Virus and Potential Host Microbes from Viral-Enriched Metagenomic Characterization in the High-Altitude Wetland, Salar de Huasco, Chile
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites
2.2. Sample Collection and Processing
2.3. Sequencing and in Silico Analysis
3. Results and Discussion
3.1. Metagenome Composition
3.2. Viral Genome Composition
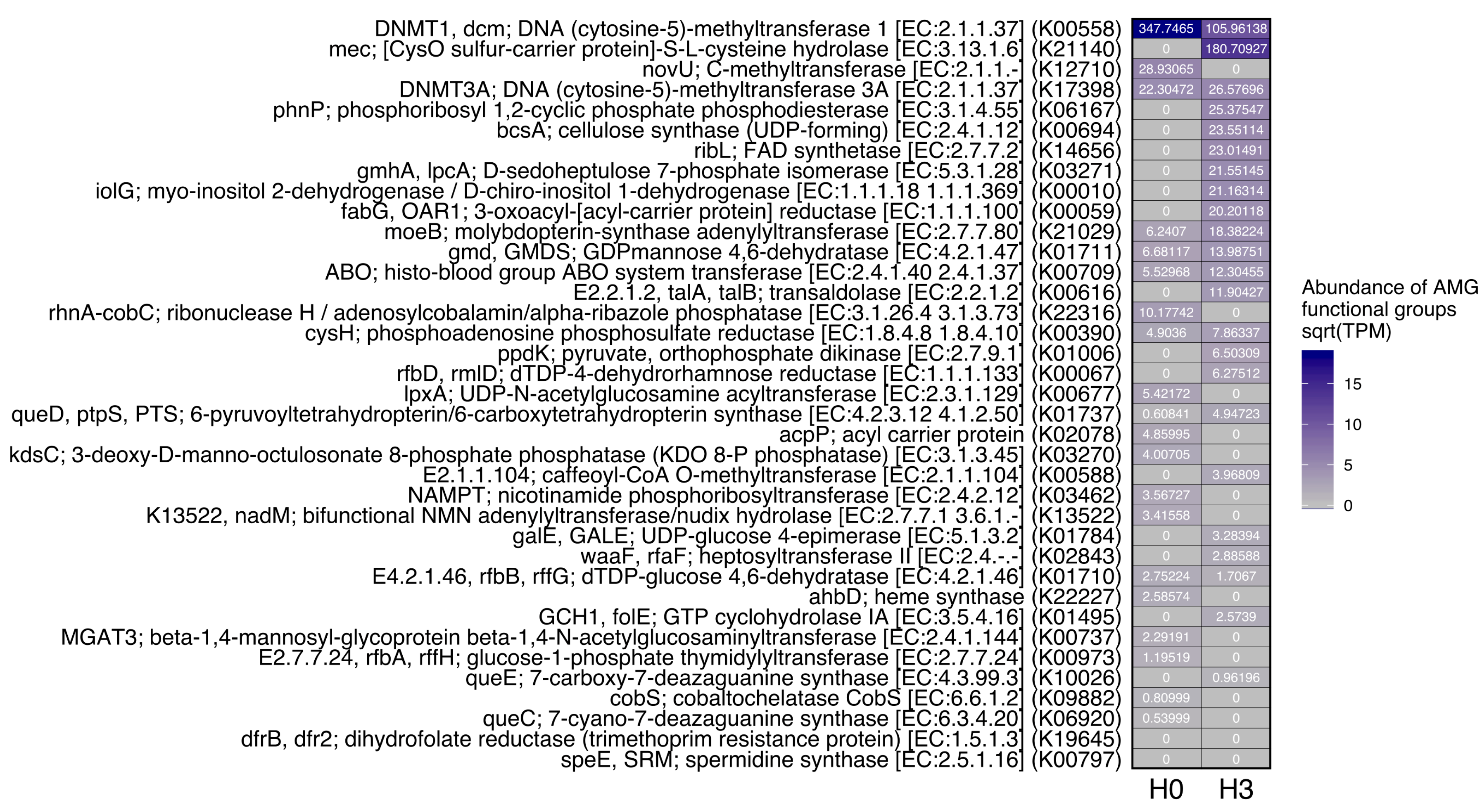
3.3. Functional Analysis of Viral Sequences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ramsar Convention Secretariat. An Introduction to the Convention on Wetlands, 5th ed.; (Previously the Ramsar Convention Manual); Ramsar Convention Secretariat: Gland, Switzerland, 2016; ISBN 2-8317-0200-3. [Google Scholar]
- Max Finlayson, C. Forty years of wetland conservation and wise use. Aquat. Conserv. Mar. Freshw. Ecosyst. 2012, 22, 139–143. [Google Scholar] [CrossRef]
- Davidson, N. How much wetland has the world lost? Long-term and recent trends in global wetland area. Mar. Freshw. Res. 2014, 65, 936–941. [Google Scholar] [CrossRef]
- Hu, S.; Niu, Z.; Chen, Y.; Li, L.; Zhang, H. Global wetlands: Potential distribution, wetland loss, and status. Sci. Total Environ. 2017, 586, 319–327. [Google Scholar] [CrossRef]
- Dorador, C.; Vila, I.; Imhoff, J.F.; Witzel, K.P. Cyanobacterial diversity in Salar de Huasco, a high altitude saline wetland in northern Chile: An example of geographical dispersion? FEMS Microbiol. Ecol. 2008, 64, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Albarracín, V.H.; Kurth, D.; Ordoñez, O.F.; Belfiore, C.; Luccini, E.; Salum, G.M.; Piacentini, R.D.; Farías, M.E. High-up: A remote reservoir of microbial extremophiles in central Andean Wetlands. Front. Microbiol. 2015, 6, 1404. [Google Scholar] [CrossRef] [PubMed]
- Dorador, C.; Vila, I.; Witzel, K.-P.; Imhoff, J.F. Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fundam. Appl. Limnol. 2013, 182, 135–159. [Google Scholar] [CrossRef]
