Abstract
Telomere comprises the ends of eukaryotic linear chromosomes and is composed of G-rich (TTAGGG) tandem repeats which play an important role in maintaining genome stability, premature aging and onsets of many diseases. Majority of the telomere are replicated by conventional DNA replication, and only the last bit of the lagging strand is synthesized by telomerase (a reverse transcriptase). In addition to replication, telomere maintenance is principally carried out by two key complexes known as shelterin (TRF1, TRF2, TIN2, RAP1, POT1, and TPP1) and CST (CDC13/CTC1, STN1, and TEN1). Shelterin protects the telomere from DNA damage response (DDR) and regulates telomere length by telomerase; while, CST govern the extension of telomere by telomerase and C strand fill-in synthesis. We have investigated both structural and biochemical features of shelterin and CST complexes to get a clear understanding of their importance in the telomere maintenance. Further, we have analyzed ~115 clinically important mutations in both of the complexes. Association of such mutations with specific cellular fault unveils the importance of shelterin and CST complexes in the maintenance of genome stability. A possibility of targeting shelterin and CST by small molecule inhibitors is further investigated towards the therapeutic management of associated diseases. Overall, this review provides a possible direction to understand the mechanisms of telomere borne diseases, and their therapeutic intervention.
1. Introduction
Naturally, the ends of eukaryotic chromosomes are prone to misrecognized as the double-strand breaks (DSBs) that pose a critical challenge for cell viability and integrity of the genome. The eukaryotic cells overcome this challenge by forming a protective structure at chromosome ends comprising a tandem array of telomeric DNA repeats and telomere-binding proteins [1]. Telomeres of humans are composed of G-rich (TTAGGG) tandem repeats. The duplex telomeric repeat sequences range from 2–14 kb in length in addition to the single-strand overhang of about 12–10 nucleotides [2,3]. The telomere length and its regulations are mainly maintained by three key players namely, telomerase, shelterin, and CST complexes.
The conventional replication mechanism cannot offer the synthesis of chromosomal end which ultimately leads to shortening of telomere at each round of cell division. In normal cells, telomere length is maintained by a ribonucleoprotein (RNP), called telomerase, which is consisting of catalytic (hTERT) and RNA (hTR) components [4,5]. Components of shelterin and CST complexes act as a shield to protect the unique genomic information as illustrated in Figure 1. These two complexes ensure complete replication of the entire genome and thus prevent senescence which is associated with telomere shortening [6,7].
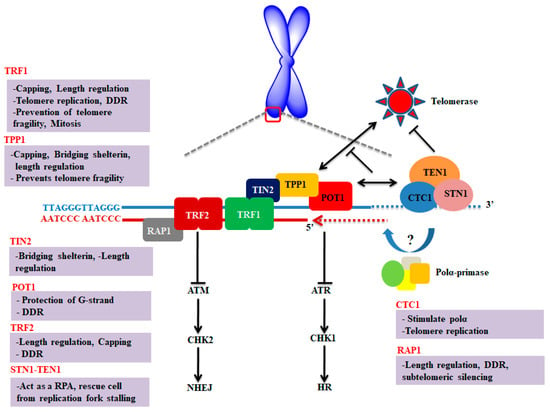
Figure 1.
Schematic representation and key molecular function of shelterin and CST complexes at the telomere. The key functions of each component of CST and shelterin complexes were briefly discussed. Shelterin complex proteins are mainly involved in suppressing DNA damage response; however, CST complex is primarily regulating the length of telomere. Figure was adapted and modified from [7,8,9].
Most of the proteins of these complexes have one or more OB (oligonucleotide/oligosaccharide binding)-fold domains. OB-fold proteins have an affinity to bind nucleic acid and belonging to the nucleic acid-binding superfamily [10,11]. Shelterin consists of six subunits including telomere repeat factor 1 and 2 (TRF1 and 2), repressor/activator protein 1 (RAP1), TRF1-interacting nuclear factor 2 (TIN2), adrenocortical dysplasia homolog (ACD, also referred to as TINT1/PTOP/PIP1 (TPP1) and protection of telomeres 1 (POT1) [6]. Among these six subunits, three components bind with DNA in sequence-specific manner such as POT1 which specifically interacts with single-stranded DNA of telomere (1TTAGGGTTAG10), whereas TRF1 and TRF2 particularly bound to the double-stranded region of telomeric DNA [12]. In addition, shelterin components perform multiple roles at telomere as they prevent chromosomal ends from being recognized as DNA double-strand breaks (DSB), regulate telomere replication, as well as monitor the telomerase access to telomere [13].
The CST complex is composed of three subunits including, conserved telomere maintenance component 1 (CTC1), suppressor of CDC thirteen homolog (STN1) and telomere length regulation protein TEN1 homolog (TEN1) [14,15,16]. The component of CST (CTC1-STN1) specifically localizes to the single-stranded telomeric DNA and limits telomerase action to prevent overextension of G-overhang. This interaction of CTC1-STN1 to telomeric DNA is stabilized by TEN1 [17]. Moreover, CTC1 originally identified to stimulate DNA polymerase alpha (Polα) [18] and plays a key role in telomere replication [19]. However, the STN1-TEN1 complex of CST acts as a replication protein, A (RPA)-like complex involved in rescuing cells from replication fork stalling during replication stress [20]. In contrast with the shelterin complex which is mainly engaged in the protection of telomere, the CST complex promotes the replication of telomere and does not show any direct role in repressing DDR at telomere [21].
Due to their key role in telomere maintenance, mutations in the CST and shelterin components causes a dramatic alteration in the cellular physiology and often leads to life-threatening diseases including, cancer, bone marrow (BM) failure, dyskeratosis congenital (DC), coats plus (CP), and premature aging syndromes [1,8,14,15,22,23,24]. In this paper, we have extensively analyzed and highlighted the structural features of each component of CST and shelterin complexes, and its importance in maintaining genome integrity. Further, we have analyzed the occurrence of about 115 clinically important mutations based on available literature and broadly discussed their implication in disease development. Finally, we have discussed possible therapeutic opportunities for each component of both complexes in detail.
2. Structure and Function of Shelterin Complex
2.1. TRF1 and TRF2
TRF1 and TRF2 are consisting of 439 (UNIPROT-P54274 (TERF1_HUMAN)) and 542 amino acid residues (UNIPROT-Q15554 (TERF2_HUMAN), respectively. Both TRF1 and TRF2 have a similar domain architecture composed of TRF homology (TRFH) at their N-terminal and a SANT/Myb (Swi3, Ada2, N-Cor, and TFIIIB/Myb) DNA-binding domain at C-terminal, which are connected together through flexible hinge region (Figure 2A,B, respectively) [25,26]. The C-terminal SANT/Myb domainof both proteins are almost similar which provide specificity to interact with the double-stranded DNA of telomere [25]. TRF1 and TRF2 interact with DNA as a homodimer or oligomer formed through the interaction of TRFH domain. The TRFH domains are responsible for dimerization and contain a specific peptide docking site for the recruitment of other proteins to the telomere [27].
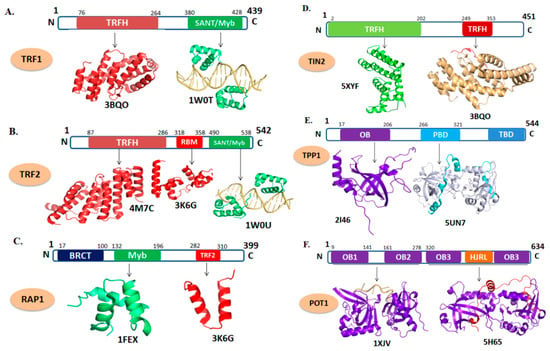
Figure 2.
Domain organization and tertiary structure of the shelterin complex. (A) telomere repeat factor 1 (TRF1) (PDB ID: 3BQO; depicts the cartoon representation of TRF homology (TRFH) domain of TRF1 in red. 1W0T; the DNA-binding domain of TRF1 shown in green in complex with telomeric DNA which is shown as light orange). (B) TRF2 (PDB ID: 4M7C; represents TRFH domain of TRF2. 3K6G; represents the repressor/activator protein 1 (RAP1)–binding motif of TRF2 in red. 1W0U; represents the DNA-binding motif of TRF2). (C) RAP1 (PDB ID: 1FEX; represent the Myb domain shown in green and 3K6G; shows RCT domain in red). (D) TIN2 (PDB ID: 5XYF; cartoon representation in green depicts TRFH domain and 3BQO; TRF1-interacting nuclear factor 2 (TIN2) peptide shown in red complexed with TRFH domain of TRF1). (E) TPP1 (PDB ID: 2I46; depicts the OB-fold domain in purple and 5I2X; represents the POT1 interacting domain of TPP1 (cyan) with C-terminal domain of POT1 (gray)). (F) POT1 (PDB ID: 1XJV; depicts the C-terminal domain of POT1 in purple in complex with telomeric DNA (light orange) 5H65; represents the POT1 interacting domain of TPP1 (red) with C-terminal domain of POT1 (purple).
Both TRF1 and TRF2 interact as a homodimer to double-stranded telomeric DNA [28], but they do not interact directly with each other [25]. Interaction of TRF1 and TRF2 to double-stranded telomeric DNA is further stabilized by RAP1 and TIN2. Analysis of the three-dimensional structure of TRF1 and TRF2 reveals that dimer formation is mainly mediated by the respective TRFH domain [29]. The interface of respective TRFH domain contains six α helices (H1, H2, and H9 of each TRFH domain). The H1 and H2 α-helices of one monomer pack against H1 and H2 of another monomer, respectively, to form a symmetrical antiparallel four-helix bundle [29]. Despite a high structural similarity in TRFH domains of both proteins, they do not interact with each other mainly due to incompatible hydrophobic networks that form the dimerization interface. The amino acid residues involved in intermolecular interactions of TRFH domains of both TRF proteins play an important role in maintaining the dimer interface [29]. Mutations of amino acid residues of the TRFH domain (dimer interface) effects protein-protein and protein-DNA activities.
Inactivation of the TRF1 gene in the mouse embryonic fibroblasts (MEFs) resulted in the stalling of DNA-replication process at the telomere which leads to the formation of gaps, called fragile telomeres [30,31]. Inactivation of the TRF1 gene and its subsequent abortive replication enhances the activation of robust DNA damage response (DDR) [32,33]. However, inactivation of the TRF2 gene resulted in the telomere fusion mediated by NHEJ events independently of replication fork stalling [34]. Up-regulation or down-regulation of TRF1 and TRF2 genes has been reported in many diseases including cancer [35,36,37].
2.2. RAP1
RAP1 consists of 399 amino acid residues (Uniprot ID-Q9NYB0), and has three distinct domains (Figure 2C). The N-terminal BRCT (BRCA1 C-terminus) domain is responsible for the recognition of phosphorylated peptide. The C-terminal TRF2 domain interacts with TRF2 protein. The Myb domain generally used to binds with telomeric DNA in budding yeast that has two copies of Myb domain. However, the mammalian RAP1 has only one Myb domain and do not involve in such interaction and therefore RAP1 dependent on TRF2 for telomeric interaction [38,39]. The architecture and topology of mammalian Myb domains are very close to that of budding yeast and TRF1; however, the surface electrostatic potential of the mammalian Myb domain is distinct from that of other Myb domains. Myb domains that have DNA-binding activity exhibit a positively charged large surface closely with the highly negatively charged backbone of DNA. Conversely, mammalian Myb domain shows no distinct positive surface, reveals its lack of DNA-binding property [40].
RAP1 interacts with telomeric DNA only through TRF2 which is essential in chromosome ends protection since its deletion from telomeres resulted in ATM-dependent DNA damage signaling and NHEJ pathway-mediated massive end-to-end fusions [41]. Inactivation of RAP1 resulted in the telomere shortening, hyperpigmentation, and enhanced DDR activation [42]. Recently, two independent studies from different groups demonstrated that mutations in the RAP1 gene are associated with chronic lymphocytic leukemia and familial melanoma [43,44].
2.3. TIN2
TIN2 protein is consists of 451 amino acids (UNIPROT-Q9BSI4 (TINF2_HUMAN)) and comprised of two distinct domains (Figure 2D). TIN2 interacts with TPP1 and TRF2 through two different interacting modules [27], one is the N-terminal domain (residues 2–202), which recognized a short TIN2-binding motif of TRF2 (residues 350–366). The other is the short TRFH-binding motif (TBM) (residues 256–276) at the C-terminal portion of TIN2, which interacts with TRFH domains of both TRF1 and TRF2 [27]. Interactions of TIN2 with these proteins develop a bridge between the components of shelterin complex and ssDNA or dsDNA of telomere [45,46]. In the ternary complex, each polypeptide of the TIN2-binding motif of TPP1 is folded into a helix–loop–helix motif. Both helices and the connecting loop make extensive contacts with TIN2 (residues 2–202). The driving force for the binding of TIN2-binding motif of TPP1 to TIN2 (residues 2–202) is van der Waals interactions, as most conserved residues of TIN2-binding motif of TPP1 are hydrophobic in nature. The core of this extended interface between TIN2-binding motif of TPP1 and TIN2 (residues 2–202) consists of a panel of hydrophobic residues from both proteins. The extensive contacts among the side chains of these residues mediate the specificity of TIN2-binding motif of TPP1 recognition by TIN2 (residues 2–202).
TIN2 act as an adaptor which plays crucial role in stabilizing the subunit of shelterin complex. It binds both TRF1 and TRF2, providing stability to these proteins at telomere [46]. In addition, it is also interacting with TPP1 and is indispensable for the recruitment of TPP1/POT1’s to the shelterin complex. As the central hub of the shelterin complex, TIN2 plays pivotal roles in telomere maintenance and end protection [46,47,48,49,50,51]. Inactivation or loss of TIN2 results in the activation of DDR, proliferative arrest, CHK1, and CHK2 phosphorylation. Thus, activation of ATM and ATR pathways take place at telomeres deficient of TIN2, and this deficiency results in the commencement of repair mechanisms which in turn activates the alternative lengthening of telomere (ALT) pathway. On the other hand, knockdown studies of the TIN2 gene in model organisms show a reduction in the TRF2 function, telomerase recruitment to the telomere [52,53], abnormal mitochondrial morphology, reduced glycolysis and increase oxidative metabolism [52]. Furthermore, the expression of a mutant form of TIN2 or deletion imposes a severe effect on the stability of the shelterin complex [46,54]. Till date, more than 25 clinically important mutations are reported which are known to alter the cell survival and ultimately resulted in DC and pulmonary fibrosis [55,56].
