ERK Dephosphorylation through MKP1 Deacetylation by SIRT1 Attenuates RAS-Driven Tumorigenesis
Abstract
:1. Introduction
2. Results
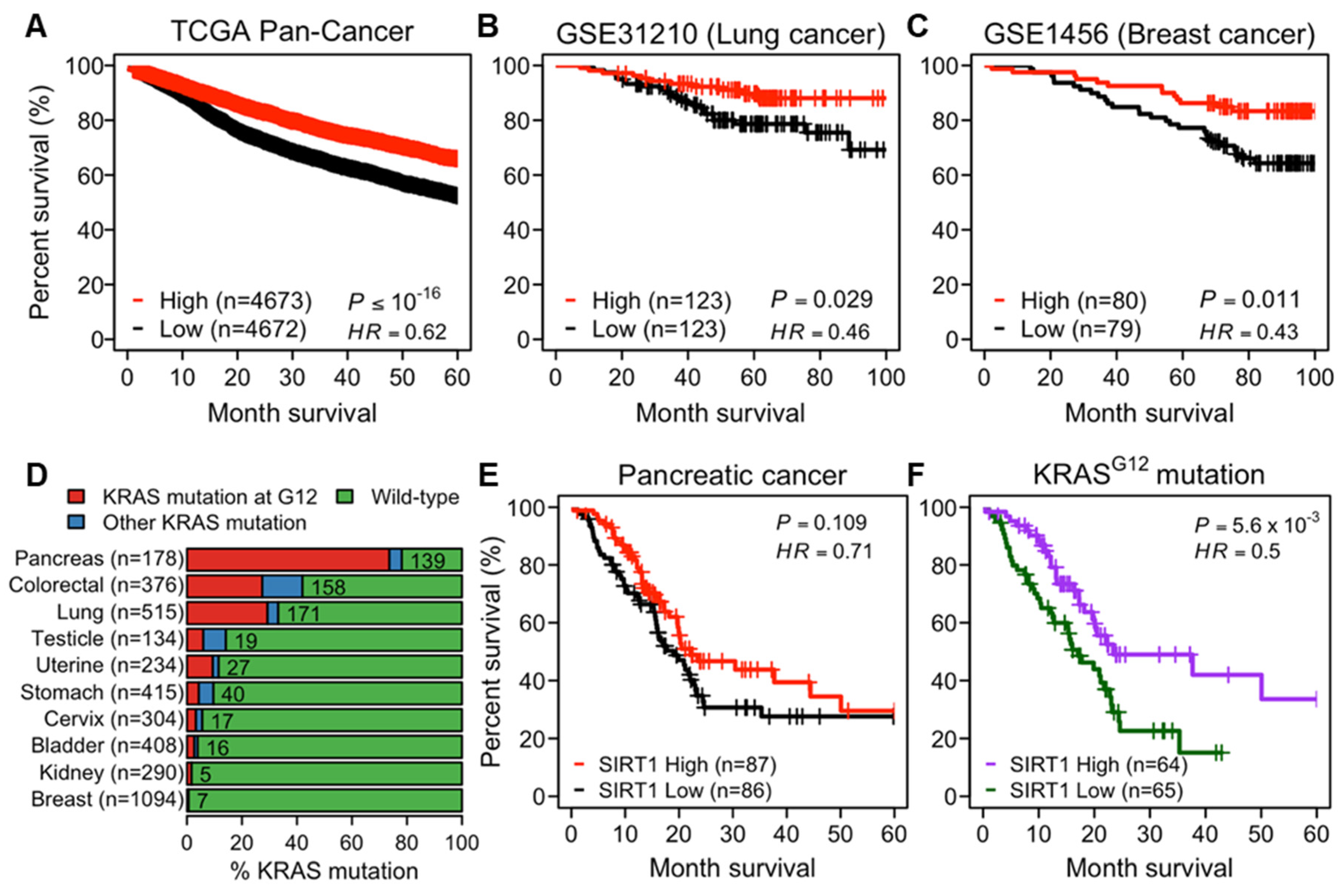
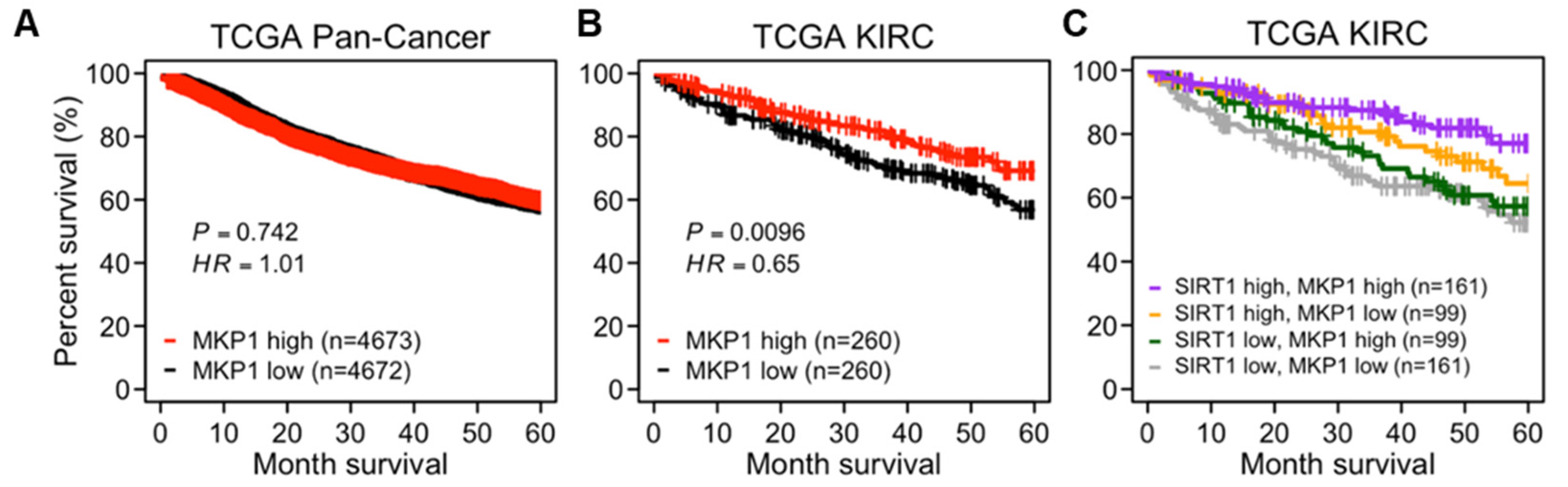
2.1. Higher Expression of SIRT1 Correlates to Better Prognosis in Human Cancers
2.2. SIRT1 Suppresses RAS-Driven Tumorigenic Activities In Vitro
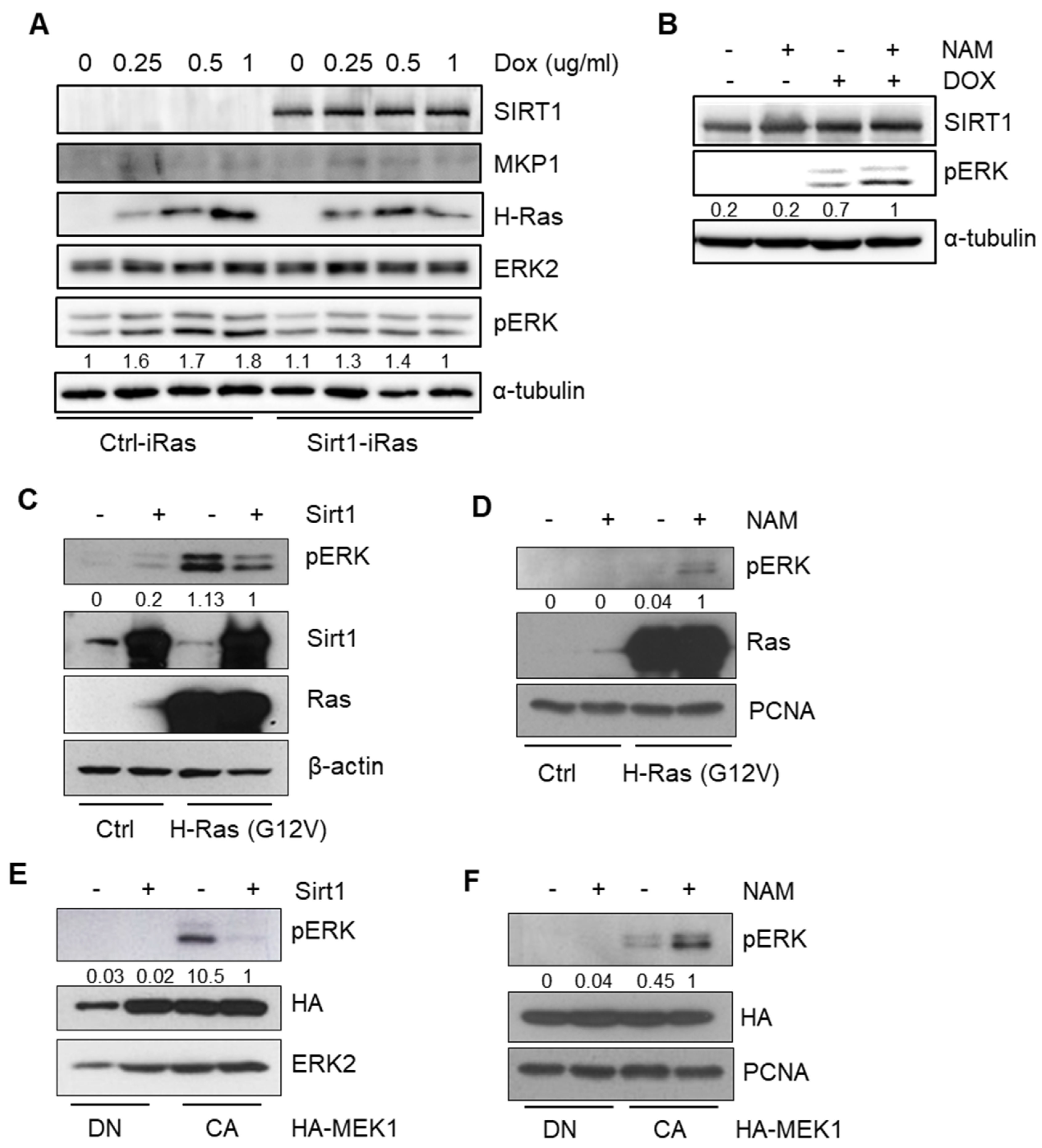
2.3. SIRT1 Inhibits RAS-MEK-ERK Axis via Dephosphorylation of ERK
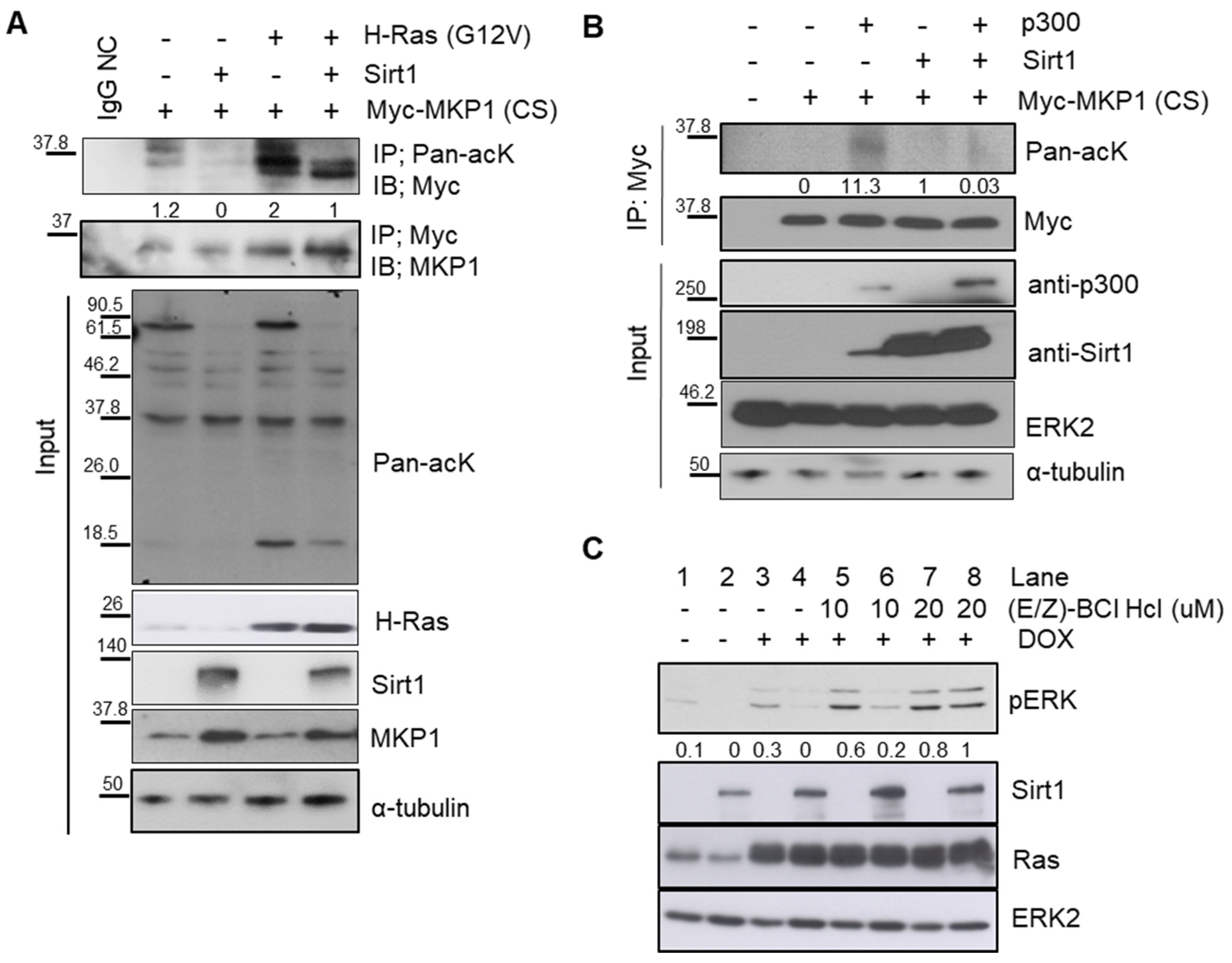
2.4. SIRT1 Dephosphorylates ERK through Deacetylation of MKP1
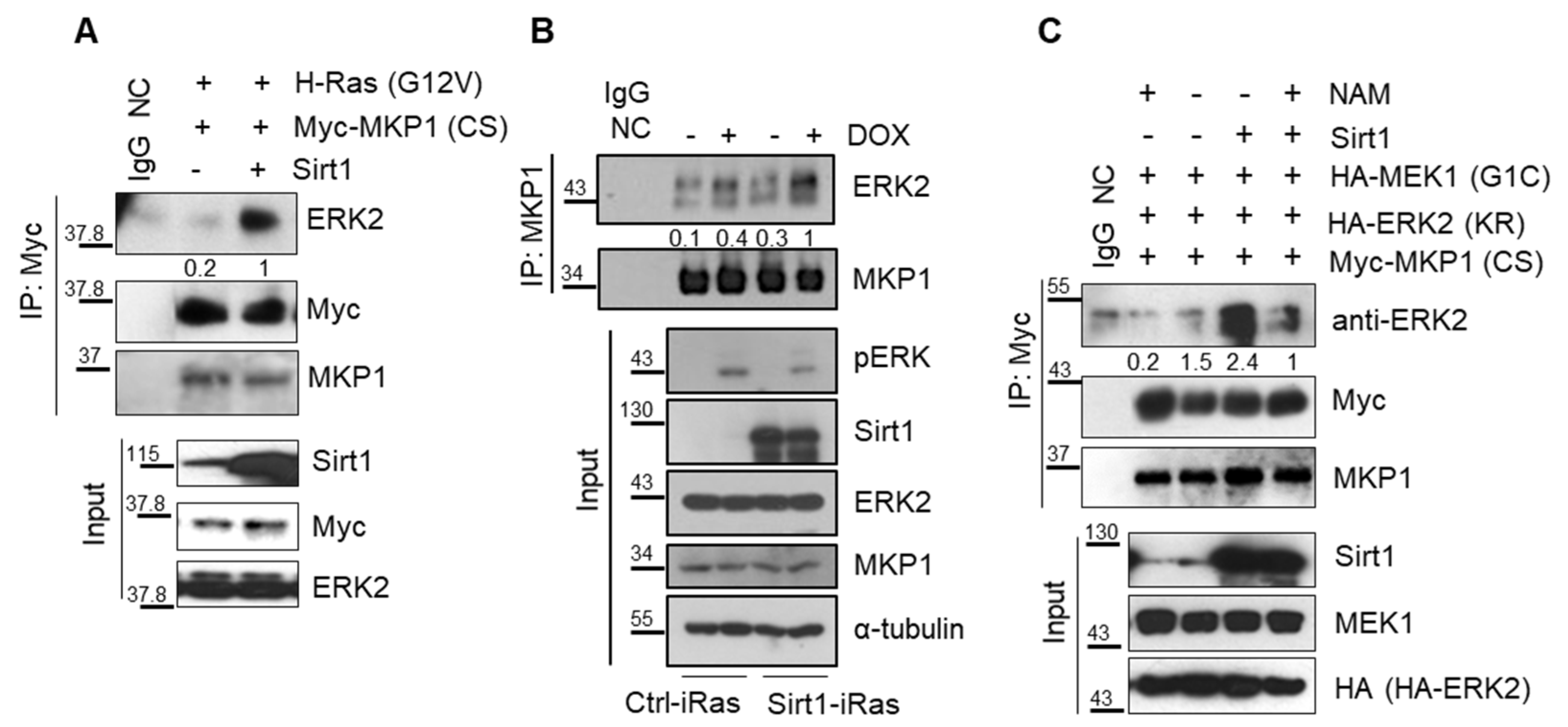
2.5. SIRT1 Promotes Interaction between MKP1 and ERK via Its Deacetylase Activity
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Establishment of Stable Cell Line
4.2. Reagents and Antibodies
4.3. Public Data Resources and Survival Analysis
4.4. Gene Set Enrichment Analysis (GSEA)
4.5. Transient Expression of Exogenous Protein
4.6. Cell Proliferation, Clonogenic Assat, and Soft-Agar Assay
4.7. RNA Preparation and Real-Time PCR
4.8. Immunoblotting and Immunoprecipitation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nemoto, S.; Fergusson, M.M.; Finkel, T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J. Biol. Chem. 2005, 280, 16456–16460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Gil, N.Y.; Zhang, X.H.; Chun, K.H.; Fang, G.; Kim, J.; Cho, H.; Jang, C.Y.; Cha, H.J. Sirt1 Regulates microtubule dynamics through negative regulation of Plk1 in mitosis. J. Cell. Biochem. 