An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications
Abstract
:1. Introduction
2. Results
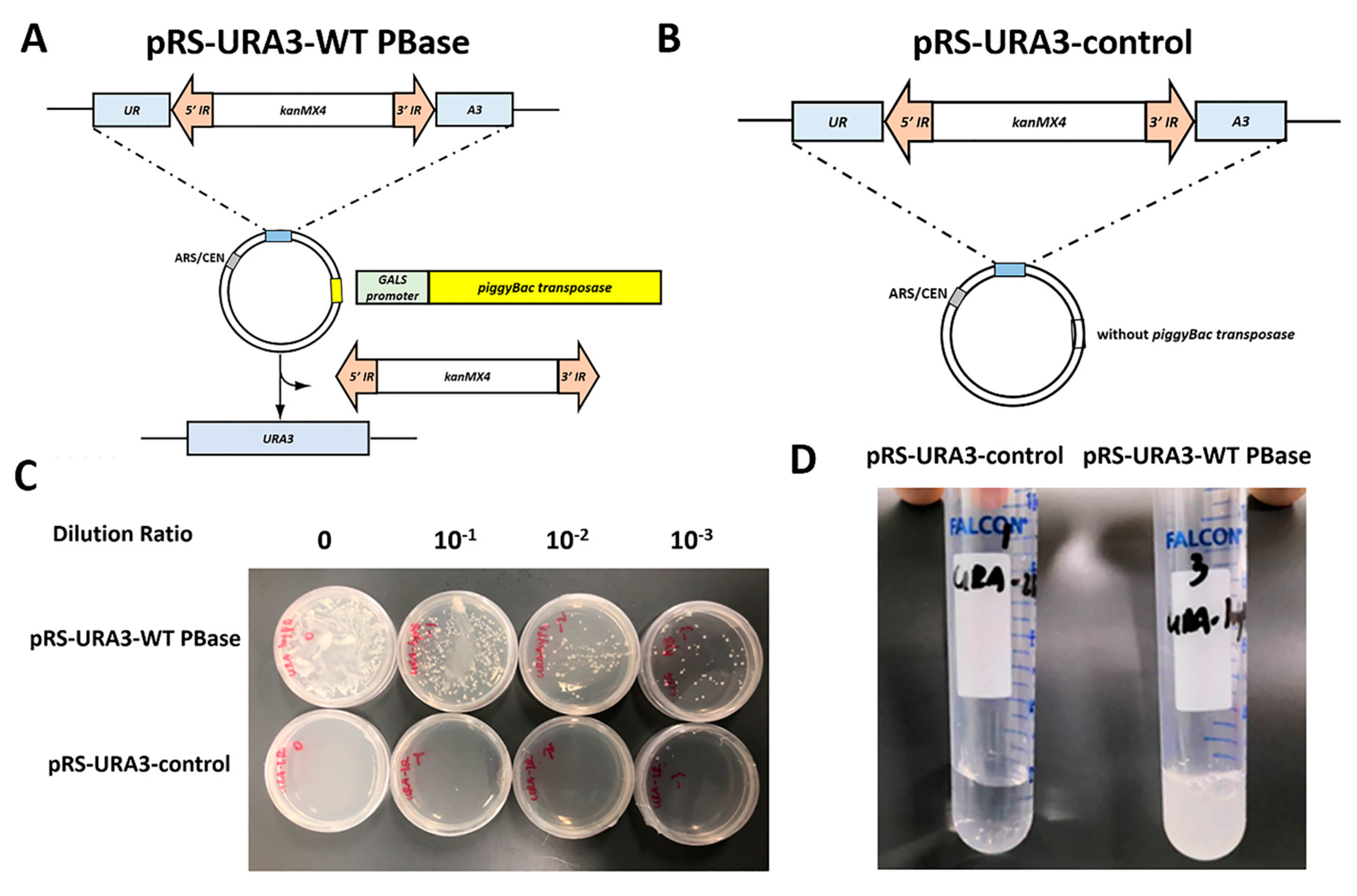
2.1. Construction of an Efficient Screening System in Yeast
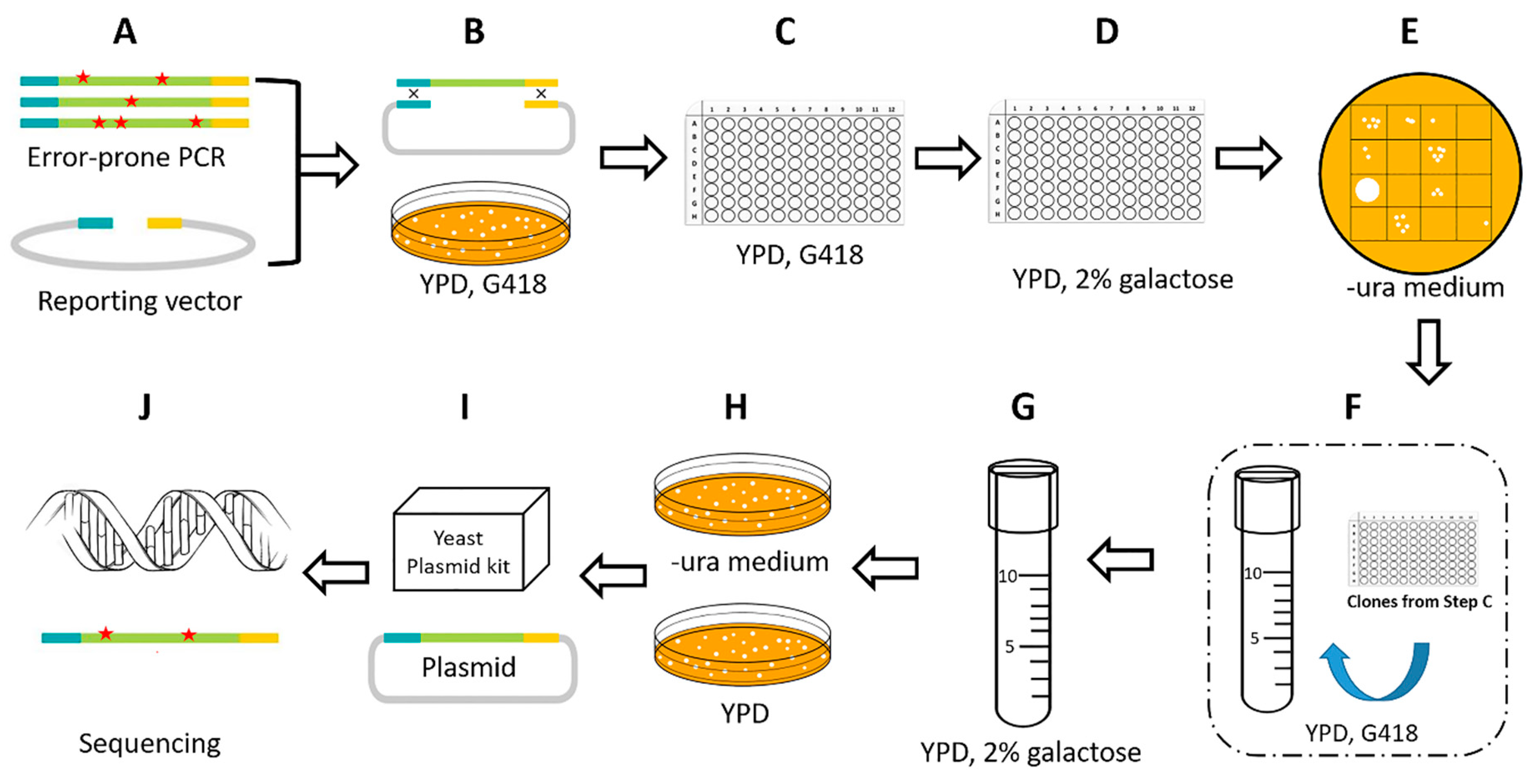
2.2. Improvement of a High-throughput Screening Process
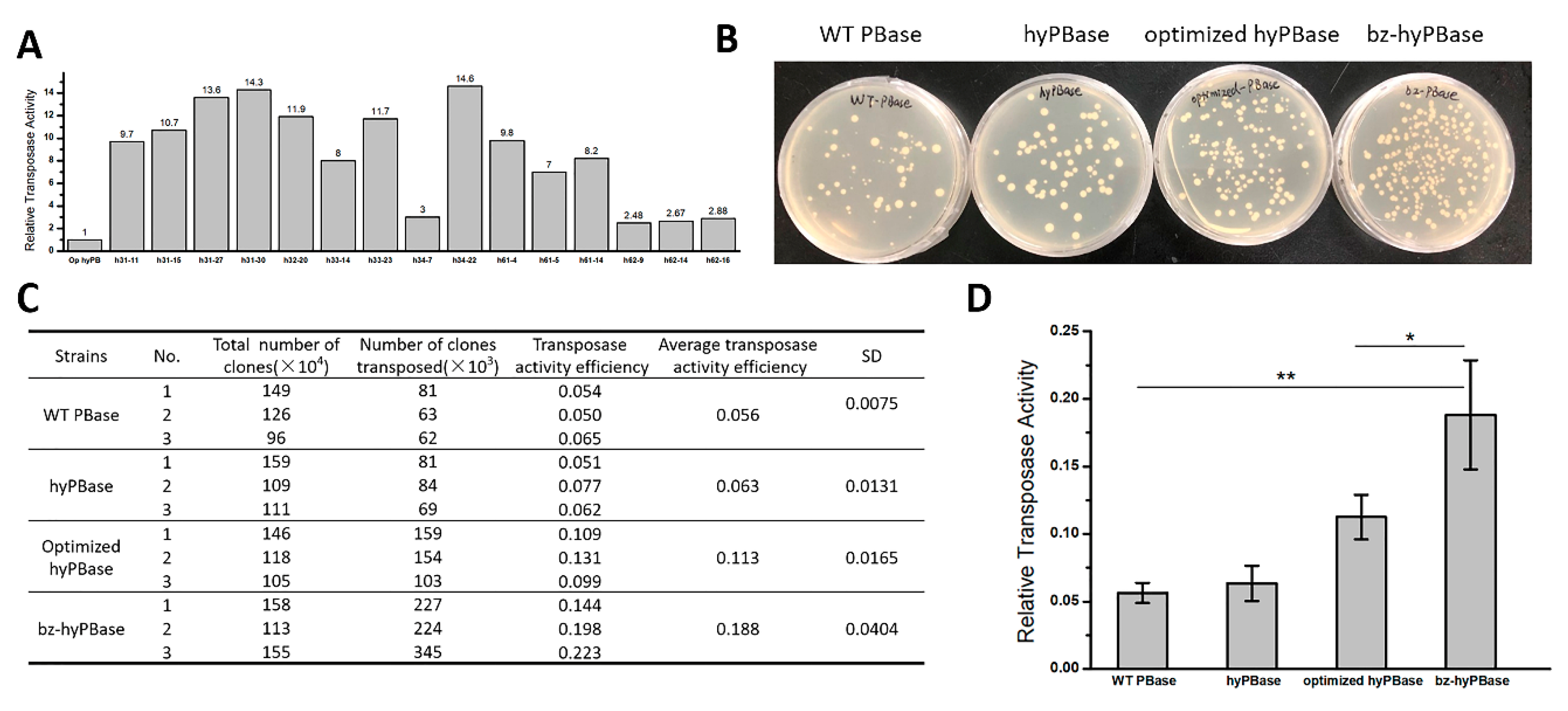
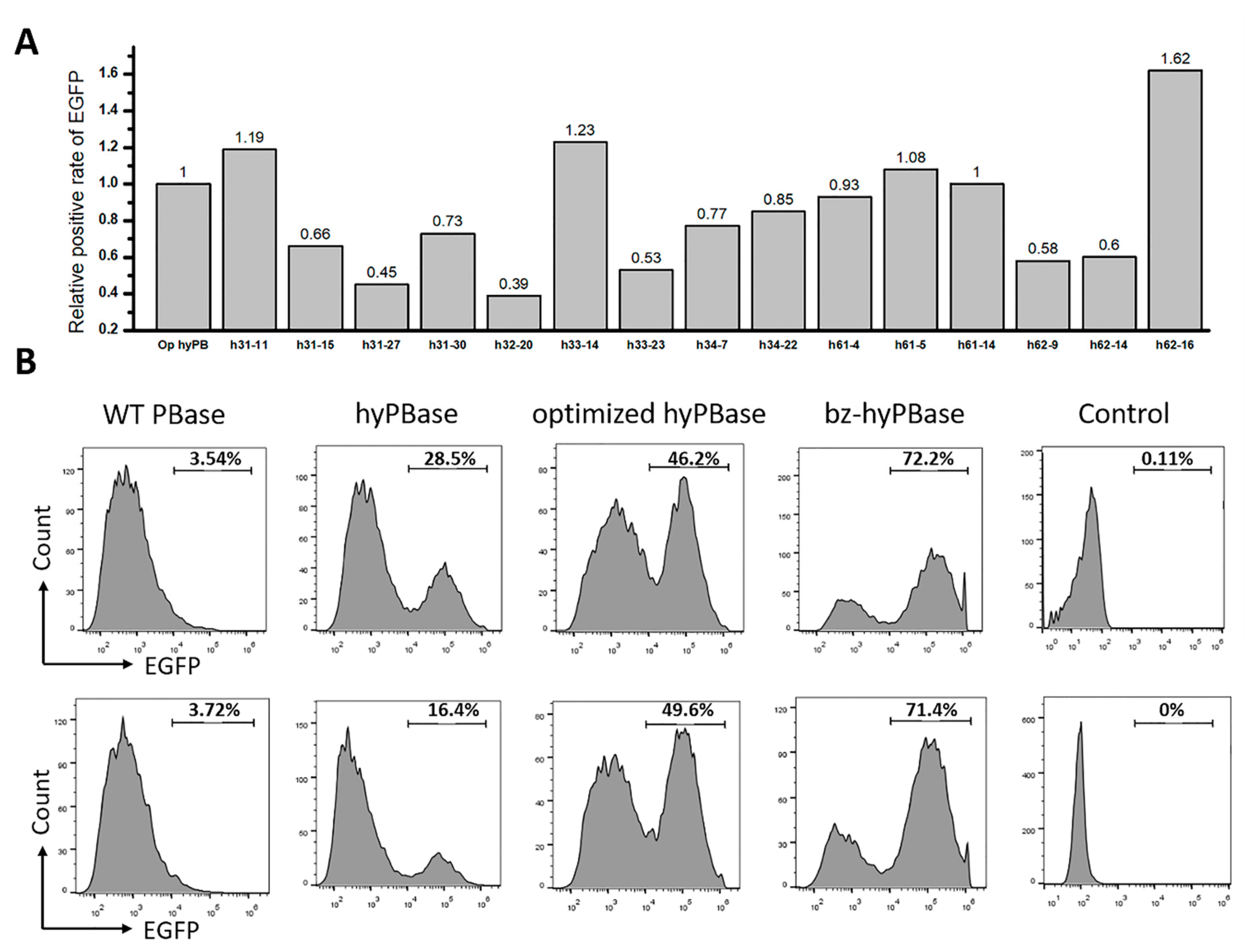
2.3. Isolation of Hyperactive piggyBac Transposase Mutants
2.4. Hyperactive Mutants in Mammalian Cells
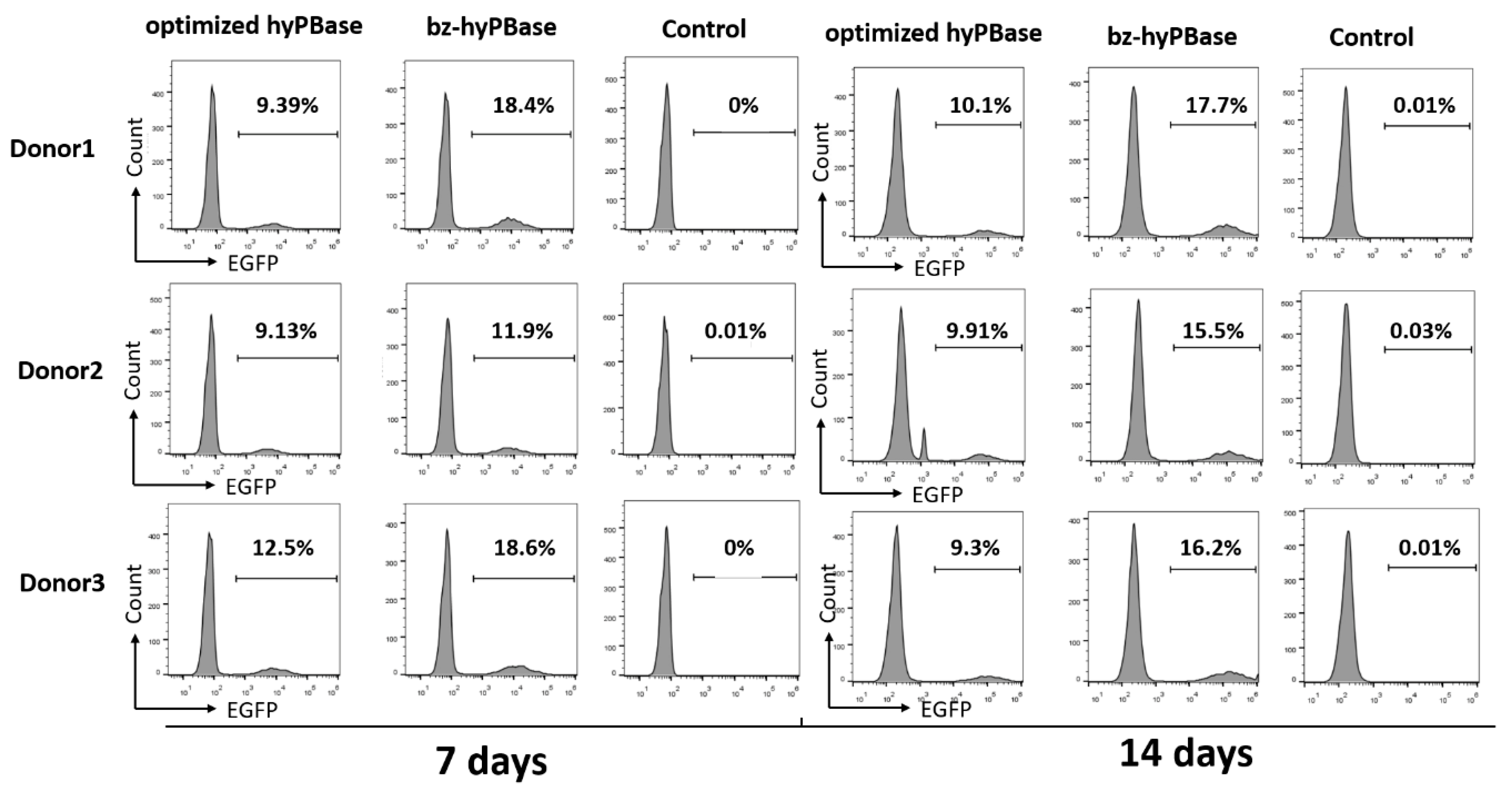
2.5. bz-hyPBase Improved the Editing Efficiency of T cells
2.6. The Effectiveness of the Screening System
3. Discussion
4. Materials and Methods
4.1. Strains, Growth Condition, and Cell Culture
4.2. Plasmid Construction
4.3. Mutant Library Construction
4.4. Screening of Yeast Mutants
4.5. Transposition Assay in Mammalian Cells
4.6. Flow Cytometry
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mcclintock, B. The Origin and Behavior of Mutable Loci in Maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, B.I.; Baker, B. Movers and shakers: Maize transposons as tools for analyzing other plant genomes. Curr. Opin. Cell Biol. 1995, 7, 406–413. [Google Scholar] [CrossRef]
- Spradling, A.C.; Stern, D.M.; Kiss, I.; Roote, J.; Laverty, T.; Rubin, G.M. Gene disruptions using P transposable elements: An integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 1995, 92, 10824–10830. [Google Scholar] [CrossRef] [Green Version]