- Molina, V.; Hernández, K.; Dorador, C.; Eissler, Y.; Hengst, M.; Pérez, V.; Harrod, C. Bacterial Active Community Cycling in Response to Solar Radiation and Their Influence on Nutrient Changes in a High-Altitude Wetland. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Molina, V.; Dorador, C.; Fernández, C.; Bristow, L.; Eissler, Y.; Hengst, M.; Hernandez, K.; Olsen, L.M.; Harrod, C.; Marchant, F.; et al. The activity of nitrifying microorganisms in a high-altitude Andean wetland. FEMS Microbiol. Ecol. 2018, 94, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aceituno, P. Aspectos generales del clima en el Altiplano Sudamericano. In El Altiplano: Ciencia y Conciencia de los Andes, Actas del 21 Simposio Internacional de Estudios Altiplánicos; Charrier, R., Aceituno, P., Castro, M., Llanos, A., Raggi, L.A., Eds.; Universidad de Chile: Santiago, Chile, 1997; Volume 21, pp. 63–69. [Google Scholar]
- Risacher, F.; Alonso, H.; Salazar, C. The origin of brines and salts in Chilean salars: A hydrochemical review. Earth Sci. Rev. 2003, 63, 249–293. [Google Scholar] [CrossRef]
- Aguilar, P.; Acosta, E.; Dorador, C.; Sommaruga, R. Large differences in bacterial community composition among three nearby extreme waterbodies of the high Andean plateau. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Eissler, Y.; Gálvez, M.J.; Dorador, C.; Hengst, M.; Molina, V. Active microbiome structure and its association with environmental factors and viruses at different aquatic sites of a high-altitude wetland. Microbiologyopen 2019, 8, 1–13. [Google Scholar] [CrossRef]
- Dorador, C.; Vila, I.; Remonsellez, F.; Imhoff, J.F.; Witzel, K.P. Unique clusters of Archaea in Salar de Huasco, an athalassohaline evaporitic basin of the Chilean Altiplano. FEMS Microbiol. Ecol. 2010, 73, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef]
- Hurwitz, B.L.; Hallam, S.J.; Sullivan, M.B. Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol. 2013, 14, R123. [Google Scholar] [CrossRef]
- Anantharaman, K.; Duhaime, M.B.; Breier, J.A.; Wendt, K.A.; Toner, B.M.; Dick, G.J. Sulfur Oxidation Genes in Diverse Deep-Sea Viruses. Science (80-). 2014, 344, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, N.A.; Fuchsman, C.A.; Rocap, G.; Fuhrman, J.A. Discovery of several novel, widespread, and ecologically distinct marine Thaumarchaeota viruses that encode amoC nitrification genes. ISME J. 2019, 13, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Castelán-Sánchez, H.G.; Lopéz-Rosas, I.; García-Suastegui, W.A.; Peralta, R.; Dobson, A.D.W.; Batista-García, R.A.; Dávila-Ramos, S. Extremophile deep-sea viral communities from hydrothermal vents: Structural and functional analysis. Mar. Genom. 2019, 46, 16–28. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef]