2015, 116, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.X. SIRT1, is it a tumor promoter or tumor suppressor? Int. J. Biol. Sci. 2009, 5, 147–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.Y.; Surh, Y.J. Janus-faced role of SIRT1 in tumorigenesis. Ann. N. Y. Acad. Sci. 2012, 1271, 10–19. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.L.; Mostoslavsky, R.; Saito, S.; Manis, J.P.; Gu, Y.; Patel, P.; Bronson, R.; Appella, E.; Alt, F.W.; Chua, K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 2003, 100, 10794–10799. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Park, J.R.; Kwon, O.S.; Lee, T.H.; Nakano, I.; Miyoshi, H.; Chun, K.H.; Park, M.J.; Lee, H.J.; Kim, S.U.; et al. SIRT1 is required for oncogenic transformation of neural stem cells and for the survival of “cancer cells with neural stemness” in a p53-dependent manner. Neuro-Oncology 2015, 17, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Motta, M.C.; Divecha, N.; Lemieux, M.; Kamel, C.; Chen, D.; Gu, W.; Bultsma, Y.; McBurney, M.; Guarente, L. Mammalian SIRT1 represses forkhead transcription factors. Cell 2004, 116, 551–563. [Google Scholar] [CrossRef] [Green Version]
- Huffman, D.M.; Grizzle, W.E.; Bamman, M.M.; Kim, J.S.; Eltoum, I.A.; Elgavish, A.; Nagy, T.R. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007, 67, 6612–6618. [Google Scholar] [CrossRef] [Green Version]
- Stunkel, W.; Peh, B.K.; Tan, Y.C.; Nayagam, V.M.; Wang, X.; Salto-Tellez, M.; Ni, B.; Entzeroth, M.; Wood, J. Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. 2007, 2, 1360–1368. [Google Scholar] [CrossRef]