- Moerman, D.G.; Benian, G.M.; Waterston, R.H. Molecular-Cloning of the Muscle Gene Unc-22 in Caenorhabditis-Elegans by Tc1 Transposon Tagging. Proc. Natl. Acad. Sci. USA 1986, 83, 2579–2583. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Sim, C.; Hong, Y.S.; Hogan, J.R.; Fraser, M.J.; Robertson, H.M.; Collins, F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genom. 2003, 270, 173–180. [Google Scholar] [CrossRef]
- Meir, Y.J.; Lin, A.; Huang, M.F.; Lin, J.R.; Weirauch, M.T.; Chou, H.C.; Lin, S.J.; Wu, S.C. A versatile, highly efficient, and potentially safer piggyBac transposon system for mammalian genome manipulations. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 4429–4443. [Google Scholar] [CrossRef]
- Cesana, D.; Volpin, M.; Secanechia, Y.N.S.; Montini, E. Safety and Efficacy of Retroviral and Lentiviral Vectors for Gene Therapy. In Safety and Efficacy of Gene-Based Therapeutics for Inherited Disorders; Springer International Publishing: Cham, Switzerland, 2017; pp. 9–35. [Google Scholar] [CrossRef]
- Vargas, J.E.; Chicaybam, L.; Stein, R.T.; Tanuri, A.; Delgado-Canedo, A.; Bonamino, M.H. Retroviral vectors and transposons for stable gene therapy: Advances, current challenges and perspectives. J. Transl. Med. 2016, 14. [Google Scholar] [CrossRef] [Green Version]