- Parikka, K.J.; Le Romancer, M.; Wauters, N.; Jacquet, S. Deciphering the virus-to-prokaryote ratio (VPR): Insights into virus–host relationships in a variety of ecosystems. Biol. Rev. 2017, 92, 1081–1100. [Google Scholar] [CrossRef]
- Wigington, C.H.; Sonderegger, D.; Brussaard, C.P.D.; Buchan, A.; Finke, J.F.; Fuhrman, J.A.; Lennon, J.T.; Middelboe, M.; Suttle, C.A.; Stock, C.; et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 2016, 1. [Google Scholar] [CrossRef]
- Jackson, E.F.; Jackson, C.R. Viruses in wetland ecosystems. Freshw. Biol. 2008, 53, 1214–1227. [Google Scholar] [CrossRef]
- De Cárcer, D.A.; López-Bueno, A.; Pearce, D.A.; Alcamí, A. Biodiversity and distribution of polar freshwater DNA viruses. Sci. Adv. 2015, 1. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Robin, A.; Ravet, V.; Personnic, S.; Theil, S.; Colombet, J.; Sime-Ngando, T.; Debroas, D. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS ONE 2012, 7, e33641. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Sun, C.; Li, S.; Cui, Y.S.; Wei, Y.L.; Ji, X.L. Genetic Diversity of Bacteriophage Communities in Napahai Wetland. In Proceedings of the International Conference on Industrial Electronics and Applications (IEA 2015), Paris, France, 27–28 June 2015. [Google Scholar] [CrossRef]
- Xiang, Y.; Wang, S.; Li, J.; Wei, Y.; Zhang, Q.; Lin, L.; Ji, X. Isolation and characterization of two lytic cold-active bacteriophages infecting Pseudomonas fluorescens from the Napahai plateau wetland. Can. J. Microbiol. 2017, 64, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Ji, X.; Zhang, C.; Ding, Y.; Kuang, A.; Zhang, S.; Zhang, Q.; Lin, L.; Wei, Y. Isolation and characterization of wetland VSW-3, a novel lytic cold-active bacteriophage of Pseudomonas fluorescens. Can. J. Microbiol. 2016, 63, 110–118. [Google Scholar] [CrossRef]
- Roux, S.; Hallam, S.J.; Woyke, T.; Sullivan, M.B. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. Elife 2015, 4, 1–20. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of virome function from genomic sequences. bioRxiv 2019, 855387. [Google Scholar] [CrossRef]
