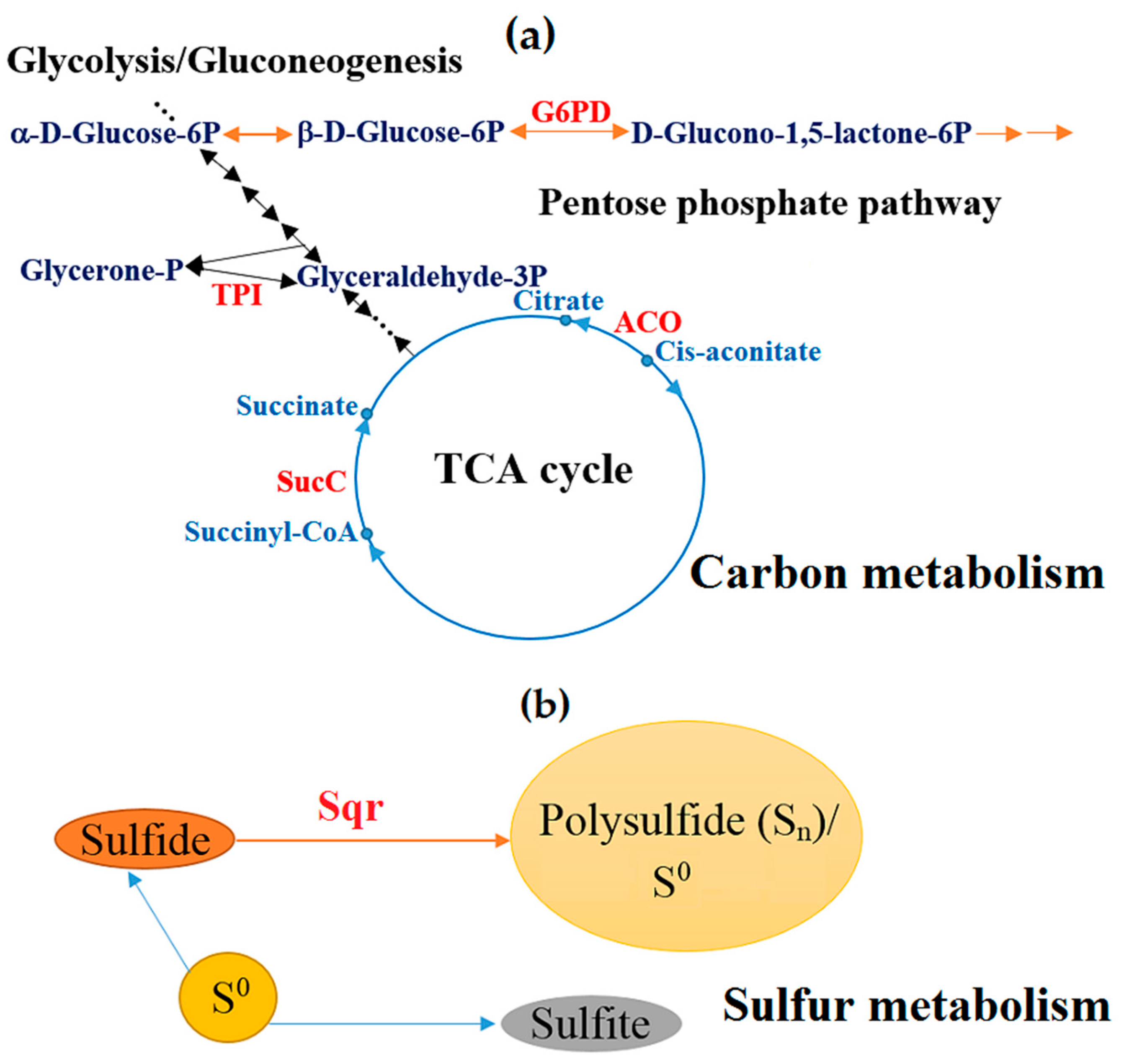
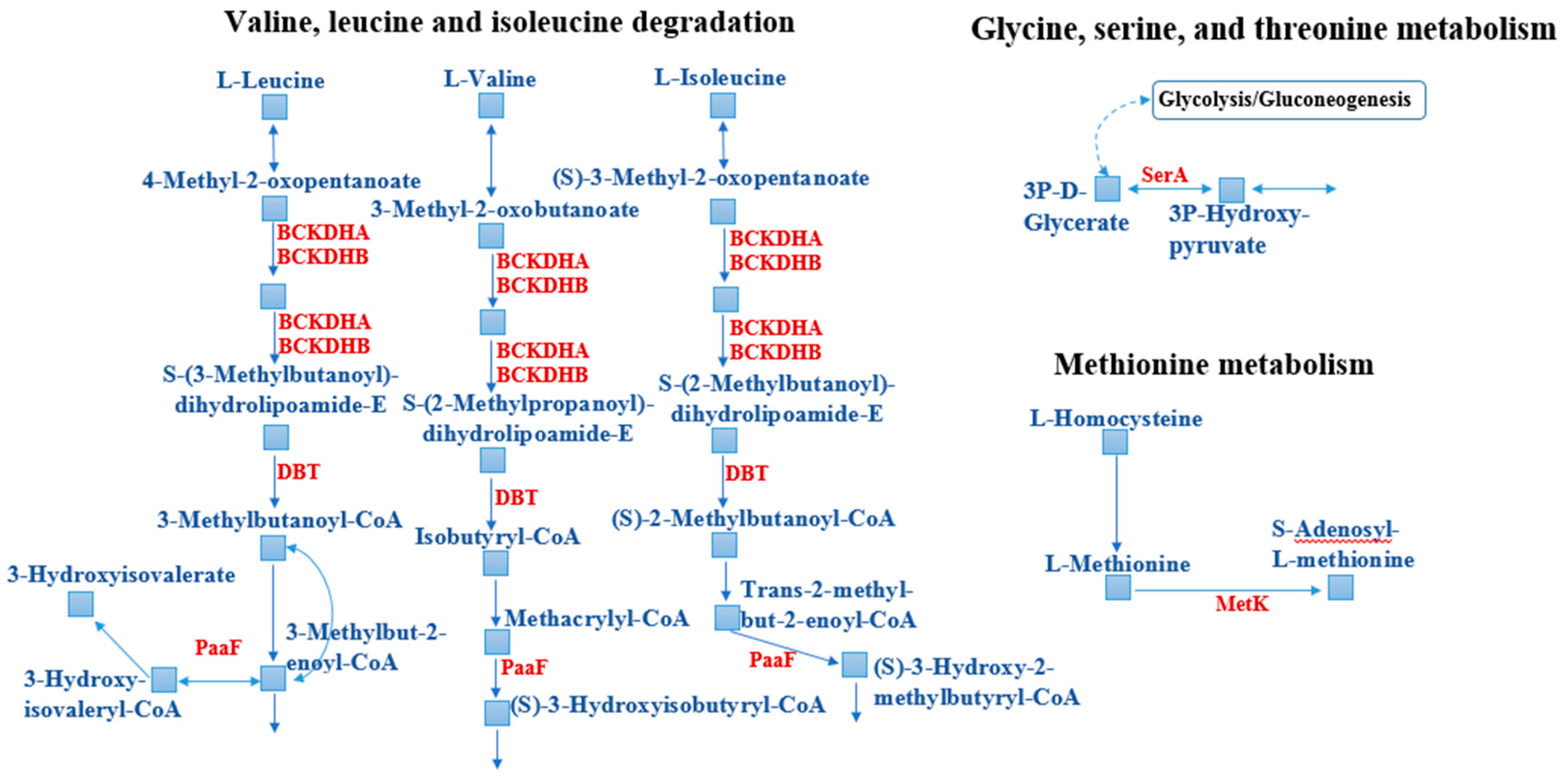
3.3.2. Metabolic Reorganization
4. Conclusions
- Sb. thermotolerans rapidly adapted to the toxic amount of the high-arsenic gold-bearing sulfide concentrate and proved to be among the most As-tolerant organisms known to date;
- although the cellular adsorption and intracellular accumulation of metal(loid)s occurred to high levels, the bacterium retained its growth and efficient substrate oxidation;
- in total, 30 upregulated proteins were involved in the response to the toxic content of the sulfide concentrate in the growth medium, and only three proteins, which, however, did not affect the vital activity of the preliminarily nonadapted Sb. thermotolerans cells, were downregulated;
- Sb. thermotolerans cells responded to adverse conditions by metabolic changes, including reinforcement of pathways of constructive and energy metabolism, and by activation of defense systems against unfavorable factors. At the same time, no specific metal-resistance components were regulated in response to metal(loid)s accumulated in the culture medium and by the cells;
- proteins of the stress response, such as the metal-related stress protein MBL fold metallo-hydrolase and GroEL chaperonin, probably, played crucial roles in the tolerance to the high-arsenic sulfide ore concentrate and arsenic, in particular;
- the markedly upregulated sulfide:quinone oxidoreductase, cysteine desulfurase, and S-adenosylmethionine synthetase were other main contributors to the bacterial response. Apart from the enzymatic function in sulfur metabolism, sulfide:quinone oxidoreductase potentially fulfilled the second function of a resistance-conferring protein in Sb. thermotolerans.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaksonen, A.H.; Boxall, N.J.; Gumulya, Y.; Khaleque, H.N.; Morris, C.; Bohu, T.; Cheng, K.Y.; Usher, K.M.; Lakaniemi, A.-M. Recent progress in biohydrometallurgy and microbial characterisation. Hydrometallurgy 2018, 180, 7–25. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Johnson, D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 2007, 153, 315–324. [Google Scholar] [CrossRef]
- Bond, P.L.; Druschel, G.K.; Banfield, J.F. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 2000, 66, 4962–4971. [Google Scholar] [CrossRef]
- d’Hugues, P.; Joulian, C.; Spolaore, P.; Michel, C.; Garrido, F.; Morin, D. Continuous bioleaching of a pyrite concentrate in stirred reactors: Population dynamics and exopolysaccharide production vs. bioleaching performance. Hydrometallurgy 2008, 94, 34–41. [Google Scholar] [CrossRef]
- Dopson, M.; Lindstrӧm, E.B. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Microb. Ecol. 2004, 48, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Bogdanova, T.I.; Tsaplina, I.A.; Kondrat’eva, T.F.; Duda, V.I.; Suzina, N.E.; Melamud, V.S.; Tourova, T.P.; Karavaiko, G.I. Sulfobacillus thermotolerans sp. nov., a thermotolerant, chemolithotrophic bacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.L.; Shu, W.S.; Hallberg, K.B.; Li, F.; Lan, C.Y.; Zhou, W.H.; Huang, L.N. Culturable and molecular phylogenetic diversity of microorganisms in an open-dumped, extremely acidic Pb/Zn mine tailings. Extremophiles 2008, 12, 657–664. [Google Scholar] [CrossRef]
- Tsaplina, I.A.; Krasil’nikova, E.N.; Zhuravleva, A.E.; Egorova, M.A.; Zakharchuk, L.M.; Suzina, N.E.; Bogdanova, T.I.; Kondrat’eva, T.F. Phenotypic properties of Sulfobacillus thermotolerans: Comparative aspects. Microbiology 2008, 77, 654–664. [Google Scholar] [CrossRef]