- Morgan, R.A.; Boyerinas, B. Genetic Modification of T Cells. Biomedicines 2016, 4, 9. [Google Scholar] [CrossRef]
- Wu, S.C.; Meir, Y.J.; Coates, C.J.; Handler, A.M.; Pelczar, P.; Moisyadi, S.; Kaminski, J.M. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. USA 2006, 103, 15008–15013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, L.C.; Goebel, M.; Corsaro, B.G.; Wang, H.G.; Rosen, E.; Fraser, M.J. Transposon Mutagenesis of Baculoviruses—Analysis of Trichoplusia-Ni Transposon Ifp2 Insertions within the Fp-Locus of Nuclear Polyhedrosis Viruses. Virology 1989, 172, 156–169. [Google Scholar] [CrossRef]
- Galvan, D.L.; Nakazawa, Y.; Kaja, A.; Kettlun, C.; Cooper, L.J.N.; Rooney, C.M.; Wilson, M.H. Genome-wide Mapping of PiggyBac Transposon Integrations in Primary Human T Cells. J. Immunother. 2009, 32, 837–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.B.; Zhuang, Y.; Han, M.; Xu, T.; Wu, X.H. piggyBac as a high-capacity transgenesis and gene-therapy vector in human cells and mice. Dis. Models Mech. 2013, 6, 828–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadinanos, J.; Bradley, A. Generation of an inducible and optimized piggyBac transposon systemy. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Guo, H.F.; Tammana, S.; Jung, Y.C.; Mellgren, E.; Bassi, P.; Cao, Q.; Tu, Z.J.; Kim, Y.C.; Ekker, S.C.; et al. Gene Transfer Efficiency and Genome-Wide Integration Profiling of Sleeping Beauty, Tol2, and PiggyBac Transposons in Human Primary T Cells (vol. 18, pg 1803, 2010). Mol. Ther. 2010, 18, 2038. [Google Scholar] [CrossRef]