- Atlas, E.L.; Gordon, L.I.; Hager, S.W.; Park, P.K. A Practical Manual for Use of the Technicon AutoAnalyzer in Seawater Nutrient Analyses (Revised); Tech. Rep. 215; Department of Oceanography, School of Science, Oregon State University: Corvallis, OR, USA, 1971. [Google Scholar]
- Chen, F.; Lu, J.-R.; Binder, B.J.; Liu, Y.-C.; Hodson, R.E. Application of Digital Image Analysis and Flow Cytometry To Enumerate Marine Viruses Stained with SYBR Gold Application of Digital Image Analysis and Flow Cytometry To Enumerate Marine Viruses Stained with SYBR Gold. Appl. Environ. Microbiol. 2001, 67, 539–545. [Google Scholar] [CrossRef]
- Wommack, K.E.; Sime-Ngando, T.; Winget, D.M.; Jamindar, S.; Helton, R.R. Filtration-based methods for the collection of viral concentrates from large water samples. In Manual of Aquatic Viral Ecology; American Society of Limnology and Oceanography: Waco, TX, USA, 2010; pp. 110–117. ISBN 9780984559107. [Google Scholar]
- Colombet, J.; Robin, A.; Lavie, L.; Bettarel, Y.; Cauchie, H.M.; Sime-Ngando, T. Virioplankton “pegylation”: Use of PEG (polyethylene glycol) to concentrate and purify viruses in pelagic ecosystems. J. Microbiol. Methods 2007, 71, 212–219. [Google Scholar] [CrossRef]
- Lasken, R.S. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem. Soc. Trans. 2009, 37, 450–453. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Bowtie2. Nat. Methods 2013, 9, 357–359. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.5-4; R Development Core Team: Vienna, Austria, 2019. [Google Scholar]
- Cowardin, L.M.; Carter, V.; Golet, F.C.; LaRoe, E.T. Classification of Wetlands and Deepwater Habitats of the United States; U.S. Department of the Interior Fish and Wildlife Service Office of Biological Services: Washington, DC, USA, 1979. [Google Scholar]
- Molina, V.; Eissler, Y.; Cornejo, M.; Galand, P.E.; Dorador, C.; Hengst, M.; Fernandez, C.; Francois, J.P. Distribution of greenhouse gases in hyper-arid and arid areas of northern Chile and the contribution of the high altitude wetland microbiome (Salar de Huasco, Chile). Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2018, 111, 1421–1432. [Google Scholar] [CrossRef]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef]