- Fomchenko, N.V.; Muravyov, M.I. Two-step biohydrometallurgical technology of copper-zinc concentrate processing as an opportunity to reduce negative impacts on the environment. J. Environ. Manag. 2018, 226, 270–277. [Google Scholar] [CrossRef]
- Muravyov, M.; Panyushkina, A. Distinct roles of acidophiles in complete oxidation of high-sulfur ferric leach product of zinc sulfide concentrate. Microorganisms 2020, 8, 386. [Google Scholar] [CrossRef]
- Hao, C.; Wang, L.; Dong, H.; Zhang, H. Succession of acidophilic bacterial community during bio-oxidation of refractory gold-containing sulfides. Geomicrobiol. J. 2010, 27, 683–691. [Google Scholar] [CrossRef]
- Tsaplina, I.A.; Panyushkina, A.E.; Melamud, V.S.; Grigor’eva, N.V.; Kondrat’eva, T.F. Leaching of pyrite–arsenopyrite concentrate in bioreactors during continuous cultivation of a thermoacidophilic microbial community. Microbiology 2014, 83, 568–576. [Google Scholar] [CrossRef]
- Bulaev, A.; Belyi, A.; Panyushkina, A.; Solopova, N.; Pivovarova, T. Microbial population of industrial biooxidation reactors. Solid State Phenom. 2017, 262, 48–52. [Google Scholar] [CrossRef]
- Tsaplina, I.A.; Zhuravleva, A.E.; Egorova, M.A.; Bogdanova, T.I.; Krasil’nikova, E.N.; Zakharchuk, L.M.; Kondrat’eva, T.F. Response to oxygen limitation in bacteria of the genus Sulfobacillus. Microbiology 2010, 79, 13–22. [Google Scholar] [CrossRef]
- Panyushkina, A.E. Metabolic potential of Sulfobacillus thermotolerans: Pathways for assimilation of nitrogen compounds and the possibility of lithotrophic growth in the presence of molecular hydrogen. Microbiology 2019, 13, 759–763. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Babenko, V.V.; Nikitina, A.S.; Selezneva, O.V.; Tsaplina, I.A.; Letarova, M.A.; Kostryukova, E.S.; Letarov, A.V. Sulfobacillus thermotolerans: New insights into resistance and metabolic capacities of acidophilic chemolithotrophs. Sci. Rep. 2019, 9, 15069. [Google Scholar] [CrossRef]
- Tuffin, I.M.; Hector, S.B.; Deane, S.M.; Rawlings, D.E. Resistance determinants of a highly arsenic-resistant strain of Leptospirillum ferriphilum isolated from a commercial biooxidation tank. Appl. Environ. Microbiol. 2006, 72, 2247–2253. [Google Scholar] [CrossRef]
- Navarro, C.A.; von Bernath, D.; Jerez, C.A. Heavy metal resistance strategies of acidophilic bacteria and their acquisition: Importance for biomining and bioremediation. Biol. Res. 2013, 46, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Navarro, C.A.; Arancibia, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 2010, 28, 839–848. [Google Scholar] [CrossRef]
- Orellana, L.H.; Jerez, C.A. A genomic island provides Acidithiobacillus ferrooxidans ATCC 53993 additional copper resistance: A possible competitive advantage. Appl. Microbiol. Biotechnol. 2011, 92, 761–767. [Google Scholar] [CrossRef]
- Salazar, C.; Acosta, M.; Galleguillos, P.; Shmaryahu, A.; Quatrini, R.; Holmes, D.S.; Demergasso, C. Analysis of gene expression in response to copper stress in Acidithiobacillus ferrooxidans strain D2, isolated from a copper bioleaching operation. Adv. Mat. Res. 2013, 825, 157–161. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Z.; Liu, L.; Deng, F.; Liu, X.; Qiu, G. Metal resistance-related genes are differently expressed in response to copper and zinc ion in six Acidithiobacillus ferrooxidans strains. Curr. Microbiol. 2014, 69, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Almárcegui, R.J.; Navarro, C.A.; Paradela, A.; Albar, J.P.; von Bernath, D.; Jerez, C.A. New copper resistance determinants in the extremophile Acidithiobacillus ferrooxidans: A quantitative proteomic analysis. J. Proteome Res. 2014, 13, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Dekker, L.; Arsène-Ploetze, F.; Santini, J.M. Comparative proteomics of Acidithiobacillus ferrooxidans grown in the presence and absence of uranium. Res. Microbiol. 