- Ford, J.; Jiang, M.; Milner, J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005, 65, 10457–10463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.-H.; Lee, M.-O.; Lee, J.-S.; Oh, J.-S.; Cho, S.-U.; Cha, H.-J. Sirt1 promotes DNA damage repair and cellular survival. Biomol. Ther. 2011, 19, 282–287. [Google Scholar] [CrossRef] [Green Version]
- Uhl, M.; Csernok, A.; Aydin, S.; Kreienberg, R.; Wiesmuller, L.; Gatz, S.A. Role of SIRT1 in homologous recombination. DNA Repair (Amsterdam) 2010, 9, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Yuan, J.; Pei, H.; Liu, T.; Ann, D.K.; Lou, Z. KAP1 Deacetylation by SIRT1 promotes non-homologous end-joining repair. PLoS ONE 2015, 10, e0123935. [Google Scholar] [CrossRef]
- Wang, R.H.; Sengupta, K.; Li, C.; Kim, H.S.; Cao, L.; Xiao, C.; Kim, S.; Xu, X.; Zheng, Y.; Chilton, B.; et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 2008, 14, 312–323. [Google Scholar] [CrossRef] [Green Version]
- Costa-Machado, L.F.; Martin-Hernandez, R.; Sanchez-Luengo, M.A.; Hess, K.; Vales-Villamarin, C.; Barradas, M.; Lynch, C.; de la Nava, D.; Diaz-Ruiz, A.; de Cabo, R.; et al. Sirt1 protects from K-Ras-driven lung carcinogenesis. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef] [Green Version]