- Clasen, J.L.; Brigden, S.M.; Payet, J.P.; Suttle, C.A. Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshw. Biol. 2008, 53, 1090–1100. [Google Scholar] [CrossRef]
- Hernández, K.L.; Yannicelli, B.; Olsen, L.M.; Dorador, C.; Menschel, E.J.; Molina, V.; Remonsellez, F.; Hengst, M.B.; Jeffrey, W.H. Microbial activity response to solar radiation across contrasting environmental conditions in Salar de Huasco, northern Chilean altiplano. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Sommaruga, R.; Obernosterer, I.; Herndl, G.J.; Psenner, R. Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Appl. Environ. Microbiol. 1997, 63, 4178–4184. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Bertilsson, S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 2004, 6, 1228–1243. [Google Scholar] [CrossRef]
- Sarmento, H.; Casamayor, E.O.; Auguet, J.-C.; Vila-Costa, M.; Felip, M.; Camarero, L.; Gasol, J.M. Microbial food web components, bulk metabolism, and single-cell physiology of piconeuston in surface microlayers of high-altitude lakes. Front. Microbiol. 2015, 6, 361. [Google Scholar] [CrossRef] [PubMed]
- Lasken, R.S.; Stockwell, T.B. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007, 7, 1–11. [Google Scholar] [CrossRef]
- Bruder, K.; Malki, K.; Cooper, A.; Sible, E.; Shapiro, J.W.; Watkins, S.C.; Putonti, C. Freshwater metaviromics and bacteriophages: A current assessment of the state of the art in relation to Bioinformatic challenges. Evol. Bioinforma. 2016, 12, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, M.K.; Atanasova, N.S.; Oksanen, H.M.; Bamford, D.H. Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environ. Microbiol. 2013, 15, 1674–1686. [Google Scholar] [CrossRef]
- Skvortsov, T.; De Leeuwe, C.; Quinn, J.P.; McGrath, J.W.; Allen, C.C.R.; McElarney, Y.; Watson, C.; Arkhipova, K.; Lavigne, R.; Kulakov, L.A. Metagenomic characterisation of the viral community of lough neagh, the largest freshwater lake in Ireland. PLoS ONE 2016, 11, e150361. [Google Scholar] [CrossRef]
- Menouni, R.; Hutinet, G.; Petit, M.A.; Ansaldi, M. Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiol. Lett. 2015, 362, 1–10. [Google Scholar] [CrossRef]