2.4. TPP1
TPP1 is encoded by ACD gene and consisting of 544 amino acids (UNIPROT ID-Q96AP0 (ACD_HUMAN)). TPP1 is a multidomain protein with confined regions that act as interacting sites for other shelterin proteins and telomerase [57,58,59,60]. The N-terminal of TPP1 comprises an OB-fold (amino acid residues, 87–250) domain, a centrally positioned POT1 binding domain (PBD) (residues, 250–334), and a C-terminal TIN2 binding domain (TBD) (residues 510–544) were also reported (Figure 2E). The structural analysis of the OB-fold domain reveals a typical OB-fold architecture that is common among many DNA-binding proteins [57]. OB-fold domains have been implicated in coordinating protein–protein interactions within multi-component complexes [61,62]. In this regard, the OB-fold domain of TPP1 helps in the establishment of interactions with the telomerase facilitating its recruitment to the telomere [53,58,63,64]. In addition, the OB-fold domain of TPP1 interacts with the TERT component of telomerase and involved in regulation or recruitment of telomerase to the telomere [46,65]. The TEL (TPP1’s glutamate (E) and leucine-rich (L)) patch of this domain is responsible for telomerase recruitment at telomere [58,64,66]. Several mutations in TPP1 gene have been found that are coupled with many diseases including cancer [43].
2.5. POT1
POT1 is consisting of 634 amino acid residues (UNIPROT-Q9NUX5 (POTE1_HUMAN)). It has four distinct domains, including two N-terminal OB-fold (OB1 and OB2) domains and a third OB-fold (OB3) along with Holiday junction resolvase (HJR) domains at C-terminal of POT1 (Figure 2F) [67]. POT1 and telomeric DNA interaction are mediated by the two OB-fold domains of POT1, while the C-terminal region binds with TPP1 [65,68]. POT1 binding to TPP1 enhances its DNA binding properties by 10-fold [57,60,69]. The POT1–TPP1 complex recruits telomerase to the ends of the chromosome [57,65] through direct contacts of telomerase with the TEL patch situated at the N-terminal OB-fold of TPP1 [58]. In addition, POT1 interaction to the telomeric DNA overhang helps in resolving G-quadruplexes and allows telomerase loading to telomeres for telomere extension [70].
The N-terminal of POT1 adopts an elongated conformation and is composed of two OB-fold domains closely connected by a short linker [71,72]. The ssDNA of telomere in the complex spans both OB1 and OB2, binding in the continuous concave groove [73]. Further, it is reported that OB1 of POT1 makes much more extensive contact with the ssDNA than OB2 [22,73].
The C-terminal of POT1 consists of 330–634 amino acid residues which interact with the central OB-fold domain of TPP1 (255–337 amino acid residues). The C-terminal of POT1 consists of an OB-fold and a HJR domain. The canonical OB-fold of C-terminus is structurally like Telomere End-Binding Protein α (TEBPα, PDB ID: 1OTC) of Oxytricha Nova. The TPP1 helix α1 is leucine/valine rich and the majority of contacts with the POT1C, HJR are hydrophobic in nature [67].
POT1 is essential for the inhibition of DDR-mediated ATR pathway and implicated in the regulation of 3′ G-strand overhang [74]. By its strong affinity to the 3′ overhang at telomere, POT1 acts as a natural inhibitor of telomerase. Any alteration in the DNA-binding domain of POT1 is leading to excessive telomere elongation, signifying unregulated access of telomerase to the telomere [75]. Silencing of either POT1 or TPP1 gene through the RNA-interference (RNAi) technology increases the telomere length and chromosomal instability [76], indicating its role in the maintenance of telomerase access to the telomere overhang [73]. Since the POT1 plays a key role in the protection of telomere concerning telomere elongation and cell immortality, it has been considered as a promising drug target for cancer therapy [77]. Several mutations in the POT1 gene have been identified in different types of melanoma and CP patients [78].
3. Structure and Function of CST Complex
3.1. CTC1
CTC1 is composed of 1217 amino acid residues (UniProtKB-Q2NKJ3 (CTC1_HUMAN)). The CDC13 is a yeast homolog of mammalian CTC1 which has four OB-folds domains (Figure 3A). Recently, a high-resolution structure of an OB-fold domain of CTC1 has been solved which is found at the C-terminal region of the protein (Figure 3B). In mice, deletion of the CTC1 gene resulted in a rapid loss of C-strand telomeric DNA which causes catastrophic telomere loss and premature death [21]. Taken together, it has been proposing that the CTC1 is vital for telomere length maintenance by promoting efficient telomere replication and C-strand protection. As CTC1 have critical role in CST complex formation its mutation brings lethality in the cellular environment [79]. Several naturally occurring mutations are known in CTC1 which are associated with CP and DC [80,81].
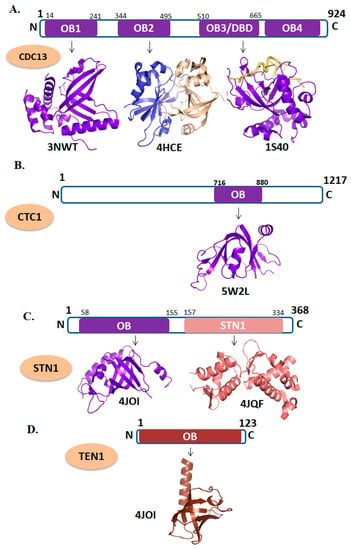
Figure 3.
Domain organizations and tertiary structures of CST (CTC1-STN1) complex. (A) CDC13 (PDB ID: 3NWT; OB1 domain shown in purple, 4HCE; represents the dimer of OB2 domain, 1S40; depicts the DNA-binding domain of CDC13 in complex with telomeric DNA). (B) CTC1 (PDB ID: 5W2L; represent the OB-fold domain of CTC1 in purple. (C) STN1 (PDB ID: 4JOI; cartoon representation of N-terminal OB-fold domain of STN1and 4JQF depicts the N-terminal domain of STN1 in salmon color). (D) TEN1 (PDB ID: 4JOI; depicts the OB-fold domain of TEN1 in chocolate color).
3.2. STN1-TEN1
The STN1 (UniProt KB-Q9H668) and TEN1 (UniProt KB-Q86WV5) are comprised of 368 and 123 amino acids residues, respectively. STN1 is consist of two domains, the N-terminal OB-fold and C-terminal STN1domain (Figure 3B) [15], whereas, TEN1 is composed of a single OB-fold domain (Figure 3C). Human TEN1 does not interact with ssDNA of telomere which is in contrast to the yeast TEN1 [16,82]. Conversely, human STN1 binds with ssDNA with high affinity but lacking specificity [82]. High affinity and specificity of the human CST complex for single-stranded telomeric DNA are provided by the larger component of CST, CDC13/CTC1 [83]. Furthermore, STN1-TEN1 complex formation is vital for the proper functioning of CST complex. Mutations in the STN1 gene have been associated with CP [15,84] in contrast to the mutations in the TEN1 gene [16,82].
4. Diseases
Several human diseases are associated with the telomere shortening including, liver cirrhosis, ulcerative colitis, atherosclerosis, cardiovascular disease, cancer and many premature aging syndromes [85,86,87,88]. Here, we have extensively analyzed the mutations in CST and shelterin complexes and their association with disease development. Some clinically important mutations, associated genes, and their corresponding cellular fitness are shown in the Table 1.

Table 1.
Some clinically important mutations, associated genes, and their corresponding cellular pathologies. Among both complexes, protection of telomeres 1 (POT1) and conserved telomere maintenance component 1 (CTC1) appear as most recurrent mutated genes.
4.1. Role of Shelterin Complex in Cancer
The structures of telomeric DNA is comprised of tandem repeats of G-rich (TTAGGG) sequences protected with shelterin complex. Shelterin complex is comprised of six proteins (TRF1, TRF2, TIN2, POT1, TPP1, and RAP1) which is essential for telomere protection, chromosomal stability and regulation of telomere length. Further, the shelterin complex implicated in the modulation of telomerase activity at chromosome ends recognizes telomeric DNA and remodels it into a t-loop. This process shelters the 3′ overhang from being recognized as DNA damage. Changes in the structure and function of any of the components of this complex may lead to undesirable DDR that often leads to the development of tumorigenesis and cancer progression [110,111,112].
4.1.1. RAP1
Several mutations are reported in all three domains of RAP1 which are associated with cancer. Some of the critically important mutations are, p.M5I, p.D10H, p.Q191R, and p.R364X, found in the patient of familial melanoma, whereas mutations including p.A104P, p.R133Q are reported in the CLL [43,44]. In addition, a nonsense mutation p.R364X in the RAP1 gene leading to the formation of a C-terminal truncated protein that disrupts the TRF2-binding domain, and subsequently diminishes the binding to shelterin complex, which possibly leads to melanoma susceptibility [43]. Additionally, p.Q191R and p.M5I mutations are noticed in the patients of cutaneous malignant melanoma (CMM), sporadic melanoma, and ovarian cancer. These mutations are found in the Myb and BRCT domains, respectively, and considerably affect the interaction of RAP1 to telomeric overhang. Due to this, RAP1 is unable to regulate telomere homeostasis and disturb cellular fitness [43]. Recently, two novel mutations (p.A104P and p.R133Q) were also reported in CLL patients [44].
4.1.2. TPP1
Interaction of shelterin complex and hTERT has been carried out by TPP1 and POT1 complex [107]. Mutations or inhibition of the TPP1/POT1 subunit increases the telomere length, which clearly shows that TPP1/POT1 subunit is essentially required to inhibit the elongation of telomere [64,67]. A nonsense mutation p.Q320X in the TPP1 gene was recently identified which disrupts the POT1 and TIN2 binding, which ultimately resulted in the formation of a non-functional shelterin complex [113]. In consistence, mutations such as p.V272M and p.N249S were identified in the lower-density melanomas, whereas p.A200T and p.I322F mutations were observed in the CMM patients. Among the five mutations discussed above, four mutations were found in the POT1-binding domain of TPP1 that is critical for the proper functioning of the shelterin complex. More recently, a novel mutation, p.G223V was also identified next to the TEL patch in the OB-fold domain of TPP1 that interacts directly with the catalytic subunit of telomerase. TPP1, as a subunit of shelterin complex was particularly shown to binds with POT1 to shield telomeres and recruit telomerase to the chromosome ends [58,63,64]. Further extensive analysis of p.G223V mutants shows an increase in the cell survival of childhood pre-B acute lymphoblastic leukemia (cALL), and protect leukemia cell from apoptosis and increase the telomere length [94]. Further studies are needed to translate the underlying molecular mechanisms implicating TPP1 in apoptosis inhibition of leukemia cells. Altogether, these data strongly support a role for TPP1 p.G223V mutant in promoting leukemia cell maintenance. Interestingly, recurrent somatic mutations in the OB-fold domains of POT1 reported causing telomere dysfunction in CLL signifying that alteration of TPP1-telomere binding could lead to genomic instability and cancer [114].
4.1.3. POT1
POT1 was the first identified member of the shelterin complex which is mutated in cancer. Mutation in POT1 mainly found in OB-fold domain which plays critical role in telomeric DNA binding [101]. POT1 mutations are not confined to the OB-fold domain only, instead it spread through entire length of POT1 [78,97,101,102], and likely causative of many disease including CLL [101,102], familial melanoma [96], CMM [97], CP [100], cardiac angiosarcoma [99], and familial glioma [98]. Most of the mutation is clustered in the OB-fold domain which disrupts the POT1′s ability to interact with telomeric DNA and does not able to regulate the length of the telomere. Patients carrying these mutations have a very high probability of sister chromatid fusions or sometimes chromosome fusion or may have fragile chromosomal ends [8,101].
In two independent studies, researchers found many significant mutations in the OB-fold region of the POT1 gene which are coupled with CLL [101,102]. Cells from CLL patients have telomeric and chromosomal abnormalities which indicated that mutations in POT1 gene support the acquisition of the malignant features. Due to the discovery of POT1 as a commonly mutated gene in CLL which perhaps facilitates novel approaches for the clinical management of this disease [101,102]. Currently, three more mutations (p.Y36C, p.Q376R, p.Q358S) in the POT1 gene have been identified from CLL patients, predicted to disrupt the interaction of POT1 with TPP1 and telomere overhang which consequently contributed to the CLL pathogenesis [44].
Robles-Espinoza and co-worker [96] identified p.Y89C, p.Q94E and p.R273L mutations in the POT1 gene associated with familial melanoma. Most of the mutations either effects the splicing of POT1 mRNA or alter key residues in OB-fold domains [96]. These mutations disrupted the protein-telomere interaction and lead to telomere lengthening [96]. In consistence, three novel mutations (p.G95C, p.E450X, p.D617Efs) have been identified in the POT1 gene from glioma patients [115,116,117]. Out of three, two mutations in the POT1 gene (p.G95C, p.E450X) are predicted to destabilize the POT1 structure, thus shelterin complex. Mutation in this region affects the DNA-binding as well as TPP1 binding property of POT1 [98]. Along this line, p.D617Efs mutation is predicted to disrupt the TPP1-binding [98]. Moreover, p.R117C in the POT1 gene is associated with cardiac angiosarcoma (CAS), a rare malignant tumor, whose genetic basis is not fully understood [99].
4.2. Role of Shelterin Complex in DC and CP
Recently, several reports highlighted the importance of shelterin complex in hematopoiesis, DC and CP [8,118]. DC is characterized by a disordered human telomere with many pleiotropic manifestations that often lead to the BM failure [119,120]. However, CP is a rare autosomal disorder with intracellular calcification [100]. In addition to the mutations in shelterin complex genes, a large number of mutations in other genes are also reported as a major cause for DC [121,122]. Here, we briefly discuss only the shelterin complex genes implicated in DC and CP.