- Khalaf, W.F.; White, H.; Wenning, M.J.; Orazi, A.; Kapur, R.; Ingram, D.A. K-Ras is essential for normal fetal liver erythropoiesis. Blood 2005, 105, 3538–3541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, J.L. Ras oncogenes in human cancer: A review. Cancer Res. 1989, 49, 4682–4689. [Google Scholar]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambard, J.C.; Lefloch, R.; Pouyssegur, J.; Lenormand, P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta 2007, 1773, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Braig, M.; Schmitt, C.A. Oncogene-induced senescence: Putting the brakes on tumor development. Cancer Res. 2006, 66, 2881–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulavin, D.V.; Fornace, A.J., Jr. p38 MAP kinase’s emerging role as a tumor suppressor. Adv. Cancer Res. 2004, 92, 95–118. [Google Scholar] [CrossRef]
- Cuenda, A.; Rousseau, S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 2007, 1773, 1358–1375. [Google Scholar] [CrossRef] [Green Version]
- Westermarck, J.; Li, S.P.; Kallunki, T.; Han, J.; Kahari, V.M. p38 mitogen-activated protein kinase-dependent activation of protein phosphatases 1 and 2A inhibits MEK1 and MEK2 activity and collagenase 1 (MMP-1) gene expression. Mol. Cell. Biol. 2001, 21, 2373–2383. [Google Scholar] [CrossRef] [Green Version]
- Li, S.P.; Junttila, M.R.; Han, J.; Kahari, V.M.; Westermarck, J. p38 Mitogen-activated protein kinase pathway suppresses cell survival by inducing dephosphorylation of mitogen-activated protein/extracellular signal-regulated kinase kinase1,2. Cancer Res. 2003, 63, 3473–3477. [Google Scholar] [CrossRef] [Green Version]