2016, 167, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, F.; Miri, S.; Jahani, S. Effect of metal sulfide pulp density on gene expression of electron transporters in Acidithiobacillus sp. FJ2. Arch. Microbiol. 2017, 199, 521–530. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Dopson, M.; Wexler, M.; Sawers, R.G.; Bond, P.L. Molecular insight into extreme copper resistance in the extremophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Microbiology 2005, 151, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Remonsellez, F.; Arancibia, R.; Jerez, C.A. Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea 2013, 2013, 289236. [Google Scholar] [CrossRef]
- Mangold, S.; Potrykus, J.; Björn, E.; Lövgren, L.; Dopson, M. Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 2013, 17, 75–85. [Google Scholar] [CrossRef]
- Martínez-Bussenius, C.; Navarro, C.A.; Jerez, C.A. Microbial copper resistance: Importance in biohydrometallurgy. Microb. Biotechnol. 2017, 10, 279–295. [Google Scholar] [CrossRef]
- Soto, D.F.; Recalde, A.; Orell, A.; Albers, S.-V.; Paradela, A.; Navarro, C.A.; Jerez, C.A. Global effect of the lack of inorganic polyphosphate in the extremophilic archaeon Sulfolobus solfataricus: A proteomic approach. J. Proteomics. 2019, 191, 143–152. [Google Scholar] [CrossRef]
- Dopson, M.; Holmes, D. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 2014, 98, 8133–8144. [Google Scholar] [CrossRef]
- Dopson, M.; Ossandon, F.J.; Lövgren, L.; Holmes, D.S. Metal resistance or tolerance? Acidophiles confront high metal loads via both abiotic and biotic mechanisms. Front. Microbiol. 2014, 5, 157. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, J.; Mi, S.; Lin, J. Arsenic resistance operon structure in Leptospirillum ferriphilum and proteomic response to arsenic stress. Bioresour. Technol. 2010, 101, 9811–9814. [Google Scholar] [CrossRef]
- Jiang, H.; Liang, Y.; Yin, H.; Xiao, Y.; Guo, X.; Xu, Y.; Hu, Q.; Liu, H.; Liu, X. Effects of arsenite resistance on the growth and functional gene expression of Leptospirillum ferriphilum and Acidithiobacillus thiooxidans in pure culture and coculture. BioMed Res. Int. 2015, 2015, 203197. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, P.; Guiliani, N.; Valenzuela, L.; Beard, S.; Jerez, C.A. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 2004, 70, 4491–4498. [Google Scholar] [CrossRef] [PubMed]
- Felício, A.P.; Oliveira, E.; Odena, M.A.; Garcia, O., Jr.; Bertolini, M.C.; Ferraz, L.F.C.; Ottoboni, L.M.M.; Novo, M.T.M. Differential proteomic analysis of Acidithiobacillus ferrooxidans cells maintained in contact with bornite or chalcopyrite: Proteins involved with the early bacterial response. Process Biochem. 2011, 46, 770–776. [Google Scholar] [CrossRef]
- Bellenberg, S.; Huynh, D.; Poetsch, A.; Sand, W.; Vera, M. Proteomics reveal enhanced oxidative stress responses and metabolic adaptation in Acidithiobacillus ferrooxidans biofilm cells on pyrite. Front. Microbiol. 2019, 10, 592. [Google Scholar] [CrossRef]
- Carlos, C.; Reis, F.C.; Vicentini, R.; Madureira, D.J.; Ottoboni, L.M.M. The rus operon genes are differentially regulated when Acidithiobacillus ferrooxidans LR is kept in contact with metal sulfides. Curr. Microbiol. 2008, 57, 375–380. [Google Scholar] [CrossRef]
- Christel, S.; Herold, M.; Bellenberg, S.; Hajjami, M.E.; Buetti-Dinh, A.; Pivkin, I.V.; Sand, W.; Wilmes, P.; Poetsch, A.; Dopson, M. Multi-omics reveals the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilumT. Appl. Environ. Microbiol. 2018, 84, e02091-17. [Google Scholar] [CrossRef]