- Wang, W.; Lin, C.; Lu, D.; Ning, Z.; Cox, T.; Melvin, D.; Wang, X.; Bradley, A.; Liu, P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 9290–9295. [Google Scholar] [CrossRef] [Green Version]
- Yusa, K. piggyBac Transposon. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Nakanishi, H.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. Comparison of piggyBac transposition efficiency between linear and circular donor vectors in mammalian cells. J. Biotechnol. 2011, 154, 205–208. [Google Scholar] [CrossRef]
- Woodard, L.E.; Wilson, M.H. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015, 33, 525–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mates, L.; Chuah, M.K.L.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.; Schmitt, A.; Becker, K.; Matrai, J.; et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009, 41, 753–761. [Google Scholar] [CrossRef]
- Querques, I.; Mades, A.; Zuliani, C.; Miskey, C.; Alb, M.; Grueso, E.; Machwirth, M.; Rausch, T.; Einsele, H.; Ivics, Z.; et al. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat. Biotechnol. 2019, 37, 1502–1512. [Google Scholar] [CrossRef]
- Yusa, K.; Zhou, L.; Li, M.A.; Bradley, A.; Craig, N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA 2011, 108, 1531–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, R.; Fain-Thornton, J.; Craig, N.L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008, 27, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchholz, F.; Angrand, P.O.; Stewart, A.F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998, 16, 657–662. [Google Scholar] [CrossRef]
- Yusa, K.; Takeda, J.; Horie, K. Enhancement of Sleeping Beauty transposition by CpG methylation: Possible role of heterochromatin formation. Mol. Cell. Biol. 2004, 24, 4004–4018. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, R.; Kokubu, C.; Yusa, K.; Keng, V.W.; Horie, K.; Takeda, J. Sleeping Beauty transposase has an affinity for heterochromatin conformation. Mol. Cell. Biol. 2007, 27, 1665–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; Wu, X.H.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggyBac resource (PB) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Doherty, J.E.; Huye, L.E.; Yusa, K.; Zhou, L.Q.; Craig, N.L.; Wilson, M.H. Hyperactive piggyBac Gene Transfer in Human Cells and In Vivo. Hum. Gene Ther. 2012, 23, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, Y.; Huye, L.E.; Dotti, G.; Foster, A.E.; Vera, J.F.; Manuri, P.R.; June, C.H.; Rooney, C.M.; Wilson, M.H. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J. Immunother. (Hagerstown MD 1997) 2009, 32, 826–836. [Google Scholar] [CrossRef] [Green Version]