4.2.1. TIN2
TIN2 was the first identified component of shelterin, implicated in DC. Patients harboring mutation in the TIN2 coding gene usually has a shorter telomere with symptoms appear at an early age [92,123]. Mutation in this gene is mainly associated with variants of DC called Hoyeraal-Hreidarsson (HH) [124] and Revesz syndromes [92,125]. Currently, more than 25 mutations are reported from DC patients and its variants [89,90,91,92]. All mutations reported are clustered in the exon 6a, corresponding to the amino acid 269–298 [91,92,125,126]. Domain organization of TIN2 shows that all of the mutations occurs in the TRFH domain which is responsible for the interaction of TIN2 with TRF1 [27]. Shortening of telomere occurring due to TIN2 mutations has been explained by two mechanisms that may affect the maintenance of telomere length [51,127]. These mechanisms are based on the reduction in recruitment of telomerase in TPP1-dependent where telomere shortening was not accompanied by changes in total telomerase activity, TIN2 localization, or telomere end protection. Interestingly, TIN2 participates in the TPP1-dependent recruitment of telomerase activity and compromise the telomere recruitment of telomerase, leading to telomere shortening and the associated pathogenesis [127]. However, another report shows that telomere shortening also occur through TPP1-independent manners, where they indicate that TIN2-R282H mutation elongates telomeres at a reduced frequency as this mutation affects the telomerase-telomere co-localization, separable from its role in telomere protection [51]. In addition, studies on the p.K280E mutation are associated with telomere shortening and DDR activation [91,128]. In addition, a study has also shown that DC-linked mutation disrupts the binding of TIN2 with HP1γ (a heterochromatin protein) which affects the cohesion of normal sister telomere [129].
4.2.2. TPP1
Mutation in the TPP1 gene is found to be associated with the inherited BM failure and DC [95,130]. p.K170X mutation is associated with telomere shortening as this mutation brings structural changes in TEL patches (a small region in OB-fold of TPP1). Structural changes in TEL patches inhibit the TPP1’s ability to bind telomerase and finally results in the DC pathogenesis. While, mutation substituting threonine for proline at position p.T491P in the TIN2-interacting domain of TPP1 does not affect the recruitment of telomerase but resulted in modest disruption of the TPP1/TIN2 interaction, the consequence of which still unknown [130].
In consistence, p.K170X mutation in DC causes cancer progression [130]. These findings suggest that TEL patches of TPP1’s are important in maintaining telomere homeostasis. In addition, Zhong and co-worker [64] generated several mutations (single and double mutant) in TEL patch in order to see the effect of these mutations on efficacy of TPP1’s to recruit telomerase to the telomere. They identified a putative interaction surface that monitors the interaction between telomerase and TPP1 and hence mutation in TEL patch is no longer able to regulate the telomere homeostasis [64].
Mutation in TEL patch causes defective recruitment of telomerase to telomeric ends and this defect is supposed to result in telomere shortening as observed in affected individuals [95,130]. These findings supported the study of engineered mutations in embryonic stem cells [131]. Interestingly, mutations in the TEL patch have not been translated in vivo, suggesting the pathogenic mechanism remains to be fully investigated.
4.2.3. POT1
Apart from a reported POT1 gene mutation in cancer, its p.S322L mutation causes CP. Mutation in the POT1 gene probably disrupts the POT1/CST-dependent telomere fill-in and this deficiency in the fill-in step generates truncated telomeres that halt the proliferation of cells lacking telomerase [100].
4.3. CST Complex and Diseases
4.3.1. Role of CTC1 in CP and DC
There are several naturally occurring mutations in CTC1 gene that resulted in a range of rare genetic disorders such as CP and DC. Several patients with CP have shortened telomeres, suggesting that telomere dysfunction/telomere shortening possibly plays an important role in CP disease development. Mutations in CTC1 gene are responsible for CP spread throughout the genome, and their associations with human disease are typically biallelic [7,104,105]. Approximately 20 mutations have been reported in the CTC1 gene which either abrogates the interaction of CTC1 with ssDNA of telomere or DNA polα or to the STN1-TEN1 subunit of CST complex [104,132].
Interestingly, three naturally occurring mutations at the N-terminal and central region of CTC1 (p.A227V, p.V259M, and p.V665G) interrupt CTC1/polα-primase binding to the telomere. Particularly, the p.V259M mutation resulted in a significant accumulation of telomere-free ends. Furthermore, mutation including p.L1142H, p.A227V, p.V259M, p.R987, and p.1196-Δ7 (deletion of amino acid residues 1196–1202) not only interrupt the association of CTC1-STN1 and polα-primase to telomere but also negatively affect the nuclear localization of this complex [7,104,105,106,133]. In addition, there are four novel mutations (p.C424G, p.L247P, p.L464V) in CTC1 gene have been identified in DC [134]. The lethality of these mutations were identified using bioinformatics tool called PolyPhen2, which shows that these mutation are probably lethal [134].
Few novel mutations in the CTC1 gene (p.H484P, p.G278V, p.Y281H) have been identified in Indian families. These mutations were clustered in the N-terminal OB-fold domain of CTC1 which might induce conformational changes in protein structure that ultimately effects the reduction in binding affinity of CTC1 with ssDNA. This change in binding affinity compromises the telomere structural integrity which ultimately leads to CP [106,107].
4.3.2. STN1-TEN1
STN1 participates in multiple aspects of telomere homeostasis and mutation in this gene is coupled with CP and DC [84,109,135]. Recently, C. Bryan and co-worker [82] have generated single and double mutants (p.D78A, p.I164A, and p.R27Q, p.Y115A, p.R119Q) in STN1 and TEN1 gene, respectively to determine the effects of these mutations on STN1-TEN1 complex formation [82]. They further reported that single mutants showed a moderate loss of binding affinity while double mutants abolished STN1-TEN1 binding. Loss of binding affinity between STN1 and TEN1 occurs due to the disruption of a key salt bridge and hydrophobic interactions between these two CST subunits. This disruption in STN1-TEN1 complex formation results in a non-functional CST complex which leads the formation of elongated telomere and chromosomal abnormalities [82].
A mutational study on the OB-fold domain of STN1 was performed to address the effect of mutations on DNA-binding properties of STN1. Bhattacharjee and co-workers [109] generated three mutations (p.W89A, p.R139L, p.Y141A) where p.W89A and p.Y141A mutations were chosen because of the equivalent mutations in mouse STN1 decrease DNA-binding approximately 60% in pull-down assays [135] which is well translated through in vivo experiments [109]. However, from in vitro studies, they found that STN1 mutation disrupts binding to short DNA substrates only [109], suggesting that CST actually binds to DNA in a dynamic fashion which could provide a mechanistic explanation for how CST helps to resolve a diverse array of replication problems to preserve genome stability [109]. Two novel mutations (p.R135T, p.D157Y) are reported in the STN1 gene and these mutations were found to be pathogenic in nature [84].
5. Therapeutic Strategies against Telomere Maintaining Components
Here we try to highlight the telomerase, telomere, CST, and shelterin complexes as a therapeutic target to address a large number of diseases associated with telomere malfunctions. Telomere instability fails to maintain the integrity of the genome and often coupled with a shortening of telomere and its fusion [31,136]. Instability of telomere occurs due to several reasons, one of the well-known is the shortening of telomere with each cell division [137]. Furthermore, instability of telomere can arise when telomeric DNA form G-quadruplexes (G4) which interferes with the synthesis of telomeric DNA and result in telomere fragility and possibly rapid loss of telomere [138]. In addition to telomere shortening and formation of G4 structure, telomere instability can also results from the deprotection of telomere which is enhanced due to unavailability of telomere binding proteins, and this unavailability leads to the loss of DDR suppression and enhancement of genomic rearrangements [139,140]. Since, the stability of telomere contributes to the replication immortality in cancer cells, thus targeting telomere stability through interfering with the telomere synthesis carried out by telomerase or protection may provide a newer approach for the treatment of cancer and other telomere diseases. Key molecular targets, which play a direct role in maintaining telomere integrity, are shown in Figure 4. Some important small molecules inhibitors which inhibit the function of these molecular targets are shown in Figure 5. Details of these inhibitors and their mechanism of action are listed in Table 2.
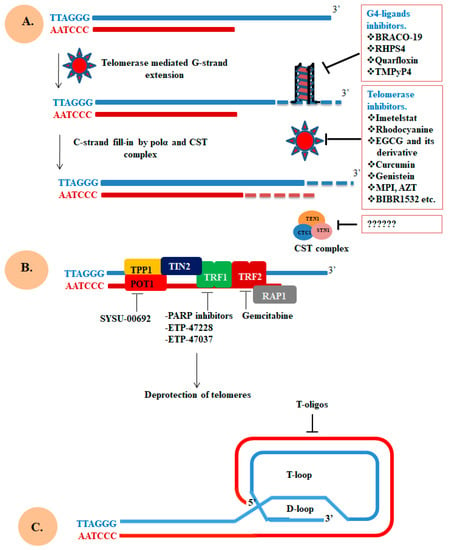
Figure 4.
Molecular target directly affecting the integrity of telomere. (A) G4-ligands bind with telomeric end to stabilize or promote the formation of G4 structure which abrogates the telomeric end extension. Telomerase inhibitors target either hTERT or hTR and block telomere extension, consequently disrupting telomere integrity. The CST complex is crucial for C-strand synthesis and defect in this complex dysregulate telomerase action and telomere loss. (B) Targeting components of the shelterin complex involved in the protection of telomeric end. (C) Use of T-oligos technology to block the lengthening of telomere. Figure is adapted and modified from the papers [192,206,207].
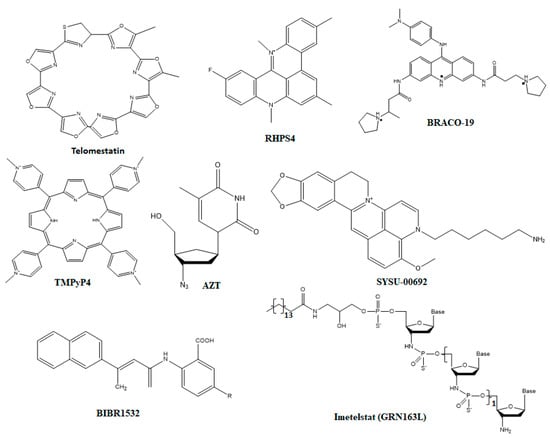
Figure 5.
Structures of small molecule inhibitors that target different molecular interactions responsible for the telomere integrity. The chemical structures compounds depicted here have binding affinity against protection of telomeres 1 (POT1).

Table 2.
List of important small molecule inhibitors and their mechanism of action.
5.1. Telomerase
Therapeutic targeting of telomerase is a continuously growing and evolving approach to treat telomere borne diseases, as telomerase overexpression is associated with almost all types of cancer [184,185]. As we discussed earlier, human telomerase is consisting of two core components: the hTERT and hTR. Presently, there are many classes of compounds tested that either target hTERT or hTR for their ability to suppress or inhibit the growth of tumor [186,187,188]. A large number of small molecules including those screened from chemical library of reverse transcriptase show direct interaction with hTERT. Along this line, a compound (BIBR1532) binds directly to the hTERT and inhibits the catalytic function of telomerase [141,146], promote shortening of telomere and senescence in human cancer cells [142]. However, further studies show that BIBR1532 could be promising when given in combination with traditional chemotherapeutic agents [144,145].
Other effective inhibitors such as isothiazolone (TMPI) [148], epicatechin (EGCG and MST312) [149,150] and derivative of bisindole are reported to be selective against telomerase. Also, quinoxaline [189], nitrostyrene [190], rubromycin analogs [191], chrolactomycin, and helenalin have been demonstrated to block the action of telomerase [192,193].
Moreover, other molecules including inhibitors of cyclooxygenase-2 (COX-2) [194], histone deacetylase (HDAC) inhibitors [195,196], and tyrosine kinase inhibitors [197] show inhibitory effect on telomerase along with several other key physiological processes [198,199]. Together with these compounds, other molecules that specifically inhibit phosphorylation of telomerase such as bis-indolylmaleimide I and H-7,231 antioxidants (vitamin E) [200], anti-inflammatory nonsteroidal drugs (aspirin) are reported to have inhibitory effects on telomerase [192]. In consistence, several other nucleoside analogs are considered as promising drugs for the inhibition of telomerase [201,202]. These inhibitors include azidothymidine (AZT) [151], l-enantiomers (L-dTTP and L-dGTP) [203], 6-thio-2′-deoxyguanosine 5′-triphosphate (TDG-TP) [204]. Carbovir 5′-triphosphate, 2′,3′-Dideoxyguanosine 5′-triphosphate and D-carbocyclic-2′-deoxyguanosine 5′-triphosphate have an inhibitory effect on the telomerase [205].
In addition to targeting hTERT, some novel chemical approaches has been implemented to target the RNA component of telomerase (hTR) while giving a special emphasis on the 11-mer template region (5′-CUAACCCUAAC-3′). Such molecules are called small interfering RNA (siRNA) or ribozymes, one such oligonucleotide known as Imetelstat (GRN163L) inhibit hTR to forming an active complex with hTERT, and hence interferes with telomerase activity, promote telomere shortening, senescence, or apoptosis [208]. Interestingly, observation from several clinical trials support for the potential use of Imetelstat to treat patients harboring hematological cancers and also for those which have solid lung tumors with short telomere [209]. Since this compound has been tested back since 2005 and recently has completed phase III clinical trials and announced for the treatment against intermediate and high-risk patients of myelodysplastic syndromes [207]. Details of other modified nucleotide approach and peptide nucleic acid (PNA) approach targeted against telomerase are available in the literature [210,211,212]. Various approaches regarding the development of effective telomerase immunotherapy have led to the development of useful products. The most advanced products along this line are GRNVAC1 and GV1001 (telomerase peptide vaccine). Both peptide vaccines were particularly designed to enhance an immune response to cancer cell but their efficacy in patients has not been evaluated completely [160,161]. Thus, there is an urgent need to complete evaluation of these compounds to use in combination or alone for certain types of cancer [209].
The strategies listed above offered their prospective advantages and disadvantages. The merit of targeting telomerase is the high specificity for cancer cells due to its high occurrence. Considering as a bearable risk of targeting of normal telomerase positive somatic cells, the prospect to pursue a broad-spectrum cancer therapy propel the scientific researcher over the past twenty years. Somatic cells that express telomerase, such as hematopoietic cells, greatly affected by targeting telomerase activity in progenitor cells as normal hematopoietic stem cells should be less affected by telomerase inhibition therapy than cancerous cells, which almost always have a higher rate of proliferation and thus have higher amount of telomerase activity [213].