- Sand, W.; Gehrke, T. Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 2006, 157, 49–56. [Google Scholar] [CrossRef]
- Vera, M.; Krok, B.; Bellenberg, S.; Sand, W.; Poetsch, A. Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics 2013, 13, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Belnap, C.P.; Pan, C.; VerBerkmoes, N.C.; Power, M.E.; Samatova, N.F.; Carver, R.L.; Hettich, R.L.; Banfield, J.F. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 2010, 4, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Bowen, B.P.; Thomas, B.C.; Mueller, R.S.; Denef, V.J.; VerBerkmoes, N.C.; Hettich, R.L.; Northen, T.R.; Banfield, J.F. Metabolome-proteome differentiation coupled to microbial divergence. MBio 2010, 1, e00246-10. [Google Scholar] [CrossRef] [PubMed]
- Mosier, A.C.; Li, Z.; Thomas, B.C.; Hettich, R.L.; Pan, C.; Banfield, J.F. Elevated temperature alters proteomic responses of individual organisms within a biofilm community. ISME J. 2015, 9, 180–194. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Jensen, O.N.; Wilm, M.; Shevchenko, A.; Mann, M. Peptide sequencing of 2-DE gel-isolated proteins by nanoelectrospray tandem mass spectrometry. Methods Mol. Biol. 1999, 112, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–462. [Google Scholar] [CrossRef] [PubMed]
- Melamud, V.S.; Pivovarova, T.A.; Kondrat’eva, T.F.; Karavaiko, G.I. Study of oxidation by bacteria of a difficult-to-dress gold-containing pyrite-arsenopyrite concentrate under moderately thermophilic conditions. Appl. Biochem. Microbiol. 1999, 35, 161–167. [Google Scholar]
- Goyal, N.; Jain, S.C.; Banerjee, U.C. Comparative studies on the microbial adsorption of heavy metals. Adv. Environ. Res. 2003, 7, 311–319. [Google Scholar] [CrossRef]
- Pagani, M.A.; Casamayor, A.; Serrano, R.; Atrian, S.; Ariño, J. Disruption of iron homeostasis in Saccharomyces cerevisiae by high zinc levels: A genome-wide study. Mol. Microbiol. 2007, 65, 521–537. [Google Scholar] [CrossRef]
- Watling, H.R.; Perrot, F.A.; Shiers, D.W. Comparison of selected characteristics of Sulfobacillus species and review of their occurrence in acidic and bioleaching environments. Hydrometallurgy 2008, 93, 57–65. [Google Scholar] [CrossRef]
- Dave, S.R.; Gupta, K.H.; Tipre, D.R. Characterization of arsenic resistant and arsenopyrite oxidizing Acidithiobacillus ferrooxidans from Hutti gold leachate and effluents. Bioresour. Technol. 2008, 99, 7514–7520. [Google Scholar] [CrossRef]
- Casas-Flores, S.; Gómez-Rodríguez, E.Y.; García-Meza, J.V. Community of thermoacidophilic and arsenic resistant microorganisms isolated from a deep profile of mine heaps. AMB Express 2015, 5, 54. [Google Scholar] [CrossRef] [PubMed]
- Kotze, A.A.; Tuffin, I.M.; Deane, S.M.; Rawlings, D.E. Cloning and characterization of the chromosomal arsenic resistance genes from Acidithiobacillus caldus and enhanced arsenic resistance on conjugal transfer of ars genes located on transposon TnAtcArs. Microbiology 2006, 152, 3551–3560. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Baker-Austin, C.; Koppineedi, P.R.; Bond, P.L. Growth in sulfidic mineral environments: Metal resistance mechanisms in acidophilic micro-organisms. Microbiology 2003, 149, 1959–1970. [Google Scholar] [CrossRef]
- Banerjee, S.; Datta, S.; Chattyopadhyay, D.; Sarkar, P. Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J. Environ. Sci. Health A. Tox. Hazard. Subst. Environ. Eng. 2011, 46, 1736–1747. [Google Scholar] [CrossRef]
- Yin, X.X.; Wang, L.H.; Bai, R.; Huang, H.; Sun, G.X. Accumulation and transformation of arsenic in the blue-green alga Synechocystis sp. PCC6803. Water Air Soil Pollut. 2012, 223, 1183–1190. [Google Scholar] [CrossRef]