- Prigozhin, D.M.; Mavrici, D.; Huizar, J.P.; Vansell, H.J.; Alber, T. Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-l-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling. J. Biol. Chem. 2013, 288, 31549–31555. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, S.; Kumar, D.; Mishra, A.; Dewangan, R.P.; Shrivastava, P.; Ramachandran, S.; Taneja, B. The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis. Acta Crystallogr. Sect. D 2013, 69, 2543–2554. [Google Scholar] [CrossRef] [PubMed]
- Donovan, D.M.; Foster-Frey, J.; Dong, S.; Rousseau, G.M.; Moineau, S.; Pritchard, D.G. The Cell Lysis Activity of the Streptococcus agalactiae Bacteriophage B30 Endolysin relies on the Cysteine, Histidine-Dependent Amidohydrolase/Peptidase Domain. Appl. Environ. Microbiol. 2006, 72, 5108–5112. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.H.; Mann, N.H. Lysogenic and lytic viral production in marine microbial communities. Aquat. Microb. Ecol. 1997, 13, 95–100. [Google Scholar] [CrossRef]
- Pradeep Ram, A.S.; Sime-Ngando, T. Resources drive trade-off between viral lifestyles in the plankton: Evidence from freshwater microbial microcosms. Environ. Microbiol. 2010, 12, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Berdjeb, L.; Sime-Ngando, T.; Jacquet, S. Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environ. Microbiol. 2011, 13, 616–630. [Google Scholar] [CrossRef]
- Carlson, R.E. A trophic state index for lakes. Limnol. Oceanogr. 1977, 22, 361–369. [Google Scholar] [CrossRef]
- Sullivan, M.B.; Coleman, M.L.; Weigele, P.; Rohwer, F.; Chisholm, S.W. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005, 3, e144. [Google Scholar] [CrossRef]
- Breitbart, M. Marine viruses: Truth or dare. Ann. Rev. Mar. Sci. 2012, 4. [Google Scholar] [CrossRef]
- Zimmerman, A.E.; Howard-Varona, C.; Needham, D.M.; John, S.G.; Worden, A.Z.; Sullivan, M.B.; Waldbauer, J.R.; Coleman, M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020, 18, 21–34. [Google Scholar] [CrossRef]