Telomerase targeting offers two major apprehensions. The first is the time required for telomerase inhibition. Several studies in cultured cells show that effective inhibition of telomerase requiring several weeks to months to stop the proliferation of cells [214]. The second issue raised is resistance mechanisms to telomerase inhibition which lead to progressive shortening of telomere and when telomeres of cancer cells attain a critical length, a crisis phase occur, where strong selective pressure probably favor the growth of resistant cells such as those which start using alternative mechanism of telomere maintenance such as ALT [215,216,217].
5.2. Telomeres
The restrictions and prospective challenges to target telomerase enzyme while knowing the fact that inhibition of telomerase would not able to target the one-sixth of melanoma which uses ALT pathway. These challenges further propel researchers to explore telomeres as a healing target [218]. Folding of telomeric DNA into G4 structure blocks the action of telomerase through locking the telomeric single-stranded region into an inactive conformation which is no more available to recognized nor elongated by telomerase [219]. This incapability of telomerase to reach the telomere can activate telomere length-independent damage signal causing instant cell arrest or death. Formation of G4 structure not only affects the function of telomerase but it also inhibits the binding of component of both complexes (shelterin and CST) with single-stranded telomeric DNA [220,221]. For the complete replication of chromosomal ends, it requires unwinding of higher-order G4 structures carried out by a number of helicases such as BLM, WRN, and RecQ along with POT1 [222]. In normal condition, G4 function as a capping structure and its stabilization through G4 ligands block the unwinding which ultimately affects the length of telomere [223].
The G4 structures emerge as a novel therapeutic target to repress telomerase-mediated telomere regulation. The chemical compounds which can block interactions between telomerase and telomere, include G4 ligands that stabilize the G4 structures [224]. In vitro and in vivo studies of G4 ligands have shown to have a promising anticancer activity which leads to the search for small molecules that specifically interact and stabilize the G4 structure [225,226]. There are two key limitations on the use of G4 stabilizing molecules are, one is the lack of potency and second is poor selectivity between G4 and DNA duplex [227]. Telomestatin is a widely studied inhibitors reported to promote telomere shortening and to block proliferation of cancer cells in vitro and in model organisms [228]. In 2013, the first NMR structure of G4 in complex with Telomestatin was solved by Wan Jun Chung and co-worker [229]. This observation pushes forward researcher for the design and synthesis of topology specific G4-targeting compounds valuable for the development of effective anticancer drugs [229].
In consistence, RHPS4 is a pentacyclic acridinium ligand form complex with G4 through end- stacking mechanism. The π-system of RHPS4 overlaps mainly with two bases of each tetrad through stacking interactions with the G-tetrads [230,231]. Further studies show that the RHPS4 abrogates the function of telomerase and subsequently promotes telomere uncapping and finally leads to senescence [167]. The crystal structure of a 3,6,9-trisubstituted acridine, BRACO-19, in complex with G4 were solved by Campbell et al. [232]. BRACO-19 is sandwiched between two quadruplexes to form a biological unit in a way such that the G4 are uniquely stacked 5′–3′ direction. Moreover, a number of modifications on BRACO-19 have been made and those modified molecules shows strong binding affinity with G4 along with telomerase inhibitory activity and exhibits anti-proliferation property on cancer cell lines [166,192,233]. There are number of other important G4 ligands such as quinacridine analogs (MMQ1), quarfloxin, naphthalene diimides (ND), porphyrin analogue (TMPyP4) that show a strong binding with G4 together with anti-cancer activity [169,234,235,236,237]. In addition to targeting both components of telomerase and G4 structures, targeting components of shelterin and CST complexes is also useful in the treatment of telomeropathies.
Compounds targeting the telomere are actually obstructs its stability. Telomeres are found in normal as well in cancerous cells, and therefore the risk of cytotoxicity in the track of such approaches is concrete. Reports on the effects of telomere targeting in normal cells are existing mainly for G4 ligands which reveal a higher resistance in normal cells compared to cancer cells [167,238,239]. Another concern raised by the telomeres targeting by G4 ligands is that even the most specific telomeric G4 ligand retains the ability to bind other G4 structure in regulatory regions or other gene promoters of the genome [240,241].
5.3. Shelterin Components
Targeting shelterin components for cancer therapy has emerged a few years ago. POT1 is targeted by a berberine derivative (Sysu-00692) which disrupts its interaction POTI with telomeric DNA [77]. Another report suggested that TRF2 as a potential therapeutic target of cancer. A well-known anti-cancerous compound, gemcitabine, a nucleoside analog approved as anti-cancer agent [156,174,242]. Since gemcitabine is a nucleoside analog which acts via incorporating into DNA in place of cytosine and consequently inhibits DNA replication and promotes telomere loss through TRF2 stabilization [156]. Further studies reveal that a metronomic treatment with gemcitabine has anti-angiogenic effects in a pancreatic cancer model [173,243]. However, care must be taken when using drugs molecule targeting components of shelterin in anti-cancer therapy as the shelterin complex is crucial for monitoring telomere stability in normal cells as well.
Recently, two novel compounds (ETP-47228 and ETP-47037) have been identified which abrogate the function of one of the important telomeres uncapping protein, TRF1. This abrogation effectively hampers the growth of previously established lung carcinomas without disturbing tissue viability [175]. From this study, targeting of shelterin components got strong support and provides proof of the concept that abrogation of shelterin component could be a useful therapeutic approach to inhibit the growth of various types of carcinomas.
Another strategy to target telomere-associated disease is to manipulate the t-loop, which forms by the invasion of the telomere single-stranded 3′ G-overhang into the duplex chromosomal end through nucleotide base pairing [6]. One of the shelterin components, TRF2 is sufficient and makes the process easy to form t-loop [244,245]. According to three state models (hypothetical), it assumed that t-loop formation hides telomeric end to recognize as double-stranded breaks [155]. Loss of t-loop formation resulted int the activation of ATM, p53 and many other downstream molecules which finally form TIF (telomere-dysfunction induced foci) and initiate many growth inhibitory responses including senescence, cell cycle arrest, or apoptosis [246]. In addition, it has been demonstrated that the use of oligonucleotide or t-oligos having homology to the G-rich region of t-loop able to induce apoptosis, senescence, or autophagy in a range of melanoma consistent with disruption of telomere loop [177,179,247,248,249]. Even though much work has been done but still there is a strong need to understand and explore the mechanism of t-loops in chromosomal end protection in a model system.
Another but the unexplored approach in targeting telomere binding proteins subject to a range of post-translational modification (PTM) including phosphorylation, ubiquitylation, SUMOylation, PARsylation. These PTM carried out by different types of kinases such as CK2, Cdkl, Fbx4, and MMS21 Tankyrase-1 and 2), which eventually control and regulate the activity and function of telomere binding protein. The above four types of PTM (phosphorylation, ubiquitylation, SUMOylation, and PARsylation) have been found on TRF1 and TRF2, except methylation which primarily found on TRF2 protein. These modifications of TRF1 and TRF2 regulate key aspects of TRF1 and TRF2 function at the chromosomal ends such as stability of TRF1 or TRF2, telomeric DNA binding, protein–protein interactions and priming of TRF1 for subsequent modification [33,250].
Interestingly, TANK1 and 2 modifies TRF1 and start the release of TRF1 from telomere which has emerged as a therapeutic target for cancer [181,251,252]. It was demonstrated that acetylation of TRF2 through p300 control TRF2 stability and thus telomere binding. High expression of TRF2 mutant (lacking acetyl modification) promotes distorted telomeres, telomeric DDR and senescence [253]. Similarly, TPP1 also shows a strong link between PTM of TPP1 and telomere regulation [254]. As small molecule inhibitors of many key enzymes which catalyzes these modifications are available commercially. In-depth understanding of these modifications control and regulate telomere protection and synthesis which may help the researcher to change the telomere status and immortalized cancer cells through inhibiting the crucial enzymes important for PTM of telomere binding proteins. Although, the CST complex facilitates the replication of telomeric DNA, mediate C-strand fill-in, and inhibit unnecessary elongation of G-strand [83].Currently, there are no cancer therapies available that exclusively target CST complex. To come up with novel possible ways to target these complexes, some hypothetical approaches are outlined in Figure 6.
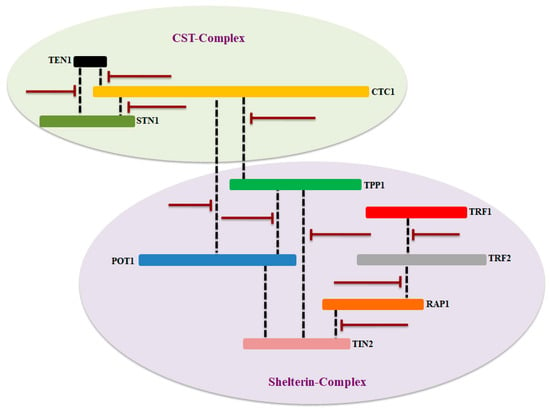
Figure 6.
Proposed key target site in CST and shelterin complex that can be targeted for therapeutic applications. Some of the key interacting partners of these complexes have been considering as an emerging targets for telomere borne diseases, while others are being investigation for therapeutic targets. The dashes line in black indicates the interaction between components of both complexes and bar-headed lines showing possible inhibition site in red.
Telomerase/telomere or inhibitors of telomere maintaining proteins might not only directly limit the growth of the cancerous cells, but could become more effective using in a synergistic fashion with existing therapeutic modalities such as with chemotherapeutic agents and anti-angiogenic agents [213,255,256].
6. Conclusions
To identify potential drug targets, knowledge from structures, biochemical features and mechanistic studies of telomerase, CST and shelterin complexes must be comprehensively employed to evaluate potential targets for translational setting against telomere borne diseases. After extensive analysis of each component of both complexes, we have identified approximately 115 clinically important mutations. Most of these mutations either affect the telomere protection or its replication process. Majority of the mutations in both complexes are implicated in several diseases including cancer, idiopathic pulmonary fibrosis, DC, CP bone marrow failure and premature aging syndromes. In addition, identification of mutations in both complexes and its association with disease raises the question that how mutation in these genes does affect the pathogenesis of different disease? Targeting telomerase with specific telomere structure (G4) to inhibit its function is a conventional strategy to fight cancer and other associated diseases. However, a great lacuna of potential drug is essentially required to address the components of CST and shelterin complexes in broader aspects. Taken together, we conclude that the emergence of innovative drug development strategies that addresses novel targets could provide a roadmap for the development of potential inhibitors with the capacity for simultaneous disruption of multiple tumor cell dependencies including those promoted by telomerase and telomere regulatory proteins (CST and shelterin complex). In addition, targeting protein–protein interaction could be useful to interrupt the interaction between components of these complexes.
Author Contributions
Conceptualization, M.A., P.K. and M.I.H.; methodology, M.A. and A.Q.; software, R.D. and A.I.; validation, P.K., A.H. and M.I.H.; formal analysis, M.F.A., F.A.; investigation, M.A. and P.K.; resources, M.F.A. and A.H.; data curation, M.A. and R.D.; writing—original draft preparation, M.A., P.K. and M.I.H.; writing—review and editing, R.D., A.I., F.A.; visualization, M.A., M.I.H.; supervision, R.D. and M.I.H.; project administration, F.A. and M.I.H.; funding acquisition, M.F.A. and A.H. All authors have read and agreed to the published version of the manuscript.
Funding
MIH is sincerely thankful to the Science & Engineering Research Board, Department of Science and Technology, Government of India, for the financial assistance (Project no: EMR/2015/002372). The APC is funded by the King Saud University, Riyadh, KSA.