- Mayzel, B.; Aizenberg, J.; Ilan, M. The elemental composition of Demospongiae from the Red Sea, Gulf of Aqaba. PLoS ONE 2014, 9, e95775. [Google Scholar] [CrossRef]
- Keren, R.; Mayzel, B.; Lavy, A.; Polishchuk, I.; Levy, D.; Fakra, S.C.; Pokroy, B.; Ilan, M. Sponge-associated bacteria mineralize arsenic and barium on intracellular vesicles. Nat. Commun. 2017, 8, 14393. [Google Scholar] [CrossRef]
- Dorian, A.B.H.; Landy, I.R.B.; Enrique, D.-P.; Luis, F.-L. Zinc and lead biosorption by Delftia tsuruhatensis: A bacterial strain resistant to metals isolated from mine tailings. J. Water Resour. Prot. 2012, 4, 207–216. [Google Scholar] [CrossRef]
- Celaya, R.J.; Noriega, J.A.; Yeomans, J.H.; Ortega, L.J.; Ruiz-Manríquez, A. Biosorption of Zn(II) by Thiobacillus ferrooxidans. Bioprocess Eng. 2000, 22, 539–542. [Google Scholar] [CrossRef]
- Liu, H.-L.; Chen, B.-Y.; Lan, Y.-W.; Cheng, Y.-C. Biosorption of Zn(II) and Cu(II) by the indigenous Thiobacillus thiooxidans. Chem. Eng. J. 2004, 97, 195–201. [Google Scholar] [CrossRef]
- Aston, J.E.; Apel, W.A.; Lee, B.D.; Peyton, B.M. Effects of cell condition, pH, and temperature on lead, zinc, and copper sorption to Acidithiobacillus caldus strain BC13. J. Hazard. Mater. 2010, 184, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Frees, D.; Chastanet, A.; Qazi, S.; Soerensen, K.; Hill, P.; Msadek, T.; Ingmer, H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 2004, 54, 1445–1462. [Google Scholar] [CrossRef] [PubMed]
- Sitthisak, S.; Kitti, T.; Boonyonying, K.; Wozniak, D.; Devreese, B.; Mongkolsuk, S.; Jayaswal, R.K. McsA and the roles of metal binding motif in Staphylococcus aureus. FEMS Microbiol. Lett. 2012, 327, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Morgante, V.; Mirete, S.; de Figueras, C.G.; Cacho, M.P.; González-Pastor, J.E. Exploring the diversity of arsenic resistance genes from acid mine drainage microorganisms. Environ. Microbiol. 2015, 17, 1910–1925. [Google Scholar] [CrossRef]
- Easton, J.A.; Thompson, P.; Crowder, M.W. Time-dependent translational response of E. coli to excess Zn(II). J. Biomol. Tech. 2006, 17, 303–307. [Google Scholar]
- Ferianc, P.; Farewell, A.; Nystrom, T. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology 1998, 144, 1045–1050. [Google Scholar] [CrossRef]
- Babu, M.M.G.; Sridhar, J.; Gunasekaran, P. Global transcriptome analysis of Bacillus cereus ATCC 14579 in response to silver nitrate stress. J. Nanobiotechnol. 2011, 9, 49. [Google Scholar] [CrossRef]
- Khan, A.L.; Ullah, I.; Hussain, J.; Kang, S.M.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.J. Regulations of essential amino acids and proteomics of bacterial endophytes Sphingomonas sp. Lk11 during cadmium uptake. Environ. Toxicol. 2016, 31, 887–896. [Google Scholar] [CrossRef]
- Siripornadulsil, S.; Thanwisai, L.; Siripornadulsil, W. Changes in the proteome of the cadmium-tolerant bacteria Cupriavidus taiwanensis KKU2500-3 in response to cadmium toxicity. Can. J. Microbiol. 2014, 60, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Bukau, B.; Kramer, G. Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 2010, 1803, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Damare, S. Differential protein expression in a marine-derived Staphylococcus sp. NIOSBK35 in response to arsenic(III). 3Biotech 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Wu, W.; Zhang, X.; Gu, T.; Zhu, M.; Tan, W. Oxidative stress induced by metal ions in bioleaching of LiCoO2 by an acidophilic microbial consortium. Front. Microbiol. 2020, 10, 3058. [Google Scholar] [CrossRef]