- Wancket, L.M.; Frazier, W.J.; Liu, Y. Mitogen-activated protein kinase phosphatase (MKP)-1 in immunology, physiology, and disease. Life Sci. 2012, 90, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Kidger, A.M.; Keyse, S.M. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs). Semin. Cell Dev. Biol. 2016, 50, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, P.S.; Ahn, N.G. Feedback regulation of Raf-1 and mitogen-activated protein kinase (MAP) kinase kinases 1 and 2 by MAP kinase phosphatase-1 (MKP-1). J. Biol. Chem. 1998, 273, 1788–1793. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Tonks, N.K.; Bar-Sagi, D. Inhibition of Ras-induced DNA synthesis by expression of the phosphatase MKP-1. Science 1994, 266, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Kwon, J.H.; Kang, S.H.; Kim, J.W.; Yang, Y.C. Increased MAPK activity and MKP-1 overexpression in human gastric adenocarcinoma. Biochem. Biophys. Res. Commun. 1998, 250, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Manzano, R.G.; Montuenga, L.M.; Dayton, M.; Dent, P.; Kinoshita, I.; Vicent, S.; Gardner, G.J.; Nguyen, P.; Choi, Y.H.; Trepel, J.; et al. CL100 expression is down-regulated in advanced epithelial ovarian cancer and its re-expression decreases its malignant potential. Oncogene 2002, 21, 4435–4447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zhang, Y.; Yu, H.; Shen, B.; Liang, Y.; Jin, R.; Liu, X.; Shi, L.; Cai, X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016, 5, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.J.; Bennett, A.M. Essential role for mitogen-activated protein (MAP) kinase phosphatase-1 in stress-responsive MAP kinase and cell survival signaling. J. Biol. Chem. 2005, 280, 16461–16466. [Google Scholar] [CrossRef] [Green Version]
- Moncho-Amor, V.; Ibanez de Caceres, I.; Bandres, E.; Martinez-Poveda, B.; Orgaz, J.L.; Sanchez-Perez, I.; Zazo, S.; Rovira, A.; Albanell, J.; Jimenez, B.; et al. DUSP1/MKP1 promotes angiogenesis, invasion and metastasis in non-small-cell lung cancer. Oncogene 2011, 30, 668–678. [Google Scholar] [CrossRef] [Green Version]
- Liao, Q.; Guo, J.; Kleeff, J.; Zimmermann, A.; Buchler, M.W.; Korc, M.; Friess, H. Down-regulation of the dual-specificity phosphatase MKP-1 suppresses tumorigenicity of pancreatic cancer cells. Gastroenterology 2003, 124, 1830–1845. [Google Scholar] [CrossRef]
- Vicent, S.; Garayoa, M.; Lopez-Picazo, J.M.; Lozano, M.D.; Toledo, G.; Thunnissen, F.B.; Manzano, R.G.; Montuenga, L.M. Mitogen-activated protein kinase phosphatase-1 is overexpressed in non-small cell lung cancer and is an independent predictor of outcome in patients. Clin. Cancer Res. 2004, 10, 3639–3649. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.Y.; Cheng, Z.; Malbon, C.C. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 2003, 191, 229–237. [Google Scholar] [CrossRef]
- Cao, W.; Bao, C.; Padalko, E.; Lowenstein, C.J. Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits toll-like receptor signaling. J. Exp. Med. 2008, 205, 1491–1503. [Google Scholar] [CrossRef] [Green Version]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef] [Green Version]