Acknowledgments
M.A. is thankful to Council of Scientific and Industrial Research, India for the award of senior research fellowship (Grant number 09/466(0197)2K18 EMR-7). Aarfa Queen is thankful to Indian Council of Medical Research (ICMR) (F.No.45/63/2018-PHA/BMS/OL). PK thanks to the Department of Biotechnology (DBT) for financial support. Authors sincerely thank to the Department of Science and Technology, Government of India for the FIST support (FIST program No. SR/FST/LSI-541/2012). MFA and AH acknowledge the generous support from Research Supporting Project (No. RSP-2019-122) by King Saud University, Riyadh, Kingdom of Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T.; Shiue, L.; Myers, R.; Cox, D.; Naylor, S.; Killery, A.; Varmus, H. Structure and variability of human chromosome ends. Mol. Cell. Biol. 1990, 10, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hoshiyama, H.; Shay, J.W.; Wright, W.E. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2007, 36, e14. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, C.; Jansson, L.I.; Stone, M.D. New perspectives on telomerase RNA structure and function. Wiley Interdiscip. Rev. RNA 2018, 9, e1456. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feigon, J. Structural biology of telomerase and its interaction at telomeres. Curr. Opin. Struct. Biol. 2017, 47, 77–87. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Rice, C.; Skordalakes, E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167. [Google Scholar] [CrossRef]
- Jones, M.; Bisht, K.; Savage, S.A.; Nandakumar, J.; Keegan, C.E.; Maillard, I. The shelterin complex and hematopoiesis. J. Clin. Investig. 2016, 126, 1621–1629. [Google Scholar] [CrossRef]
- Maciejowski, J.; de Lange, T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017, 18, 175. [Google Scholar] [CrossRef]
- Amir, M.; Kumar, V.; Dohare, R.; Rehman, M.T.; Hussain, A.; Alajmi, M.F.; El-Seedi, H.R.; Hassan, H.M.A.; Islam, A.; Ahmad, F. Investigating architecture and structure-function relationships in cold shock DNA-binding domain family using structural genomics-based approach. Int. J. Biol. Macromol. 2019, 133, 484–494. [Google Scholar] [CrossRef]
- Amir, M.; Kumar, V.; Dohare, R.; Islam, A.; Ahmad, F.; Hassan, M.I. Sequence, structure and evolutionary analysis of cold shock domain proteins, a member of ob fold family. J. Evol. Biol. 2018, 31, 1903–1917. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, K.T.; Compton, S.A.; Barrientos, K.S.; Ozgur, S.; Griffith, J.D.; Counter, C.M. Tethering telomeric double-and single-stranded DNA-binding proteins inhibits telomere elongation. J. Biol. Chem. 2008, 283, 6935–6941. [Google Scholar] [CrossRef] [PubMed]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Wang, Y.; Ackerson, S.M.; Schuck, P.L. Emerging roles of cst in maintaining genome stability and human disease. Front. Biosci. (Landmark Ed.) 2018, 23, 1564–1586. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Mohammad, T.; Kumar, V.; Alajmi, M.F.; Rehman, M.T.; Hussain, A.; Alam, P.; Dohare, R.; Islam, A.; Ahmad, F. Structural analysis and conformational dynamics of stn1 gene mutations involved in coat plus syndrome. Front. Mol. Biosci. 2019, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Kumar, V.; Mohammad, T.; Dohare, R.; Rehman, M.T.; Alajmi, M.F.; Hussain, A.; Ahmad, F.; Hassan, M.I. Structural and functional impact of non-synonymous snps in the cst complex subunit ten1: Structural genomics approach. Biosci. Rep. 2019, 39, BSR20190312. [Google Scholar] [CrossRef]
- Feng, X.; Hsu, S.-J.; Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Ctc1-stn1 terminates telomerase while stn1-ten1 enables c-strand synthesis during telomere replication in colon cancer cells. Nat. Commun. 2018, 9, 2827. [Google Scholar] [CrossRef]
- Goulian, M.; Heard, C.J.; Grimm, S.L. Purification and properties of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990, 265, 13221–13230. [Google Scholar]
- Surovtseva, Y.V.; Churikov, D.; Boltz, K.A.; Song, X.; Lamb, J.C.; Warrington, R.; Leehy, K.; Heacock, M.; Price, C.M.; Shippen, D.E. Conserved telomere maintenance component 1 interacts with stn1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 2009, 36, 207–218. [Google Scholar] [CrossRef]
- Stewart, J.A.; Wang, F.; Chaiken, M.F.; Kasbek, C.; Chastain, P.D.; Wright, W.E.; Price, C.M. Human cst promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012, 31, 3537–3549. [Google Scholar] [CrossRef]
- Gu, P.; Min, J.N.; Wang, Y.; Huang, C.; Peng, T.; Chai, W.; Chang, S. Ctc1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012, 31, 2309–2321. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Ahamad, S.; Mohammad, T.; Jairajpuri, D.S.; Hasan, G.M.; Dohare, R.; Islam, A.; Ahmad, F.; Hassan, M.I. Investigation of conformational dynamics of tyr89cys mutation in protection of telomeres 1 gene associated with familial melanoma. J. Biomol. Struct. Dyn. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Ahmad, S.; Ahamad, S.; Kumar, V.; Mohammad, T.; Dohare, R.; Alajmi, M.F.; Rehman, T.; Hussain, A.; Islam, A. Impact of gln94glu mutation on the structure and function of protection of telomere 1, a cause of cutaneous familial melanoma. J. Biomol. Struct. Dyn. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Kumar, V.; Mohammad, T.; Dohare, R.; Hussain, A.; Rehman, M.T.; Alam, P.; Alajmi, M.F.; Islam, A.; Ahmad, F. Investigation of deleterious effects of nssnps in the pot1 gene: A structural genomics-based approach to understand the mechanism of cancer development. J. Cell. Biochem. 2018, 120, 10281–10294. [Google Scholar] [CrossRef]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct myb–related proteins, trf1 and trf2. Nat. Genet. 1997, 17, 231. [Google Scholar] [CrossRef]
- Chapman, L.; Fairall, L.; Rhodes, D. How the human telomeric proteins trf1 and trf2 recognize telomeric DNA: A view from high-resolution crystal structures. EMBO Rep. 2005, 6, 39–45. [Google Scholar]
- Chen, Y.; Yang, Y.; van Overbeek, M.; Donigian, J.R.; Baciu, P.; de Lange, T.; Lei, M. A shared docking motif in trf1 and trf2 used for differential recruitment of telomeric proteins. Science 2008, 319, 1092–1096. [Google Scholar] [CrossRef]
- Van Steensel, B.; De Lange, T. Control of telomere length by the human telomeric protein trf1. Nature 1997, 385, 740–743. [Google Scholar] [CrossRef]
- Fairall, L.; Chapman, L.; Moss, H.; de Lange, T.; Rhodes, D. Structure of the trfh dimerization domain of the human telomeric proteins trf1 and trf2. Mol. Cell 2001, 8, 351–361. [Google Scholar] [CrossRef]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require trf1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef]
- Martínez, P.; Thanasoula, M.; Muñoz, P.; Liao, C.; Tejera, A.; McNees, C.; Flores, J.M.; Fernández-Capetillo, O.; Tarsounas, M.; Blasco, M.A. Increased telomere fragility and fusions resulting from trf1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009, 23, 2060–2075. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S.; Zhou, X.Z.; Ziv, Y.; Khoo, C.; Hill, D.E.; Shiloh, Y.; Lu, K.P. Telomeric protein pin2/trf1 as an important atm target in response to double strand DNA breaks. J. Biol. Chem. 2001, 276, 29282–29291. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.R.; Zhu, X.-D. Post-translational modifications of trf1 and trf2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012, 133, 421–434. [Google Scholar] [CrossRef]
- Konishi, A.; de Lange, T. Cell cycle control of telomere protection and nhej revealed by a ts mutation in the DNA-binding domain of trf2. Genes Dev. 2008, 22, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, Y.; Zou, M.; Yang, S.; Liang, X.-Q. Expression of trf1, trf2, tin2, tert, ku70, and brca1 proteins is associated with telomere shortening and may contribute to multistage carcinogenesis of gastric cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 1407–1414. [Google Scholar] [CrossRef]
- Raynaud, C.; Jang, S.-J.; Nuciforo, P.; Lantuejoul, S.; Brambilla, E.; Mounier, N.; Olaussen, K.; André, F.; Morat, L.; Sabatier, L. Telomere shortening is correlated with the DNA damage response and telomeric protein down-regulation in colorectal preneoplastic lesions. Ann. Oncol. 2008, 19, 1875–1881. [Google Scholar] [CrossRef]
- Yamada, M.; Tsuji, N.; Nakamura, M.; Moriai, R.; Kobayashi, D.; Yagihashi, A.; Watanabe, N. Down-regulation of trf1, trf2 and tin2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2002, 22, 3303–3307. [Google Scholar]
- Martínez, P.; Gómez-López, G.; Pisano, D.G.; Flores, J.M.; Blasco, M.A. A genetic interaction between rap 1 and telomerase reveals an unanticipated role for rap 1 in telomere maintenance. Aging Cell 2016, 15, 1113–1125. [Google Scholar] [CrossRef]
- Li, B.; Oestreich, S.; De Lange, T. Identification of human rap1: Implications for telomere evolution. Cell 2000, 101, 471–483. [Google Scholar] [CrossRef]
- Hanaoka, S.; Nagadoi, A.; Yoshimura, S.; Aimoto, S.; Li, B.; De Lange, T.; Nishimura, Y. Nmr structure of the hrap1 myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of myb DNA-binding domains. J. Mol. Biol. 2001, 312, 167–175. [Google Scholar] [CrossRef]
- Pardo, B.; Marcand, S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005, 24, 3117–3127. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; Gómez-López, G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.G.; Tarsounas, M.; Blasco, M.A. Mammalian rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Aoude, L.G.; Pritchard, A.L.; Robles-Espinoza, C.D.; Wadt, K.; Harland, M.; Choi, J.; Gartside, M.; Quesada, V.; Johansson, P.; Palmer, J.M. Nonsense mutations in the shelterin complex genes acd and terf2ip in familial melanoma. J. Natl. Cancer Inst. 2015, 107, dju408. [Google Scholar] [CrossRef] [PubMed]
- Speedy, H.E.; Kinnersley, B.; Chubb, D.; Broderick, P.; Law, P.J.; Litchfield, K.; Jayne, S.; Dyer, M.J.; Dearden, C.; Follows, G.A. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 2016, 128, 2319–2326. [Google Scholar] [CrossRef]
- Kim, S.-h.; Kaminker, P.; Campisi, J. Tin2, a new regulator of telomere length in human cells. Nat. Genet. 1999, 23, 405. [Google Scholar] [CrossRef]
- Ye, J.Z.-S.; Donigian, J.R.; van Overbeek, M.; Loayza, D.; Luo, Y.; Krutchinsky, A.N.; Chait, B.T.; de Lange, T. Tin2 binds trf1 and trf2 simultaneously and stabilizes the trf2 complex on telomeres. J. Biol. Chem. 2004, 279, 47264–47271. [Google Scholar] [CrossRef]
- Frescas, D.; de Lange, T. Binding of tpp1 protein to tin2 protein is required for pot1a, b protein-mediated telomere protection. J. Biol. Chem. 2014, 289, 24180–24187. [Google Scholar] [CrossRef]
- Frescas, D.; de Lange, T. Trf2-tethered tin2 can mediate telomere protection by tpp1/pot1. Mol. Cell. Biol. 2014, 34, 1349–1362. [Google Scholar] [CrossRef]
- O’Connor, M.S.; Safari, A.; Xin, H.; Liu, D.; Songyang, Z. A critical role for tpp1 and tin2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11874–11879. [Google Scholar] [CrossRef]
- Takai, K.K.; Kibe, T.; Donigian, J.R.; Frescas, D.; de Lange, T. Telomere protection by tpp1/pot1 requires tethering to tin2. Mol. Cell 2011, 44, 647–659. [Google Scholar] [CrossRef]
- Frank, A.K.; Tran, D.C.; Qu, R.W.; Stohr, B.A.; Segal, D.J.; Xu, L. The shelterin tin2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 2015, 11, e1005410. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Zhang, Y.; Zhang, Q.; Li, H.; Luo, Z.; Fang, H.; Kim, S.H.; Qin, L.; Yotnda, P.; Xu, J. Mitochondrial localization of telomeric protein tin2 links telomere regulation to metabolic control. Mol. Cell 2012, 47, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Abreu, E.; Aritonovska, E.; Reichenbach, P.; Cristofari, G.; Culp, B.; Terns, R.M.; Lingner, J.; Terns, M.P. Tin2-tethered tpp1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010, 30, 2971–2982. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Beausejour, C.; Davalos, A.R.; Kaminker, P.; Heo, S.-J.; Campisi, J. Tin2 mediates functions of trf2 at human telomeres. J. Biol. Chem. 2004, 279, 43799–43804. [Google Scholar] [CrossRef] [PubMed]
- Panichareon, B.; Seedapan, T.; Thongnoppakhun, W.; Limwongse, C.; Pithukpakorn, M.; Limjindaporn, T. Novel mutation of the tinf2 gene in a patient with dyskeratosis congenita. Case Rep. Dermatol. 2015, 7, 212–219. [Google Scholar] [CrossRef]
- Hoffman, T.; van der Vis, J.; van Oosterhout, M.; van Es, H.; van Kessel, D.; Grutters, J.; van Moorsel, C. Tinf2 gene mutation in a patient with pulmonary fibrosis. Case Rep. Pulmonol. 2016, 2016, 1310862. [Google Scholar]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The pot1–tpp1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506. [Google Scholar] [CrossRef]
- Nandakumar, J.; Bell, C.F.; Weidenfeld, I.; Zaug, A.J.; Leinwand, L.A.; Cech, T.R. The tel patch of telomere protein tpp1 mediates telomerase recruitment and processivity. Nature 2012, 492, 285. [Google Scholar] [CrossRef]
- Latrick, C.M.; Cech, T.R. Pot1–tpp1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010, 29, 924–933. [Google Scholar] [CrossRef]
- Taylor, D.J.; Podell, E.R.; Taatjes, D.J.; Cech, T.R. Multiple pot1–tpp1 proteins coat and compact long telomeric single-stranded DNA. J. Mol. Biol. 2011, 410, 10–17. [Google Scholar] [CrossRef]
- Kanamaru, S.; Leiman, P.G.; Kostyuchenko, V.A.; Chipman, P.R.; Mesyanzhinov, V.V.; Arisaka, F.; Rossmann, M.G. Structure of the cell-puncturing device of bacteriophage t4. Nature 2002, 415, 553. [Google Scholar] [CrossRef] [PubMed]