- Ehrlich, H.L. Geomicrobiology, 3rd ed.; Marcel Dekker: New York, NY, USA, 1996. [Google Scholar]
- Loenen, W.A. S-adenosylmethionine: Jack of all trades and master of everything? Biochem. Soc. Trans. 2006, 34, 330–333. [Google Scholar] [CrossRef]
- Walker, J.; Gongora, R.; Vasquez, J.-J.; Drummelsmith, J.; Burchmore, R.; Roy, G.; Ouellette, M.; Gomez, M.A.; Saravia, N.G. Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol. Biochem. Parasitol. 2012, 183, 166–176. [Google Scholar] [CrossRef]
- Dai, H.; Cao, F.; Chen, X.; Zhang, M.; Ahmed, I.M.; Chen, Z.-H.; Li, C.; Zhang, G.; Wu, F. Comparative proteomic analysis of aluminum tolerance in Tibetan wild and cultivated barleys. PLoS ONE 2013, 8, e63428. [Google Scholar] [CrossRef]
- Hego, E.; Bes, C.M.; Bedon, F.; Palagi, P.M.; Chaumeil, P.; Barré, A.; Claverol, S.; Dupuy, J.-W.; Bonneu, M.; Lalanne, C.; et al. Differential accumulation of soluble proteins in roots of metallicolous and nonmetallicolous populations of Agrostis capillaris L. exposed to Cu. Proteomics 2014, 14, 1746–1758. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, J.; Gao, Y. In silico identification of a multi-functional regulatory protein involved in Holliday junction resolution in bacteria. BMC Syst. Biol. 2012, 6, S20. [Google Scholar] [CrossRef]
- Wei, L.; Wu, Y.; Qiao, H.; Xu, W.; Zhang, Y.; Liu, X.; Wang, Q. YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol. Lett. 2018, 365, fny137. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xia, X.; Wang, D.; Zhou, Z.; Wang, G. Gene function and expression regulation of RuvRCAB in bacterial Cr(VI), As(III), Sb(III), and Cd(II) resistance. Appl. Microbiol. Biotechnol. 2019, 103, 2701–2713. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Jiang, T.; Yu, B.; Wang, L.; Gao, C.; Ma, C.; Xu, P.; Ma., Y. Escherichia coli transcription termination factor NusA: Heat-induced oligomerization and chaperone activity. Sci. Rep. 2013, 3, 2347. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M.; Wexler, M.; Sawers, R.G.; Stemmler, A.; Rosen, B.P.; Bond, P.L. Extreme arsenic resistance by the acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 2007, 11, 425–434. [Google Scholar] [CrossRef]
| Element | Content (wt%),Original Concentrate | Element Concentration (mM), Liquid Phase (after 80 h) | |
|---|---|---|---|
| Fe | Fetot a | 38.7 ± 0.51 | 39.0 ± 2.2 |
| FeS b | 34.9 ± 0.89 | ||
| S | Stot | 28.25 ± 0.76 | 27.1 ± 1.5 |
| SS | 28.1 ± 0.95 | ||
| As | Astot | 19.1± 0.75 | 35.3 ± 3.7 (4.4 As5+ and 30.9 As3+) |
| AsS | 16 ± 0.15 | ||
| Sb | 0.15 ± 0.03 | nd c (˂0.550) | |
| Zn | 0.72 ± 0.02 | 1.5 ± 0.1 | |
| Cu | 0.04 ± 0.01 | nd (˂0.077) | |
| Pb | 1.21 ± 0.06 | nd (˂0.190) | |
| Au | 135.2 ± 1.61 g/t | − d | |
| Ag | 160 ± 2.35 g/t | − | |
| Element | Growth Conditions | |||
|---|---|---|---|---|
| Optimal | Ore Concentrate, 20 g/L | |||
| Ctot, µg/g a | Cintra, µg/g b | Ctot, µg/g a | Cintra, µg/g b | |
| Fe | 60210 ± 350 | 830 ± 4.9 | 6515 ± 59 | 588 ± 6 |
| Zn | 53.5 ± 1.5 | 31.7 ± 2.5 | 1338 ± 74 | 463 ± 5.9 |
| Cu | 29 ± 0.54 | 16 ± 0.7 | 165.5 ± 18 | 125 ± 7 |
| Pb | 2.85 ± 0.36 | <0.00098 c | 370 ± 16.7 | 74 ± 2.7 |
| Ag | 1.35 ± 0.25 | <0.066 c | 135.5 ± 6.4 | 10 ± 1.4 |
| Au | 0.027 ± 0.003 | <0.0017 c | 11.8 ± 0.6 | 9.9 ± 0.5 |
| As | 1.75 ± 0.07 | 0.94 ± 0.17 | 941 ± 46 | 233.5 ± 6 |
| Sb | 1.7 ± 0.1 | 0.33 ± 0.1 | 53.5 ± 5.7 | 26.5 ± 2.3 |
| Protein Spot No. a | Putative Homolog | Protein ID, Size (a. a.) | Function(s) | Cy3/Cy5 Ratio |
|---|---|---|---|---|
| 1 | MBL fold metallo-hydrolase | BXT84_14770, 312 | Hydrolytic enzymes that include class B β-lactamases, hydroxyacylglutathione hydrolases, persulfide dioxygenases, flavodiiron proteins, insecticide hydrolases. | 6.3 ± 0.82 |