- Manuri, P.V.R.; Wilson, M.H.; Maiti, S.N.; Mi, T.J.; Singh, H.; Olivares, S.; Dawson, M.J.; Huls, H.; Lee, D.A.; Rao, P.H.; et al. piggyBac Transposon/Transposase System to Generate CD19-Specific T Cells for the Treatment of B-Lineage Malignancies. Hum. Gene Ther. 2010, 21, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Hackett, P.B.; Largaespada, D.A.; Cooper, L.J.N. A Transposon and Transposase System for Human Application. Mol. Ther. 2010, 18, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Muller, R.; Funk, M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994, 22, 5767–5768. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, Y.; Jiang, D.Q.; Cui, L.Z.; He, Z.; Wang, C.; Zhang, Z.W.; Zhu, H.L.; Ding, Y.M.; Li, L.F.; et al. Antitumor activity of EGFR-specific CAR T cells against non-small-cell lung cancer cells in vitro and in mice. Cell Death Dis. 2018, 9, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Zhang, Z.; Lv, S.; Liu, X.; Cui, L.; Jiang, D.; Zhang, Q.; Li, L.; Qin, W.; Jin, H.; et al. Engineered CAR T cells targeting mesothelin by piggyBac transposon system for the treatment of pancreatic cancer. Cell. Immunol. 2018, 329, 31–40. [Google Scholar] [CrossRef]
| Primer | Primer Oligonucleotide (5′–3′) | Application |
| pURA-F | AAGCCGCTAAAGGCATTATCCGCC | Plasmid linearization |
| pURA-R | AACTGTGCCCTCCATGGAAAAATCAGTC | Plasmid linearization |