- Bochkareva, E.; Korolev, S.; Lees-Miller, S.P.; Bochkarev, A. Structure of the rpa trimerization core and its role in the multistep DNA-binding mechanism of rpa. EMBO J. 2002, 21, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Sexton, A.N.; Youmans, D.T.; Collins, K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 2012, 287, 34455–34464. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.L.; Batista, L.F.; Freund, A.; Pech, M.F.; Venteicher, A.S.; Artandi, S.E. Tpp1 ob-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 2012, 150, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Wan, M.; Safari, A.; Kim, H.; Sun, W.; O’Connor, M.S.; Songyang, Z. Tpp1 is a homologue of ciliate tebp-β and interacts with pot1 to recruit telomerase. Nature 2007, 445, 559. [Google Scholar] [CrossRef]
- Chen, Y. The structural biology of the shelterin complex. Biol. Chem. 2019, 400, 457–466. [Google Scholar] [CrossRef]
- Rice, C.; Shastrula, P.K.; Kossenkov, A.V.; Hills, R.; Baird, D.M.; Showe, L.C.; Doukov, T.; Janicki, S.; Skordalakes, E. Structural and functional analysis of the human pot1-tpp1 telomeric complex. Nat. Commun. 2017, 8, 14928. [Google Scholar] [CrossRef]
- Lei, M.; Podell, E.R.; Baumann, P.; Cech, T.R. DNA self-recognition in the structure of pot1 bound to telomeric single-stranded DNA. Nature 2003, 426, 198. [Google Scholar] [CrossRef]
- Nandakumar, J.; Cech, T.R. DNA-induced dimerization of the single-stranded DNA binding telomeric protein pot1 from schizosaccharomyces pombe. Nucleic Acids Res. 2011, 40, 235–244. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human pot1 disrupts telomeric g-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef]
- Murzin, A.G. Ob (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861. [Google Scholar] [CrossRef] [PubMed]
- Theobald, D.L.; Mitton-Fry, R.M.; Wuttke, D.S. Nucleic acid recognition by ob-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Podell, E.R.; Cech, T.R. Structure of human pot1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004, 11, 1223. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Hockemeyer, D.; Kibe, T.; De Lange, T. Functional dissection of human and mouse pot1 proteins. Mol. Cell. Biol. 2009, 29, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Loayza, D.; De Lange, T. Pot1 as a terminal transducer of trf1 telomere length control. Nature 2003, 423, 1013. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, C.; Kurth, I.; Lingner, J. Human protection of telomeres 1 (pot1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 2005, 25, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Chen, S.; Ma, Y.; Qiu, J.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S.; Li, D. Interaction of berberine derivative with protein pot1 affect telomere function in cancer cells. Biochem. Biophys. Res. Commun. 2012, 419, 567–572. [Google Scholar] [CrossRef]
- Wong, K.; Robles-Espinoza, C.D.; Rodriguez, D.; Rudat, S.S.; Puig, S.; Potrony, M.; Wong, C.C.; Hewinson, J.; Aguilera, P.; Puig-Butille, J.A. Association of the pot1 germline missense variant p. I78t with familial melanoma. JAMA Dermatol. 2019, 155, 604–609. [Google Scholar] [CrossRef]
- Armanios, M. An emerging role for the conserved telomere component 1 (ctc1) in human genetic disease. Pediatr. Blood Cancer 2012, 59, 209–210. [Google Scholar] [CrossRef]
- Vulliamy, T.; Marrone, A.; Dokal, I.; Mason, P.J. Association between aplastic anaemia and mutations in telomerase RNA. Lancet 2002, 359, 2168–2170. [Google Scholar] [CrossRef]
- Calado, R.T.; Young, N.S. Telomere diseases. N. Engl. J. Med. 2009, 361, 2353–2365. [Google Scholar] [CrossRef] [PubMed]
- Bryan, C.; Rice, C.; Harkisheimer, M.; Schultz, D.C.; Skordalakes, E. Structure of the human telomeric stn1-ten1 capping complex. PLoS ONE 2013, 8, e66756. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Redon, S.; Lingner, J. The human cst complex is a terminator of telomerase activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.J.; Lev, A.; Zhang, Y.; Weiss, B.; Rylova, A.; Eyal, E.; Kol, N.; Barel, O.; Cesarkas, K.; Soudack, M. Mutations in stn1 cause coats plus syndrome and are associated with genomic and telomere defects. J. Exp. Med. 2016, 213, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Wang, S.C.; Prahash, A.; Sano, M.; Moravec, C.S.; Taffet, G.E.; Michael, L.H.; Youker, K.A.; Entman, M.L.; Schneider, M.D. Telomere attrition and chk2 activation in human heart failure. Proc. Natl. Acad. Sci. USA 2003, 100, 5378–5383. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; Bronner, M.P.; Brentnall, T.A.; Finley, J.C.; Shen, W.-T.; Emerson, S.; Emond, M.J.; Gollahon, K.A.; Moskovitz, A.H.; Crispin, D.A. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat. Genet. 2002, 32, 280–284. [Google Scholar] [CrossRef]
- Wiemann, S.U.; SATYANARAYANA, A.; Tsahuridu, M.; Tillmann, H.L.; Zender, L.; Klempnauer, J.; Flemming, P.; Franco, S.; Blasco, M.A.; Manns, M.P. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002, 16, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Boultby, R.; Butler, R.; Thompson, J.R.; Goodall, A.H. Telomere shortening in atherosclerosis. Lancet 2001, 358, 472–473. [Google Scholar] [CrossRef]
- Glousker, G.; Touzot, F.; Revy, P.; Tzfati, Y.; Savage, S.A. Unraveling the pathogenesis of hoyeraal–hreidarsson syndrome, a complex telomere biology disorder. Br. J. Haematol. 2015, 170, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Podlevsky, J.D.; Bley, C.J.; Omana, R.V.; Qi, X.; Chen, J.J.-L. The telomerase database. Nucleic Acids Res. 2008, 36, D339–D343. [Google Scholar] [CrossRef]
- Savage, S.A.; Giri, N.; Baerlocher, G.M.; Orr, N.; Lansdorp, P.M.; Alter, B.P. Tinf2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008, 82, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Walne, A.J.; Vulliamy, T.; Beswick, R.; Kirwan, M.; Dokal, I. Tinf2 mutations result in very short telomeres: Analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 2008, 112, 3594–3600. [Google Scholar] [CrossRef] [PubMed]
- Sasa, G.S.; Ribes-Zamora, A.; Nelson, N.D.; Bertuch, A.A. Three novel truncating tinf2 mutations causing severe dyskeratosis congenita in early childhood. Clin. Genet. 2012, 81, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Spinella, J.-F.; Cassart, P.; Garnier, N.; Rousseau, P.; Drullion, C.; Richer, C.; Ouimet, M.; Saillour, V.; Healy, J.; Autexier, C. A novel somatic mutation in acd induces telomere lengthening and apoptosis resistance in leukemia cells. BMC Cancer 2015, 15, 621. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kocak, H.; Ballew, B.J.; Bisht, K.; Eggebeen, R.; Hicks, B.D.; Suman, S.; O’Neil, A.; Giri, N.; Maillard, I.; Alter, B.P. Hoyeraal-hreidarsson syndrome caused by a germline mutation in the tel patch of the telomere protein tpp1. Genes Dev. 2014, 28, 2090–2102. [Google Scholar] [CrossRef] [PubMed]
- Robles-Espinoza, C.D.; Harland, M.; Ramsay, A.J.; Aoude, L.G.; Quesada, V.; Ding, Z.; Pooley, K.A.; Pritchard, A.L.; Tiffen, J.C.; Petljak, M. Pot1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014, 46, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.R.; Ballew, B.; Rotunno, M.; Calista, D.; Fargnoli, M.C.; Ghiorzo, P.; Bressac-de Paillerets, B.; Nagore, E.; Avril, M.F. Rare missense variants in pot1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 2014, 46, 482–486. [Google Scholar] [CrossRef]
- Bainbridge, M.N.; Armstrong, G.N.; Gramatges, M.M.; Bertuch, A.A.; Jhangiani, S.N.; Doddapaneni, H.; Lewis, L.; Tombrello, J.; Tsavachidis, S.; Liu, Y. Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst. 2015, 107, dju384. [Google Scholar] [CrossRef]
- Calvete, O.; Martinez, P.; Garcia-Pavia, P.; Benitez-Buelga, C.; Paumard-Hernández, B.; Fernandez, V.; Dominguez, F.; Salas, C.; Romero-Laorden, N.; Garcia-Donas, J. A mutation in the pot1 gene is responsible for cardiac angiosarcoma in tp53-negative li-fraumeni-like families. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Takai, H.; Jenkinson, E.; Kabir, S.; Babul-Hirji, R.; Najm-Tehrani, N.; Chitayat, D.A.; Crow, Y.J.; de Lange, T. A pot1 mutation implicates defective telomere end fill-in and telomere truncations in coats plus. Genes Dev. 2016, 30, 812–826. [Google Scholar] [CrossRef]
- Ramsay, A.J.; Quesada, V.; Foronda, M.; Conde, L.; Martínez-Trillos, A.; Villamor, N.; Rodríguez, D.; Kwarciak, A.; Garabaya, C.; Gallardo, M. Pot1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013, 45, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.; Conde, L.; Villamor, N.; Ordóñez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Beà, S.; Pinyol, M.; Martínez-Trillos, A. Exome sequencing identifies recurrent mutations of the splicing factor sf3b1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 47. [Google Scholar] [CrossRef]
- Anderson, B.H.; Kasher, P.R.; Mayer, J.; Szynkiewicz, M.; Jenkinson, E.M.; Bhaskar, S.S.; Urquhart, J.E.; Daly, S.B.; Dickerson, J.E.; O’Sullivan, J. Mutations in ctc1, encoding conserved telomere maintenance component 1, cause coats plus. Nat. Genet. 2012, 44, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Majerská, J.; Lingner, J. Molecular basis of telomere syndrome caused by ctc1 mutations. Genes Dev. 2013, 27, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Chang, S. Functional characterization of human ctc1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease coats plus. Aging Cell 2013, 12, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Netravathi, M.; Kumari, R.; Kapoor, S.; Dakle, P.; Dwivedi, M.K.; Roy, S.D.; Pandey, P.; Saini, J.; Ramakrishna, A.; Navalli, D. Whole exome sequencing in an indian family links coats plus syndrome and dextrocardia with a homozygous novel ctc1 and a rare hes7 variation. BMC Med. Genet. 2015, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Bisserbe, A.; Tertian, G.; Buffet, C.; Turhan, A.; Lambotte, O.; Nasser, G.; Alvin, P.; Tardieu, M.; Riant, F.; Bergametti, F. Cerebro-retinal microangiopathy with calcifications and cysts due to recessive mutations in the ctc1 gene. Rev. Neurol. 2015, 171, 445–449. [Google Scholar] [CrossRef]
- Keller, R.B.; Gagne, K.E.; Usmani, G.N.; Asdourian, G.K.; Williams, D.A.; Hofmann, I.; Agarwal, S. Ctc1 mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer 2012, 59, 311–314. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Stewart, J.; Chaiken, M.; Price, C.M. Stn1 ob fold mutation alters DNA binding and affects selective aspects of cst function. PLoS Genet. 2016, 12, e1006342. [Google Scholar] [CrossRef]
- Martínez, P.; Blasco, M.A. Role of shelterin in cancer and aging. Aging Cell 2010, 9, 653–666. [Google Scholar] [CrossRef]
- Patel, T.; Vasan, R.; Gupta, D.; Patel, J.; Trivedi, M. Shelterin proteins and cancer. Asian Pac. J. Cancer Prev. 2015, 16, 3085–3090. [Google Scholar] [CrossRef] [PubMed]
- Slavutsky, I. Shelterin genes, germ line mutations and chronic lymphocytic leukemia. Transl. Cancer Res. 2017, 6, S68–S71. [Google Scholar] [CrossRef]
- Gardner, E.; Bailey, M.; Schulz, A.; Aristorena, M.; Miller, N.; Mole, S.E. Mutation update: Review of tpp1 gene variants associated with neuronal ceroid lipofuscinosis cln2 disease. Hum. Mutat. 2019, 40, 1924–1938. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Kiselar, J.; Whited, T.L.; Hernandez-Sanchez, W.; Taylor, D.J. Pot1-tpp1 differentially regulates telomerase via pot1 his266 and as a function of single-stranded telomere DNA length. Proc. Natl. Acad. Sci. USA 2019, 116, 23527–23533. [Google Scholar] [CrossRef] [PubMed]
- Johansson, G.; Andersson, U.; Melin, B. Recent developments in brain tumor predisposing syndromes. Acta Oncol. 2016, 55, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.M.; Wiencke, J.K.; Lachance, D.H.; Wiemels, J.L.; Molinaro, A.M.; Eckel-Passow, J.E.; Jenkins, R.B.; Wrensch, M.R. Telomere maintenance and the etiology of adult glioma. Neuro-Oncology 2015, 17, 1445–1452. [Google Scholar] [CrossRef]
- Bagcchi, S. Pot1: A genetic link for familial glioma. Lancet Oncol. 2015, 16, e12. [Google Scholar] [CrossRef]
- Hosokawa, K.; Arai, F. The role of telomere binding molecules for normal and abnormal hematopoiesis. Int. J. Hematol. 2018, 107, 646–655. [Google Scholar] [CrossRef]
- Goldman, F.D.; Aubert, G.; Klingelhutz, A.J.; Hills, M.; Cooper, S.R.; Hamilton, W.S.; Schlueter, A.J.; Lambie, K.; Eaves, C.J.; Lansdorp, P.M. Characterization of primitive hematopoietic cells from patients with dyskeratosis congenita. Blood 2008, 111, 4523–4531. [Google Scholar] [CrossRef]
- Townsley, D.M.; Dumitriu, B.; Young, N.S. Bone marrow failure and the telomeropathies. Blood 2014, 124, 2775–2783. [Google Scholar] [CrossRef]
- Walne, A.J.; Vulliamy, T.; Marrone, A.; Beswick, R.; Kirwan, M.; Masunari, Y.; Al-Qurashi, F.-H.; Aljurf, M.; Dokal, I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein nop10. Hum. Mol. Genet. 2007, 16, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Ballew, B.J.; Yeager, M.; Jacobs, K.; Giri, N.; Boland, J.; Burdett, L.; Alter, B.P.; Savage, S.A. Germline mutations of regulator of telomere elongation helicase 1, rtel1, in dyskeratosis congenita. Hum. Genet. 2013, 132, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.D.; Dodson, L.M.; Escudero, L.; Sukumar, A.T.; Williams, C.L.; Mihalek, I.; Baldan, A.; Baird, D.M.; Bertuch, A.A. The c-terminal extension unique to the long isoform of the shelterin component tin2 enhances its interaction with trf2 in a phosphorylation-and dyskeratosis congenita-cluster-dependent fashion. Mol. Cell. Biol. 2018, 38, e00025-18. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahhab, M.A.; Aljawish, A.; Kenawy, A.M.; El-Nekeety, A.A.; Hamed, H.S.; Abdel-Aziem, S.H. Grafting of gallic acid onto chitosan nano particles enhances antioxidant activities in vitro and protects against ochratoxin a toxicity in catfish (clarias gariepinus). Environ. Toxicol. Pharm. 2016, 41, 279–288. [Google Scholar] [CrossRef]