- Chi, H.; Flavell, R.A. Acetylation of MKP-1 and the control of inflammation. Sci. Signal. 2008, 1, pe44. [Google Scholar] [CrossRef] [Green Version]
- Pages, G. MAP kinase phosphatase-1: A link between cell signaling and histone phosphorylation. Focus on “Histone H3 as a novel substrate for MAP kinase phosphatase-1.”. Am. J. Physiol. Cell Physiol. 2009, 296, C233–C234. [Google Scholar] [CrossRef] [PubMed]
- Tanoue, T.; Yamamoto, T.; Nishida, E. Modular structure of a docking surface on MAPK phosphatases. J. Biol. Chem. 2002, 277, 22942–22949. [Google Scholar] [CrossRef] [Green Version]
- Eberharter, A.; Becker, P.B. Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Grimes, M.; Hall, B.; Foltz, L.; Levy, T.; Rikova, K.; Gaiser, J.; Cook, W.; Smirnova, E.; Wheeler, T.; Clark, N.R.; et al. Integration of protein phosphorylation, acetylation, and methylation data sets to outline lung cancer signaling networks. Sci. Signal. 2018, 11, eaaq1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sestito, R.; Madonna, S.; Scarponi, C.; Cianfarani, F.; Failla, C.M.; Cavani, A.; Girolomoni, G.; Albanesi, C. STAT3-dependent effects of IL-22 in human keratinocytes are counterregulated by sirtuin 1 through a direct inhibition of STAT3 acetylation. FASEB J. 2011, 25, 916–927. [Google Scholar] [CrossRef]