| 2 | Sulfide:quinone oxidoreductase [EC 1.8.5.4] | BXT84_01945, 379 | Reaction of transformation of H2S to polysulfide, in which two electrons are transferred to the electron chain by quinone. Energy production and conversion. Sulfur metabolism. | 5.8 ± 0.75 |
| 3 | Cysteine desulfurase | BXT84_05690, 406 | Removal of elemental sulfur and selenium atoms from L-cysteine, L-cystine, L-selenocysteine, and L-selenocystine to produce L-alanine. Amino acid transport, metabolism, and degradation. Biosynthesis of cofactors, prosthetic groups, and carriers. | 5.4 ± 0.70 |
| 4 | Chaperonin GroEL | BXT84_08625, 548 | Productive folding of proteins. Chaperones, chaperonins, stress proteins, etc. | 5.3 ± 0.69 |
| 5 | S-adenosylmethionine synthetase [EC 2.5.1.6] | BXT84_12950, 399 | DNA methylation. Control of gene expression. Coenzyme transport and metabolism. Gene transcription. Cell proliferation. | 5.0 ± 0.65 |
| 6 | 2-Oxoisovalerate dehydrogenase subunit alpha | BXT84_00985, 331 | Oxidative decarboxylation of 4-methyl-2-oxopentanoate, 3-methyl-2-oxopentanoate, and 3-methyl-2-oxobutanoate. Amino acid transport, metabolism, and degradation. | 3.2 ± 0.42 |
| 7 | Triose-phosphate isomerase | BXT84_02930, 241 | Interconversion of dihydroxyacetone phosphate and D-glyceraldehyde-3-phosphate. Energy production and conversion. Carbon metabolism. Glycolysis/gluconeogenesis. | 3.2 ± 0.42 |
| 8 | Branched-chain α-keto acid dehydrogenase subunit E2 | BXT84_00995, 436 | Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) component. TCA cycle and valine, leucine, and isoleucine degradation. Energy production and conversion. Carbon metabolism. Amino acid transport, metabolism, and degradation. | 3.1 ± 0.40 |
| 9 | Hydroxypyruvate isomerase | BXT84_05190, 259 | Interconvertion of hydroxypyruvate and 2-hydroxy-3-oxopropanoate. Glyoxylate and dicarboxylate metabolism. Carbohydrate transport and metabolism. | 3.1 ± 0.40 |
| 10 | Peptidase C26Protein MinD | BXT84_08510, 247 | Gamma-Glutamyl bond cleavage in poly-gamma-glutamyl substrates. Amino acid transport/metabolism. | 2.6 ± 0.34 |
| BXT84_03685, 265 | MinD stimulates activity of the division inhibitor MinC. Cell biogenesis, cell division. | |||
| 11 | 2-Oxoisovalerate dehydrogenase subunit alpha | BXT84_00985, 331 | Oxidative decarboxylation of 4-methyl-2-oxopentanoate, 3-methyl-2-oxopentanoate, and 3-methyl-2-oxobutanoate. Amino acid transport, metabolism, and degradation. | 2.6 ± 0.34 |
| 12 | S-adenosylmethionine synthetase [EC 2.5.1.6] | BXT84_12950, 399 | DNA methylation. Control of gene expression. Coenzyme transport and metabolism. Gene transcription. Cell proliferation. | 2.4 ± 0.31 |
| 13 | Voltage-gated K channel | BXT84_12450, 317 | Binds NADPH and couples voltage-gated channel activity to the redox potential of the cell. | 2.4 ± 0.31 |
| 14 | Aconitate hydratase [EC 4.2.1.3] | BXT84_12440, 900 | The reversible isomerization of citrate/isocitrate (cis-2-methylaconitate/2-methylisocitrate) in the TCA/methylcitrate cycle. Energy production and conversion. Carbon metabolism. | 2.4 ± 0.31 |
| 15 | Rubrerythrin | BXT84_08520, 140 | Reduction of H2O2 (oxidative stress protection system). Energy production and conversion. | 2.3 ± 0.29 |
| 16 | Thioredoxin peroxidase (peroxiredoxin) | BXT84_15685, 178 | Homodimeric thiol-specific antioxidant protein reducing and detoxifying H2O2, peroxynitrite, and organic hydroperoxides. Detoxification. Oxidative stress defense. | 2.3 ± 0.29 |
| 17 | Enoyl-CoA hydratase | BXT84_12400, 254 | An important role in fatty acid metabolism. Lipid transport and metabolism. | 2.3 ± 0.30 |