- Touzot, F.; Gaillard, L.; Vasquez, N.; Le Guen, T.; Bertrand, Y.; Bourhis, J.; Leblanc, T.; Fischer, A.; Soulier, J.; de Villartay, J.-P. Heterogeneous telomere defects in patients with severe forms of dyskeratosis congenita. J. Allergy Clin. Immunol. 2012, 129, 473–482.e473. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2012, 2, 116. [Google Scholar] [CrossRef]
- Yang, D.; He, Q.; Kim, H.; Ma, W.; Songyang, Z. Tin2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J. Biol. Chem. 2011, 286, 23022–23030. [Google Scholar] [CrossRef] [PubMed]
- Frescas, D.; de Lange, T. A tin2 dyskeratosis congenita mutation causes telomerase-independent telomere shortening in mice. Genes Dev. 2014, 28, 153–166. [Google Scholar] [CrossRef]
- Canudas, S.; Houghtaling, B.R.; Bhanot, M.; Sasa, G.; Savage, S.A.; Bertuch, A.A.; Smith, S. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 2011, 25, 1807–1819. [Google Scholar] [CrossRef]
- Guo, Y.; Kartawinata, M.; Li, J.; Pickett, H.A.; Teo, J.; Kilo, T.; Barbaro, P.M.; Keating, B.; Chen, Y.; Tian, L. Inherited bone marrow failure associated with germline mutation of acd, the gene encoding telomere protein tpp1. Blood 2014, 124, 2767–2774. [Google Scholar] [CrossRef]
- Sexton, A.N.; Regalado, S.G.; Lai, C.S.; Cost, G.J.; O’Neil, C.M.; Urnov, F.D.; Gregory, P.D.; Jaenisch, R.; Collins, K.; Hockemeyer, D. Genetic and molecular identification of three human tpp1 functions in telomerase action: Recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014, 28, 1885–1899. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Kerr, C.M.; Przychozen, B.; Mahfouz, R.Z.; LaFramboise, T.; Nagata, Y.; Hanna, R.; Radivoyevitch, T.; Nazha, A.; Sekeres, M.A. Impact of germline ctc 1 alterations on telomere length in acquired bone marrow failure. Br. J. Haematol. 2019, 185, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chai, W. Pathogenic ctc1 mutations cause global genome instabilities under replication stress. Nucleic Acids Res. 2018, 46, 3981–3992. [Google Scholar] [CrossRef] [PubMed]
- Walne, A.J.; Bhagat, T.; Kirwan, M.; Gitiaux, C.; Desguerre, I.; Leonard, N.; Nogales, E.; Vulliamy, T.; Dokal, I.S. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 2013, 98, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Casteel, D.E.; Zhuang, S.; Zeng, Y.; Perrino, F.W.; Boss, G.R.; Goulian, M.; Pilz, R.B. A DNA polymerase-α· primase cofactor with homology to replication protein a-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 2009, 284, 5807–5818. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Zheng, H.; He, H.; Luo, Y.; Multani, A.; Carpenter, P.B.; Chang, S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010, 29, 2598–2610. [Google Scholar] [CrossRef]
- Allsopp, R.C.; Chang, E.; Kashefi-Aazam, M.; Rogaev, E.I.; Piatyszek, M.A.; Shay, J.W.; Harley, C.B. Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res. 1995, 220, 194–200. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.-q.; Chen, Z.; Zheng, K.-w.; Chen, C.-y.; Hao, Y.-h.; Tan, Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef]
- Denchi, E.L.; de Lange, T. Protection of telomeres through independent control of atm and atr by trf2 and pot1. Nature 2007, 448, 1068. [Google Scholar] [CrossRef]
- Sarek, G.; Marzec, P.; Margalef, P.; Boulton, S.J. Molecular basis of telomere dysfunction in human genetic diseases. Nat. Struct. Mol. Biol. 2015, 22, 867. [Google Scholar] [CrossRef]
- Pascolo, E.; Wenz, C.; Lingner, J.; Hauel, N.; Priepke, H.; Kauffmann, I.; Garin-Chesa, P.; Rettig, W.J.; Damm, K.; Schnapp, A. Mechanism of human telomerase inhibition by bibr1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002, 277, 15566–15572. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, A.; Manns, M.P.; Rudolph, K.L. Telomeres, telomerase and cancer: An endless search to target the ends. Cell Cycle 2004, 3, 1136–1148. [Google Scholar] [CrossRef]
- Damm, K.; Hemmann, U.; Garin-Chesa, P.; Hauel, N.; Kauffmann, I.; Priepke, H.; Niestroj, C.; Daiber, C.; Enenkel, B.; Guilliard, B. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001, 20, 6958–6968. [Google Scholar] [CrossRef] [PubMed]
- Bashash, D.; H Ghaffari, S.; Zaker, F.; Kazerani, M.; Hezave, K.; Hassani, S.; Rostami, M.; Alimoghaddam, K.; Ghavamzadeh, A. Bibr 1532 increases arsenic trioxide-mediated apoptosis in acute promyelocytic leukemia cells: Therapeutic potential for apl. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem. Anti-Cancer Agents) 2013, 13, 1115–1125. [Google Scholar]
- Meng, E.; Taylor, B.; Ray, A.; Shevde, L.A.; Rocconi, R.P. Targeted inhibition of telomerase activity combined with chemotherapy demonstrates synergy in eliminating ovarian cancer spheroid-forming cells. Gynecol. Oncol. 2012, 124, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Bashash, D.; Zareii, M.; Safaroghli-Azar, A.; Omrani, M.D.; Ghaffari, S.H. Inhibition of telomerase using bibr1532 enhances doxorubicin-induced apoptosis in pre-b acute lymphoblastic leukemia cells. Hematology 2017, 22, 330–340. [Google Scholar] [CrossRef]
- Shi, Y.; Sun, L.; Chen, G.; Zheng, D.; Li, L.; Wei, W. A combination of the telomerase inhibitor, bibr1532, and paclitaxel synergistically inhibit cell proliferation in breast cancer cell lines. Target. Oncol. 2015, 10, 565–573. [Google Scholar] [CrossRef]
- Hayakawa, N.; Nozawa, K.; Ogawa, A.; Kato, N.; Yoshida, K.; Akamatsu, K.-I.; Tsuchiya, M.; Nagasaka, A.; Yoshida, S. Isothiazolone derivatives selectively inhibit telomerase from human and rat cancer cells in vitro. Biochemistry 1999, 38, 11501–11507. [Google Scholar] [CrossRef]
- Naasani, I.; Seimiya, H.; Tsuruo, T. Telomerase inhibition, telomere shortening, and senescence of cancer cells by tea catechins. Biochem. Biophys. Res. Commun. 1998, 249, 391–396. [Google Scholar] [CrossRef]
- Seimiya, H.; Oh-hara, T.; Suzuki, T.; Naasani, I.; Shimazaki, T.; Tsuchiya, K.; Tsuruo, T. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors mst-312, mst-295, and mst-199 1 supported in part by a grant-in-aid for scientific research on priority areas from the ministry of education, culture, sports, science and technology, japan. 1. Mol. Cancer Ther. 2002, 1, 657–665. [Google Scholar]
- Gomez, D.E.; Armando, R.G.; Alonso, D.F. Azt as a telomerase inhibitor. Front. Oncol. 2012, 2, 113. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Nagai, N.; Shigemasa, K.; Ohama, K. Inhibition of telomerase activity and cell proliferation by a reverse transcriptase inhibitor in gynaecological cancer cell lines. Eur. J. Cancer 1999, 35, 1027–1034. [Google Scholar] [CrossRef]
- Armando, R.G.; Gomez, D.M.; Gomez, D.E. Azt exerts its antitumoral effect by telomeric and non-telomeric effects in a mammary adenocarcinoma model. Oncol. Rep. 2016, 36, 2731–2736. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Oshima, Y.; Yamamoto, Y.; Uochi, T.-A.; Kusaka, H.; Akinaga, S.; Yamashita, Y.; Pongracz, K.; Pruzan, R.; Wunder, E. A novel telomerase template antagonist (grn163) as a potential anticancer agent. Cancer Res. 2003, 63, 3931–3939. [Google Scholar] [PubMed]
- Cesare, A.J.; Karlseder, J. A three-state model of telomere control over human proliferative boundaries. Curr. Opin. Cell Biol. 2012, 24, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-H.; Chu, W.-C.; Lan, K.-H.; Li, C.-P.; Chao, Y.; Lin, H.-C.; Lee, S.-D.; Tsai, Y.-C.; Lee, W.-P. Gemcitabine causes telomere attrition by stabilizing trf2. Eur. J. Cancer 2012, 48, 3465–3474. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, J.; Yang, S.; Kuang, Z.; Tan, G.; Yang, G.; Wei, Q.; Guo, Z. Telomerase antagonist imetelstat increases radiation sensitivity in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 13600–13619. [Google Scholar] [CrossRef]
- Frink, R.E.; Peyton, M.; Schiller, J.H.; Gazdar, A.F.; Shay, J.W.; Minna, J.D. Telomerase inhibitor imetelstat has preclinical activity across the spectrum of non-small cell lung cancer oncogenotypes in a telomere length dependent manner. Oncotarget 2016, 7, 31639. [Google Scholar] [CrossRef]
- Salloum, R.; Hummel, T.R.; Kumar, S.S.; Dorris, K.; Li, S.; Lin, T.; Daryani, V.M.; Stewart, C.F.; Miles, L.; Poussaint, T.Y. A molecular biology and phase ii study of imetelstat (grn163l) in children with recurrent or refractory central nervous system malignancies: A pediatric brain tumor consortium study. J. Neuro-Oncol. 2016, 129, 443–451. [Google Scholar] [CrossRef]
- Chiappori, A.; Kolevska, T.; Spigel, D.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M. A randomized phase ii study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef]
- Middleton, G.; Silcocks, P.; Cox, T.; Valle, J.; Wadsley, J.; Propper, D.; Coxon, F.; Ross, P.; Madhusudan, S.; Roques, T. Gemcitabine and capecitabine with or without telomerase peptide vaccine gv1001 in patients with locally advanced or metastatic pancreatic cancer (telovac): An open-label, randomised, phase 3 trial. Lancet Oncol. 2014, 15, 829–840. [Google Scholar] [CrossRef]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Okabe, S.; Ohyashiki, J.; Ohyashiki, K. Telomerase inhibition with a novel g-quadruplex-interactive agent, telomestatin: In vitro and in vivo studies in acute leukemia. Oncogene 2006, 25, 5719–5725. [Google Scholar] [CrossRef] [PubMed]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel g-quadruplex-interactive telomerase inhibitor, telomestatin (sot-095), against human leukemia cells: Involvement of atm-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Maleki, P.; Ma, Y.; Iida, K.; Nagasawa, K.; Balci, H. A single molecule study of a fluorescently labeled telomestatin derivative and g-quadruplex interactions. Nucleic Acids Res. 2017, 45, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Artese, A.; Costa, G.; Ortuso, F.; Parrotta, L.; Alcaro, S. Identification of new natural DNA g-quadruplex binders selected by a structure-based virtual screening approach. Molecules 2013, 18, 12051–12070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Liu, X.; Li, Y.; Xu, S.; Ma, C.; Wu, X.; Cheng, Y.; Yu, Z.; Zhao, G.; Chen, Y. Telomere targeting with a novel g-quadruplex-interactive ligand braco-19 induces t-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget 2016, 7, 14925–14939. [Google Scholar] [CrossRef] [PubMed]
- Berardinelli, F.; Siteni, S.; Tanzarella, C.; Stevens, M.; Sgura, A.; Antoccia, A. The g-quadruplex-stabilising agent rhps4 induces telomeric dysfunction and enhances radiosensitivity in glioblastoma cells. DNA Repair 2015, 25, 104–115. [Google Scholar] [CrossRef]
- Lagah, S.; Tan, I.-L.; Radhakrishnan, P.; Hirst, R.A.; Ward, J.H.; O’Callaghan, C.; Smith, S.J.; Stevens, M.F.; Grundy, R.G.; Rahman, R. Rhps4 g-quadruplex ligand induces anti-proliferative effects in brain tumor cells. PLoS ONE 2014, 9, e86187. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting g-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Bidzinska, J.; Cimino-Reale, G.; Zaffaroni, N.; Folini, M. G-quadruplex structures in the human genome as novel therapeutic targets. Molecules 2013, 18, 12368–12395. [Google Scholar] [CrossRef]
- Zheng, X.-H.; Nie, X.; Liu, H.-Y.; Fang, Y.-M.; Zhao, Y.; Xia, L.-X. Tmpyp4 promotes cancer cell migration at low doses, but induces cell death at high doses. Sci. Rep. 2016, 6, 26592. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Di Antonio, M.; Chan, L.; Balasubramanian, S. G-quadruplex ligands exhibit differential g-tetrad selectivity. Chem. Commun. 2015, 51, 8048–8050. [Google Scholar] [CrossRef] [PubMed]
- Laquente, B.; Lacasa, C.; Ginesta, M.M.; Casanovas, O.; Figueras, A.; Galan, M.; Ribas, I.G.; Germa, J.R.; Capella, G.; Vinals, F. Antiangiogenic effect of gemcitabine following metronomic administration in a pancreas cancer model. Mol. Cancer Ther. 2008, 7, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-P.; Lan, K.-H.; Li, C.-P.; Chao, Y.; Hou, M.-C.; Lin, H.-C.; Lee, S.-D. The telomere-binding protein trf2 is required for metronomic therapeutic effects of gemcitabine and capecitabine. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 917–928. [Google Scholar] [CrossRef] [PubMed]
- García-Beccaria, M.; Martínez, P.; Méndez-Pertuz, M.; Martínez, S.; Blanco-Aparicio, C.; Cañamero, M.; Mulero, F.; Ambrogio, C.; Flores, J.M.; Megias, D. Therapeutic inhibition of trf1 impairs the growth of p53-deficient k-rasg12v-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 2015, 7, 930–949. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Iwado, E.; Eller, M.S.; Kondo, Y.; Fujiwara, K.; Li, G.-Z.; Hess, K.R.; Siwak, D.R.; Sawaya, R.; Mills, G.B. Telomere 3′ overhang-specific DNA oligonucleotides induce autophagy in malignant glioma cells. FASEB J. 2007, 21, 2918–2930. [Google Scholar] [CrossRef]