- Chen, Y.; Golding, I.; Sawai, S.; Guo, L.; Cox, E.C. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005, 3, e229. [Google Scholar] [CrossRef]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, G.; Trevathan-Tackett, S.M.; Carnell, P.E.; Macreadie, P.I. Implication of viral infections for greenhouse gas dynamics in freshwater wetlands: Challenges and perspectives. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
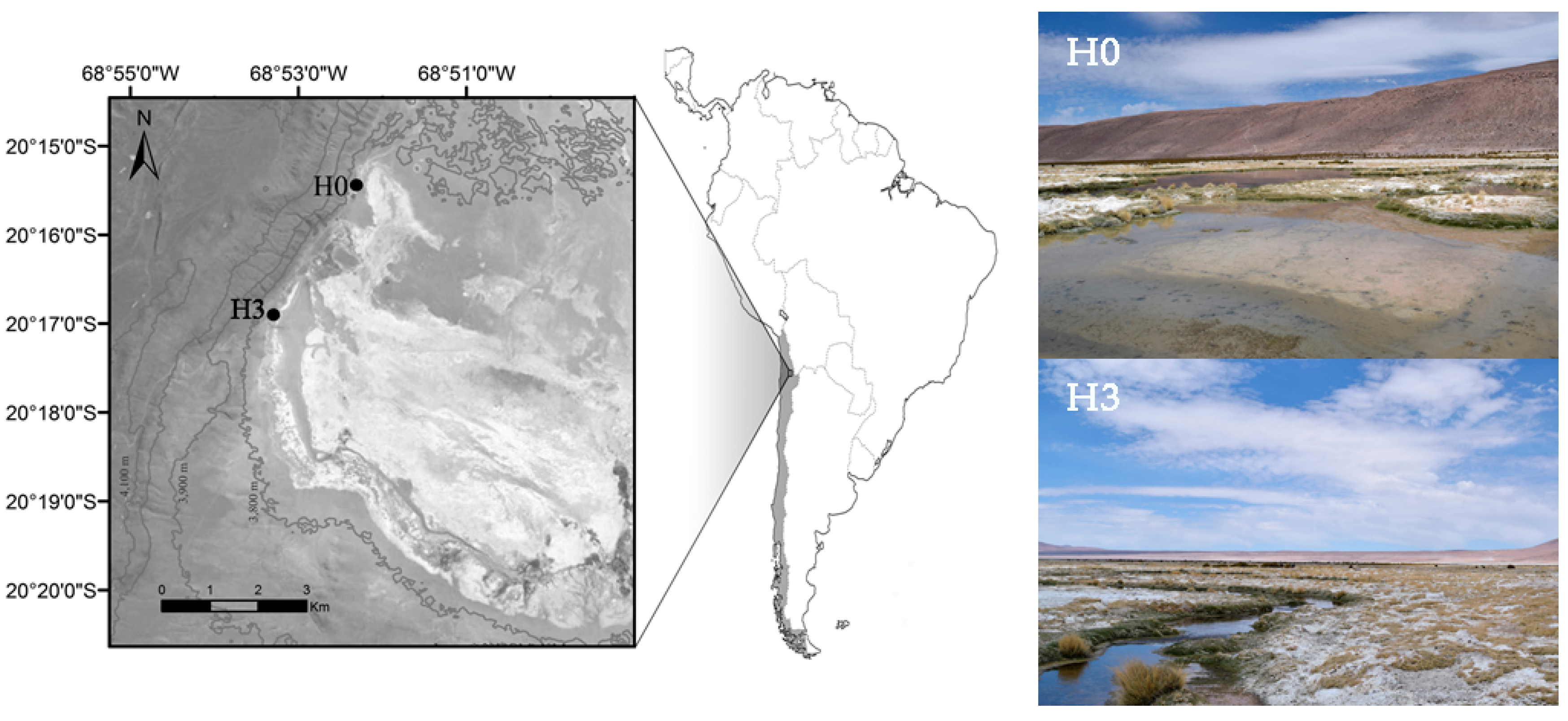
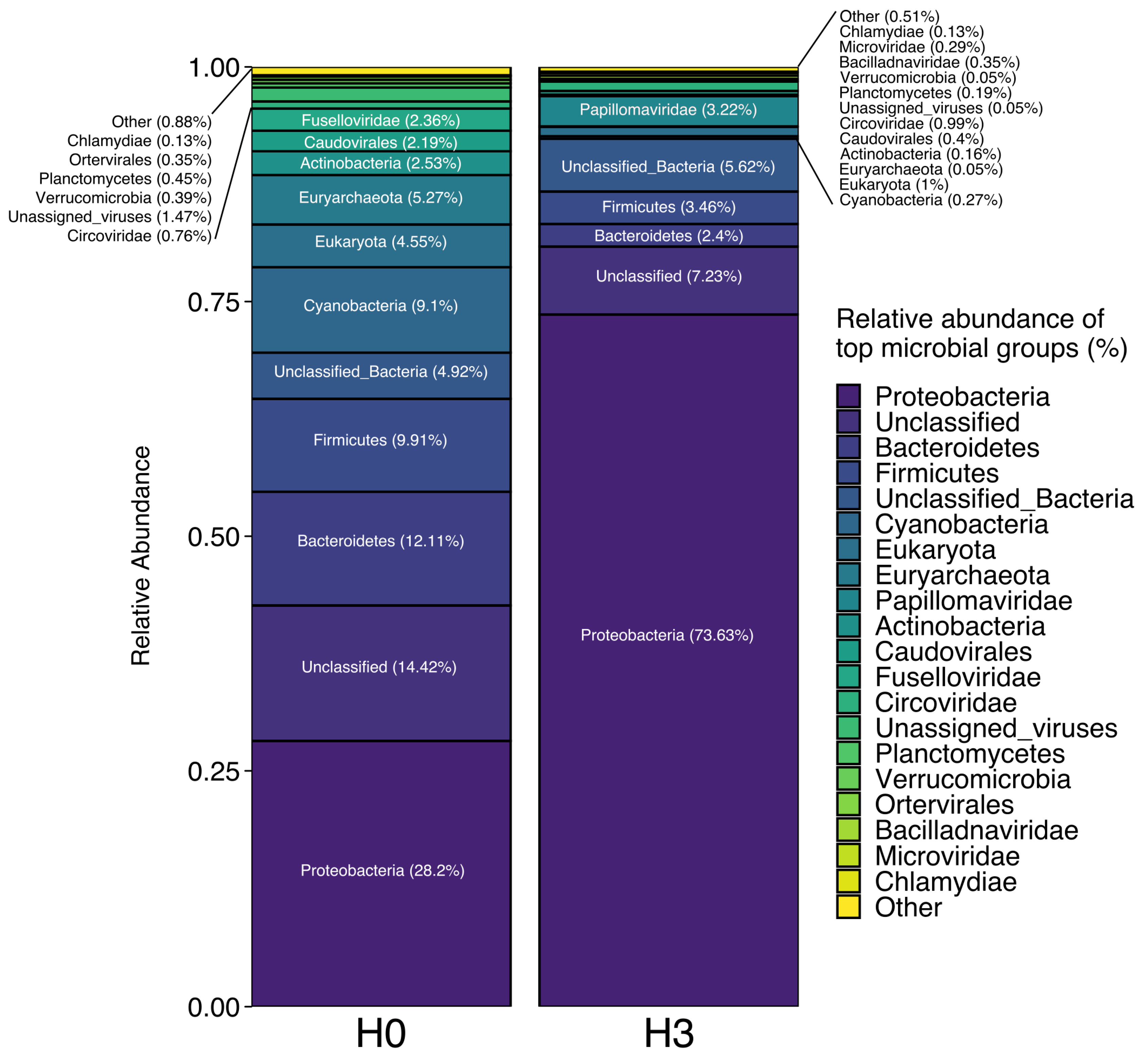
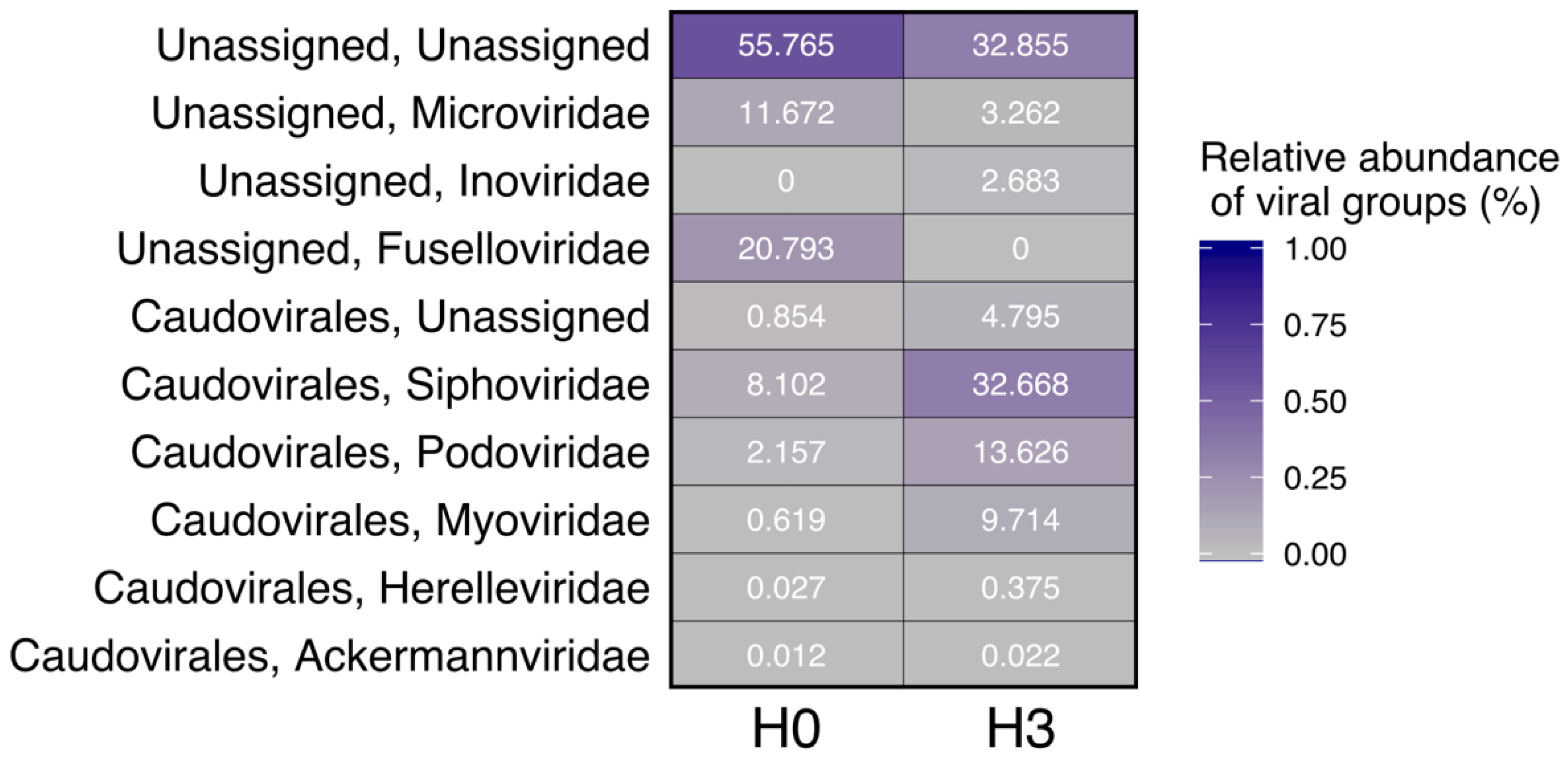
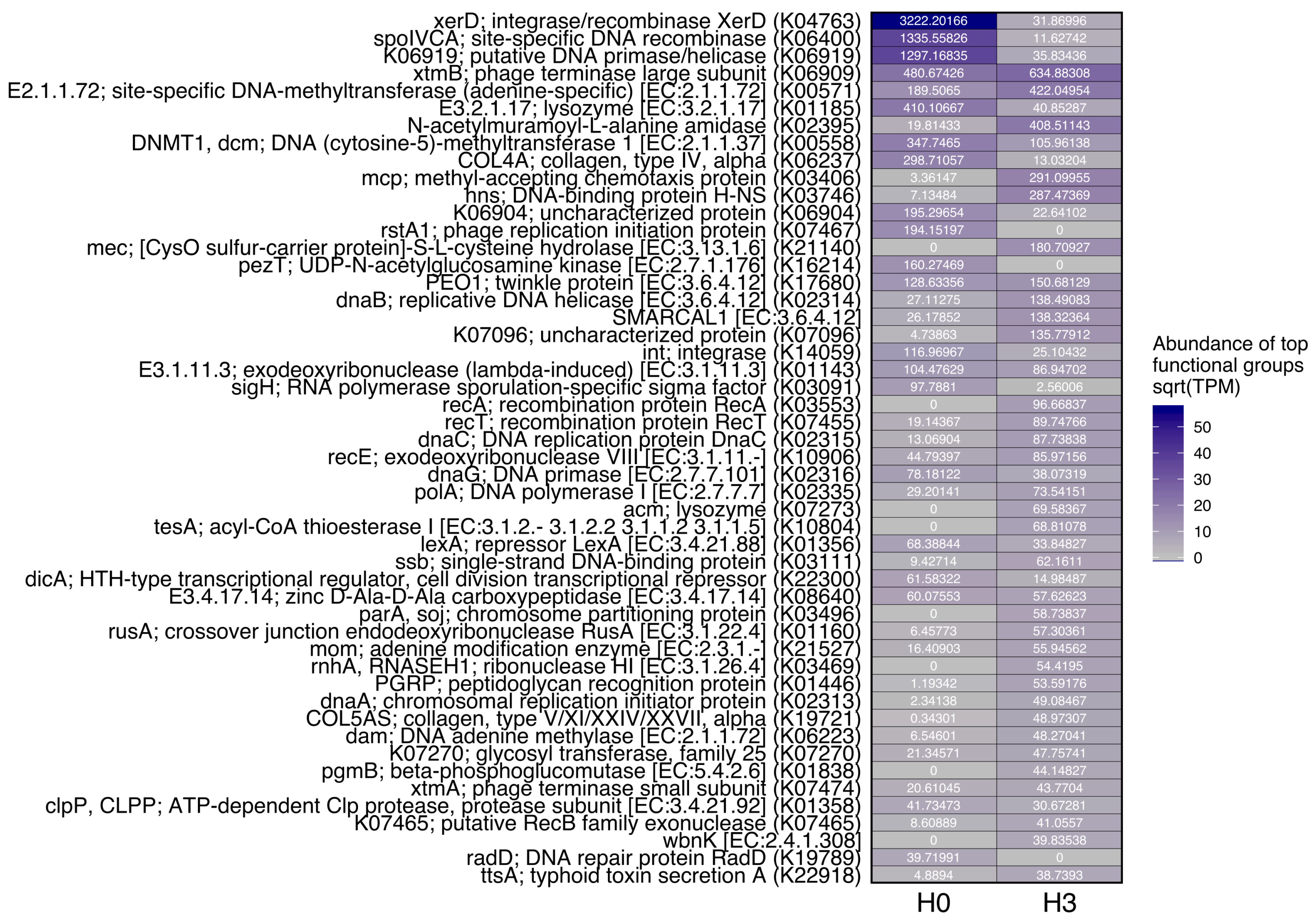
| Site | Conductivity (µS cm−1) | Water Temperature (°C) | pH | Dissolved Oxygen (mg L−1) | Phosphate (mg L−1) | Virus (VLP mL−1) | Picoplankton (cells mL−1) | VPR |
|---|---|---|---|---|---|---|---|---|
| H0 | 566.3 | 19.3 | 8.87 | 14.56 | 0.081 | 2.5 × 106 | 2.1 × 105 | 12.1 |
| H3 | 580.7 | 17.6 | 8.68 | 8.32 | 0.090 | 1.3 × 106 | 5.3 × 105 | 2.3 |
| Site | Observed | Shannon | Chao1 | Evenness |
|---|---|---|---|---|
| H0 | 2209 | 4.381 | 2346.1 | 0.00198 |
| H3 | 1905 | 3.289 | 1918.5 | 0.00173 |
| Site | Node and Category | Size (bp) 1 | Genome Structure | Features | Virus Group | Putative Host | VIBRANT ID; Viral Gene TPM |
|---|---|---|---|---|---|---|---|
| H0 | 278 and 1 | 8640 (3748) | linear | phage tail protein, phage tail tape measure protein, phage tail assembly protein, phage major tail tube protein | Caudovirales, Myoviridae, Serratia phage KSP20, 83.37% identity | Proteobacteria, Gammaproteo-bacteria (Enterobacteriaceae) | Peduovirinae; 28.39 |
| H0 | 332, 581, 2647 and 1 | 8085 (3895), 6031 (2339), 2399 (1105) | linear | several hypothetical proteins | Myovirus (Uncultured Caudovirales phage clone similar to Pseudomonas phage), 72–81.45% identity | Proteobacteria, Gammaproteo-bacteria (Pseudomonas sp.) | Peduovirinae; 12.07 Peduovirinae; 32.26 Siphoviridae; 6.68 |
| H0 | 5894 and 1 | 1425 (414, 268) | linear | terminase, portal protein, terminase large subunit | Siphoviridae (Arthobacter phage Gordon and Streptomyces phage), 69.74% and 67.52% identity | Actinobacteria | Too short; NA |
| H3 | 114 and 1 | 15881 (6652) | linear | tail fiber protein, tail length tape-measure protein, tail assembly protein K and I, tail attachment protein, tail protein, tail terminator protein, lipoprotein | Caudovirales; Siphoviridae, Pseudomonas phage phiAH14a, complete genome, 85% identity | Proteobacteria, Gammaproteo-bacteria (Pseudomonas sp.) | Siphoviridae; 1845.42 |