| pURA-IR-F | GACTGATTTTTCCATGGAGGGCACAGTTAACCCTAGAAAGATAGTCTGCGTAAAATTGACGCATGCGAC | Transposon insertion |
| pURA-IR-R | GGCGGATAATGCCTTTAGCGGCTTAACCCTAGAAAGATAATCATATTGTG | Transposon insertion |
| GR-F | TAATCAGCGAAGCGATGA | Error-prone PCR |
| GR-R | CAGCATGCCTGCTATTGTCTTCC | Error-prone PCR |
| GR-F1 | CCACTTTAACTAATACTTTC | Error-prone PCR |
| GR-R1 | CCCTCACTAAAGGGAACAAAAGCTG | Error-prone PCR |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, W.; Song, S.; Han, Y.; Chen, H.; Liu, X.; Qian, Q. An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications. Int. J. Mol. Sci. 2020, 21, 3064. https://doi.org/10.3390/ijms21093064
Wen W, Song S, Han Y, Chen H, Liu X, Qian Q. An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications. International Journal of Molecular Sciences. 2020; 21(9):3064. https://doi.org/10.3390/ijms21093064
Chicago/Turabian StyleWen, Wen, Shanshan Song, Yuchun Han, Haibin Chen, Xiangzhen Liu, and Qijun Qian. 2020. "An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications" International Journal of Molecular Sciences 21, no. 9: 3064. https://doi.org/10.3390/ijms21093064
APA StyleWen, W., Song, S., Han, Y., Chen, H., Liu, X., & Qian, Q. (2020). An efficient Screening System in Yeast to Select a Hyperactive piggyBac Transposase for Mammalian Applications. International Journal of Molecular Sciences, 21(9), 3064. https://doi.org/10.3390/ijms21093064