- Bartlett, T.E.; Zaikin, A.; Olhede, S.C.; West, J.; Teschendorff, A.E.; Widschwendter, M. Corruption of the intra-gene DNA methylation architecture is a hallmark of cancer. PLoS ONE 2013, 8, e68285. [Google Scholar] [CrossRef] [Green Version]
- Kondoh, K.; Nishida, E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim. Biophys. Acta 2007, 1773, 1227–1237. [Google Scholar] [CrossRef] [Green Version]
- Chi, H.; Barry, S.P.; Roth, R.J.; Wu, J.J.; Jones, E.A.; Bennett, A.M.; Flavell, R.A. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 2274–2279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Wang, X.; Nelin, L.D.; Yao, Y.; Matta, R.; Manson, M.E.; Baliga, R.S.; Meng, X.; Smith, C.V.; Bauer, J.A.; et al. MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. J. Exp. Med. 2006, 203, 131–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, O.-S.; Lee, H.; Kim, Y.-J.; Cha, H.-J.; Song, N.-Y.; Lee, M.-O. ERK Dephosphorylation through MKP1 Deacetylation by SIRT1 Attenuates RAS-Driven Tumorigenesis. Cancers 2020, 12, 909. https://doi.org/10.3390/cancers12040909
Kwon O-S, Lee H, Kim Y-J, Cha H-J, Song N-Y, Lee M-O. ERK Dephosphorylation through MKP1 Deacetylation by SIRT1 Attenuates RAS-Driven Tumorigenesis. Cancers. 2020; 12(4):909. https://doi.org/10.3390/cancers12040909
Chicago/Turabian StyleKwon, Ok-Seon, Haeseung Lee, Yun-Jeong Kim, Hyuk-Jin Cha, Na-Young Song, and Mi-Ok Lee. 2020. "ERK Dephosphorylation through MKP1 Deacetylation by SIRT1 Attenuates RAS-Driven Tumorigenesis" Cancers 12, no. 4: 909. https://doi.org/10.3390/cancers12040909