| H3 | 1079 and 1 | 4431 (226) | circular | major capsid protein | O. Caudovirales, Fam Microviridae, 68.92% identity | Proteobacteria | Microviridae; 247.37 |
| H3 | 1155 and 1 | 4188 (1772) | circular | terminase, head morphogenesis protein | Myovirus (Uncultured Caudovirales phage clone), 76% identity | Proteobacteria, Gammaproteo-bacteria (Pseudomonas sp.) | Siphoviridae; 22.545 |
| H3 | 300 and 2 | 10081 (10275) | linear | phage major tail sheath protein, phage major tail tube protein, phage tail protein D and E | O. Caudovirales; Fam Myoviridae; Sub Fam Peduovirinae; Peduovirus; unclassified Peduovirus, (Burkholderia phage ST79) 92% identity | Proteobacteria, Betaproteobacteria | Peduovirinae; 49.34 |
| H3 | 449 and 2 | 8201 (7268) | linear | exonuclease, DNA primase | O. Caudovirales; Siphoviridae; Pahexavirus; unclassified Pahexavirus, Propionibacterium phage pa28, complete genome, 92% identity | Actinobacteria | Siphoviridae; 107.52 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eissler, Y.; Dorador, C.; Kieft, B.; Molina, V.; Hengst, M. Virus and Potential Host Microbes from Viral-Enriched Metagenomic Characterization in the High-Altitude Wetland, Salar de Huasco, Chile. Microorganisms 2020, 8, 1077. https://doi.org/10.3390/microorganisms8071077
Eissler Y, Dorador C, Kieft B, Molina V, Hengst M. Virus and Potential Host Microbes from Viral-Enriched Metagenomic Characterization in the High-Altitude Wetland, Salar de Huasco, Chile. Microorganisms. 2020; 8(7):1077. https://doi.org/10.3390/microorganisms8071077
Chicago/Turabian StyleEissler, Yoanna, Cristina Dorador, Brandon Kieft, Verónica Molina, and Martha Hengst. 2020. "Virus and Potential Host Microbes from Viral-Enriched Metagenomic Characterization in the High-Altitude Wetland, Salar de Huasco, Chile" Microorganisms 8, no. 7: 1077. https://doi.org/10.3390/microorganisms8071077
APA StyleEissler, Y., Dorador, C., Kieft, B., Molina, V., & Hengst, M. (2020). Virus and Potential Host Microbes from Viral-Enriched Metagenomic Characterization in the High-Altitude Wetland, Salar de Huasco, Chile. Microorganisms, 8(7), 1077. https://doi.org/10.3390/microorganisms8071077