| 18 | Peptide ABC transporter substrate-binding protein | BXT84_06635, 565 | ABC-type transport system. Involved in the transport of leucine, isoleucine, and valine. Amino acid transport, metabolism, and degradation. | 2.3 ± 0.30 |
| 19 | 1,4-dihydroxy-2-naphthoyl-CoA synthase [EC 4.1.3.36] | BXT84_09740, 273 | Conversion of 2-succinylbenzoate into 1,4-di-hydroxy-2-naphthoate. Coenzyme transport and metabolism. Biosynthesis of cofactors, prosthetic groups, and carriers. | 2.2 ± 0.28 |
| 20 | Hypothetical protein | BXT84_01950, 335 | DUF1641 domain-containing uncharacterized conserved protein. Function unknown. | 2.2 ± 0.28 |
| 21 | NusA | BXT84_06895, 349 | N utilization substance protein A, a bacterial transcription termination factor. It may serve as a molecular chaperone. Transcription. Transcription factors. | 2.2 ± 0.29 |
| 22 | Pyridine nucleotide-disulfide oxidoreductase | BXT84_04315, 420 | Class Ⅰ and Ⅱ oxidoreductases and NADH oxidases and peroxidases. Energy production and conversion. | 2.2 ± 0.29 |
| 23 | Glucose-6-phosphate 1-dehydrogenase | BXT84_09125, 513 | D-glucose 6-phosphate + NADP+ ↔ 6-phospho-D-glucono-1,5-lactone + NADPH + H+. Carbon metabolism. Carbohydrate transport and metabolism. | 2.2 ± 0.29 |
| 24 | 2-Oxoisovalerate dehydrogenase subunit beta | BXT84_00990, 327 | Oxidative decarboxylation of 4-methyl-2-oxopentanoate, 3-methyl-2-oxopentanoate, and 3-methyl-2-oxobutanoate. Amino acid transport, metabolism, and degradation. | 2.0 ± 0.26 |
| ADP-ribosyl-glycohydrolase | BXT84_07905, 340 | ADP-ribosylations. Posttranslational modification, protein turnover, chaperones. | ||
| 25 | Succinate-CoA ligase subunit beta | BXT84_09905, 370 | Succinate-CoA ligase catalyzes the reversible reaction of succinyl-CoA to succinate. Energy metabolism. TCA cycle. | 2.0 ± 0.26 |
| YebC/PmpR family transcriptional regulator | BXT84_10200, 248 | Regulation of RuvABC and participation in other biological processes. Transcription, translation, ribosomal structure, and biogenesis. Regulatory functions, DNA interactions. | ||
| 26 | Phosphoglycerate dehydrogenase | BXT84_08460, 519 | Biosynthesis of L-serine from D-3-phosphoglycerate. Amino acid biosynthesis. | 2.0 ± 0.26 |
| 27 | ATP-dependent Clp protease ATP-binding subunit ClpC | BXT84_02130, 821 | ClpC ATPase of the Hsp100 family, a positive regulator of the heat shock response. Molecular chaperon. Posttranslational modification, protein turnover, chaperones. | 1.9 ± 0.25 |
| 28 | Chaperonin GroEL | BXT84_08625, 548 | Productive folding of proteins. Chaperones, chaperonins, stress proteins, etc. | 0.5 ± 0.06 |
| 29 | 0.5 ± 0.06 | |||
| 30 | Trigger factor | BXT84_03590, 437 | A ribosome-associated molecular chaperone. Protein fate, protein folding, and stabilization. | 0.4 ± 0.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panyushkina, A.; Matyushkina, D.; Pobeguts, O. Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans. Microorganisms 2020, 8, 1076. https://doi.org/10.3390/microorganisms8071076
Panyushkina A, Matyushkina D, Pobeguts O. Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans. Microorganisms. 2020; 8(7):1076. https://doi.org/10.3390/microorganisms8071076
Chicago/Turabian StylePanyushkina, Anna, Daria Matyushkina, and Olga Pobeguts. 2020. "Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans" Microorganisms 8, no. 7: 1076. https://doi.org/10.3390/microorganisms8071076
APA StylePanyushkina, A., Matyushkina, D., & Pobeguts, O. (2020). Understanding Stress Response to High-Arsenic Gold-Bearing Sulfide Concentrate in Extremely Metal-Resistant Acidophile Sulfobacillus thermotolerans. Microorganisms, 8(7), 1076. https://doi.org/10.3390/microorganisms8071076