- Gnanasekar, M.; Thirugnanam, S.; Zheng, G.; Chen, A.; Ramaswamy, K. T-oligo induces apoptosis in advanced prostate cancer cells. Oligonucleotides 2009, 19, 287–292. [Google Scholar] [CrossRef]
- Longe, H.O.; Romesser, P.B.; Rankin, A.M.; Faller, D.V.; Eller, M.S.; Gilchrest, B.A.; Denis, G.V. Telomere homolog oligonucleotides induce apoptosis in malignant but not in normal lymphoid cells: Mechanism and therapeutic potential. Int. J. Cancer 2009, 124, 473–482. [Google Scholar] [CrossRef]
- Sarkar, S.; Faller, D.V. T-oligos inhibit growth and induce apoptosis in human ovarian cancer cells. Oligonucleotides 2011, 21, 47–53. [Google Scholar] [CrossRef]
- Pitman, R.T.; Wojdyla, L.; Puri, N. Mechanism of DNA damage responses induced by exposure to an oligonucleotide homologous to the telomere overhang in melanoma. Oncotarget 2013, 4, 761–771. [Google Scholar] [CrossRef]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’connor, M.J. Inhibition of poly (adp-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, P.; Tangutoori, S.; Sridhar, S. Generation of dose–response curves and improved ic50s for parp inhibitor nanoformulations. In Cancer Nanotechnology: Methods and Protocols; Humana Press: New York, NY, USA, 2017; pp. 337–342. [Google Scholar]
- van Voss, M.R.H.; Brilliant, J.D.; Vesuna, F.; Bol, G.M.; van der Wall, E.; van Diest, P.J.; Raman, V. Combination treatment using ddx3 and parp inhibitors induces synthetic lethality in brca1-proficient breast cancer. Med. Oncol. 2017, 34, 33. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, M.M.; Wright, W.E.; Shay, J.W. Targeting telomerase-expressing cancer cells. J. Cell. Mol. Med. 2011, 15, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Marión, R.M.; de Silanes, I.L.; Mosteiro, L.; Gamache, B.; Abad, M.; Guerra, C.; Megías, D.; Serrano, M.; Blasco, M.A. Common telomere changes during in vivo reprogramming and early stages of tumorigenesis. Stem Cell Rep. 2017, 8, 460–475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Celeghin, A.; Giunco, S.; Freguja, R.; Zangrossi, M.; Nalio, S.; Dolcetti, R.; De Rossi, A. Short-term inhibition of tert induces telomere length-independent cell cycle arrest and apoptotic response in ebv-immortalized and transformed b cells. Cell Death Dis. 2016, 7, e2562. [Google Scholar] [CrossRef]
- Shi, J.B.; Chen, L.Z.; Wang, Y.; Xiou, C.; Tang, W.J.; Zhou, H.P.; Liu, X.H.; Yao, Q.Z. Benzophenone-nucleoside derivatives as telomerase inhibitors: Design, synthesis and anticancer evaluation in vitro and in vivo. Eur. J. Med. Chem. 2016, 124, 729–739. [Google Scholar] [CrossRef]
- Apte, M.S.; Cooper, J.P. Life and cancer without telomerase: Alt and other strategies for making sure ends (don’t) meet. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 57–73. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.H.; Lee, G.E.; Kim, S.W.; CHUNG, I.K. Identification of a quinoxaline derivative that is a potent telomerase inhibitor leading to cellular senescence of human cancer cells. Biochem. J. 2003, 373, 523–529. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.H.; Lee, G.E.; Lee, J.E.; Chung, I.K. Potent inhibition of human telomerase by nitrostyrene derivatives. Mol. Pharmacol. 2003, 63, 1117–1124. [Google Scholar] [CrossRef]
- Ueno, T.; Takahashi, H.; Oda, M.; Mizunuma, M.; Yokoyama, A.; Goto, Y.; Mizushina, Y.; Sakaguchi, K.; Hayashi, H. Inhibition of human telomerase by rubromycins: Implication of spiroketal system of the compounds as an active moiety. Biochemistry 2000, 39, 5995–6002. [Google Scholar] [CrossRef]
- Xu, Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Nakai, R.; Ishida, H.; Asai, A.; Ogawa, H.; Yamamoto, Y.; Kawasaki, H.; Akinaga, S.; Mizukami, T.; Yamashita, Y. Telomerase inhibitors identified by a forward chemical genetics approach using a yeast strain with shortened telomere length. Chem. Biol. 2006, 13, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Ohashi, Y.; Ishikawa, K.; Yasue, A.; Kato, R.; Achiwa, Y.; Nishio, E.; Udagawa, Y. Expression of cyclooxygenase-2 in uterine endometrial cancer and anti-tumor effects of a selective cox-2 inhibitor. Int. J. Oncol. 2005, 26, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Meng, L.; Wang, H.; Zhou, J.; Xu, G.; Wang, S.; Xi, L.; Chen, G.; Wang, B.; Zhu, T. Role of htert in apoptosis of cervical cancer induced by histone deacetylase inhibitor. Biochem. Biophys. Res. Commun. 2005, 335, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.J.; Lee, S.J.; Choi, B.T.; Park, Y.-M.; Choi, Y.H. Induction of apoptosis and inhibition of telomerase activity by trichostatin a, a histone deacetylase inhibitor, in human leukemic u937 cells. Exp. Mol. Pathol. 2007, 82, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, H.; Ishiguro, H.; Ikeda, N.; Hori, M.; Kubota, Y.; Uemura, H. Genistein induces cell growth inhibition in prostate cancer through the suppression of telomerase activity. Int. J. Urol. 2005, 12, 73–80. [Google Scholar] [CrossRef]
- Zhu, B.; Zhang, L.H.; Zhao, Y.M.; Cui, J.R.; Strada, S.J. 8-chloroadenosine induced hl-60 cell growth inhibition, differentiation, and g0/g1 arrest involves attenuated cyclin d1 and telomerase and up-regulated p21waf1/cip1. J. Cell. Biochem. 2006, 97, 166–177. [Google Scholar] [CrossRef]
- Wang, C.-M.; Jia, Z.-J.; Zheng, R.-L. The effect of 17 sesquiterpenes on cell viability and telomerase activity in the human ovarian cancer cell line ho-8910. Planta Med. 2007, 73, 180–184. [Google Scholar] [CrossRef]
- He, H.; Xia, H.H.X.; Wang, J.D.; Gu, Q.; Lin, M.; Zou, B.; Lam, S.K.; Chan, A.O.; Yuen, M.F.; Kung, H.F. Inhibition of human telomerase reverse transcriptase by nonsteroidal antiinflammatory drugs in colon carcinoma. Cancer 2006, 106, 1243–1249. [Google Scholar] [CrossRef]
- Strahl, C.; Blackburn, E.H. The effects of nucleoside analogs on telomerase and telomeres in tetrahymena. Nucleic Acids Res. 1994, 22, 893–900. [Google Scholar] [CrossRef][Green Version]
- Olivero, O.A. Mechanisms of genotoxicity of nucleoside reverse transcriptase inhibitors. Environ. Mol. Mutagenesis 2007, 48, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Yamada, R.; Tomikawa, A.; Shudo, K.; Saito, M.; Ishikawa, F.; Saneyoshi, M. Recognition of 2′-deoxy-l-ribonucleoside 5′-triphosphates by human telomerase. Biochem. Biophys. Res. Commun. 2000, 279, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, T.M.; Cathers, B.E.; Ravikumar, K.; Mamiya, B.M.; Kerwin, S.M. Inhibition of human telomerase by 7-deaza-2′-deoxyguanosine nucleoside triphosphate analogs: Potent inhibition by 6-thio-7-deaza-2′-deoxyguanosine 5′-triphosphate. Bioorg. Chem. 2001, 29, 36–55. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.D.; Chen, Y.R. Inhibition of telomerase with human telomerase reverse transcriptase antisense increases the sensitivity of tumor necrosis factor-α-induced apoptosis in prostate cancer cells. Asian J. Androl. 2007, 9, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Fadri-Moskwik, M.; Zhou, Q.; Chai, W. Beyond telomerase: Telomere instability as a novel target for cancer therapy. J. Mol. Genet. Med. Int. J. Biomed. Res. 2013, 7, 4. [Google Scholar]
- Arndt, G.M.; MacKenzie, K.L. New prospects for targeting telomerase beyond the telomere. Nat. Rev. Cancer 2016, 16, 508–524. [Google Scholar] [CrossRef]
- Hochreiter, A.E.; Xiao, H.; Goldblatt, E.M.; Gryaznov, S.M.; Miller, K.D.; Badve, S.; Sledge, G.W.; Herbert, B.-S. Telomerase template antagonist grn163l disrupts telomere maintenance, tumor growth, and metastasis of breast cancer. Clin. Cancer Res. 2006, 12, 3184–3192. [Google Scholar] [CrossRef]
- Williams, S.C. No end in sight for telomerase-targeted cancer drugs. Nat. Res. 2013, 19, 6–7. [Google Scholar] [CrossRef]
- Elayadi, A.N.; Demieville, A.; Wancewicz, E.V.; Monia, B.P.; Corey, D.R. Inhibition of telomerase by 2′-o-(2-methoxyethyl) rna oligomers: Effect of length, phosphorothioate substitution and time inside cells. Nucleic Acids Res. 2001, 29, 1683–1689. [Google Scholar] [CrossRef][Green Version]
- Kushner, D.M.; Paranjape, J.M.; Bandyopadhyay, B.; Cramer, H.; Leaman, D.W.; Kennedy, A.W.; Silverman, R.H.; Cowell, J.K. 2-5a antisense directed against telomerase rna produces apoptosis in ovarian cancer cells. Gynecol. Oncol. 2000, 76, 183–192. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Takahashi, Y.; Shinohara, A.; Wan, X.; Takahashi, S.; Niwa, K.; Tamaya, T. The 5′-end of htert mrna is a good target for hammerhead ribozyme to suppress telomerase activity. Biochem. Biophys. Res. Commun. 2000, 273, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Berardinelli, F.; Coluzzi, E.; Sgura, A.; Antoccia, A. Targeting telomerase and telomeres to enhance ionizing radiation effects in in vitro and in vivo cancer models. Mutat. Res. Rev. Mutat. Res. 2017, 773, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Cerone, M.A.; Londoño-Vallejo, J.A.; Autexier, C. Telomerase inhibition enhances the response to anticancer drug treatment in human breast cancer cells. Mol. Cancer Ther. 2006, 5, 1669–1675. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, J.; Hwang, S.S.; Liesa, M.; Gan, B.; Sahin, E.; Jaskelioff, M.; Ding, Z.; Ying, H.; Boutin, A.T.; Zhang, H. Antitelomerase therapy provokes alt and mitochondrial adaptive mechanisms in cancer. Cell 2012, 148, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Qi, B.; Zhao, B. Alternative lengthening of telomeres is induced by telomerase inhibitors in barrett’s esophageal cells. Oncol. Rep. 2013, 29, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Queisser, A.; Heeg, S.; Thaler, M.; von Werder, A.; Opitz, O.G. Inhibition of telomerase induces alternative lengthening of telomeres during human esophageal carcinogenesis. Cancer Genet. 2013, 206, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Cortez-Gonzalez, X.; Zanetti, M. Telomerase immunity from bench to bedside: Round one. J. Transl. Med. 2007, 5, 12. [Google Scholar] [CrossRef]
- Masutomi, K.; Possemato, R.; Wong, J.M.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227. [Google Scholar] [CrossRef]
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef]
- Opresko, P.L.; Mason, P.A.; Podell, E.R.; Lei, M.; Hickson, I.D.; Cech, T.R.; Bohr, V.A. Pot1 stimulates recq helicases wrn and blm to unwind telomeric DNA substrates. J. Biol. Chem. 2005, 280, 32069–32080. [Google Scholar] [CrossRef] [PubMed]
- Lansdorp, P.M. Major cutbacks at chromosome ends. Trends Biochem. Sci. 2005, 30, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by g-quartet dma structures. Nature 1991, 350, 718. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Hélène, C. G-quadruplex DNA: A target for drug design. Nat. Med. 1998, 4, 1366. [Google Scholar] [CrossRef]
- Islam, M.K.; Jackson, P.J.; Rahman, K.M.; Thurston, D.E. Recent advances in targeting the telomeric g-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. [Google Scholar] [CrossRef]
- Gowan, S.M.; Harrison, J.R.; Patterson, L.; Valenti, M.; Read, M.A.; Neidle, S.; Kelland, L.R. A g-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002, 61, 1154–1162. [Google Scholar] [CrossRef]
- Jonchhe, S.; Ghimire, C.; Cui, Y.; Sasaki, S.; McCool, M.; Park, S.; Iida, K.; Nagasawa, K.; Sugiyama, H.; Mao, H. Binding of a telomestatin derivative changes the mechanical anisotropy of a human telomeric g-quadruplex. Angew. Chem. Int. Ed. 2019, 58, 877–881. [Google Scholar] [CrossRef]
- Chung, W.J.; Heddi, B.; Tera, M.; Iida, K.; Nagasawa, K.; Phan, A.T.N. Solution structure of an intramolecular (3 + 1) human telomeric g-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013, 135, 13495–13501. [Google Scholar] [CrossRef]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.; Searle, M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d (ttagggt) 4 containing the human telomeric repeat. J. Mol. Biol. 2003, 334, 25–36. [Google Scholar] [CrossRef]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.; Searle, M.S. Recognition and stabilization of quadruplex DNA by a potent new telomerase inhibitor: Nmr studies of the 2: 1 complex of a pentacyclic methylacridinium cation with d (ttagggt) 4. Angew. Chem. Int. Ed. 2001, 40, 4749–4751. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef] [PubMed]
- Sparapani, S.; Haider, S.M.; Doria, F.; Gunaratnam, M.; Neidle, S. Rational design of acridine-based ligands with selectivity for human telomeric quadruplexes. J. Am. Chem. Soc. 2010, 132, 12263–12272. [Google Scholar] [CrossRef] [PubMed]
- Sissi, C.; Lucatello, L.; Krapcho, A.P.; Maloney, D.J.; Boxer, M.B.; Camarasa, M.V.; Pezzoni, G.; Menta, E.; Palumbo, M. Tri-, tetra-and heptacyclic perylene analogues as new potential antineoplastic agents based on DNA telomerase inhibition. Bioorg. Med. Chem. 2007, 15, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Cuenca, F.; Greciano, O.; Gunaratnam, M.; Haider, S.; Munnur, D.; Nanjunda, R.; Wilson, W.D.; Neidle, S. Tri-and tetra-substituted naphthalene diimides as potent g-quadruplex ligands. Bioorg. Med. Chem. Lett. 2008, 18, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Hounsou, C.; Guittat, L.; Monchaud, D.; Jourdan, M.; Saettel, N.; Mergny, J.L.; Teulade-Fichou, M.P. G-quadruplex recognition by quinacridines: A sar, nmr, and biological study. ChemMedChem 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Ghosh, R.; Neidle, S. Structural basis for binding of porphyrin to human telomeres. Biochemistry 2007, 46, 2390–2397. [Google Scholar] [CrossRef]
- Cookson, J.C.; Dai, F.; Smith, V.; Heald, R.A.; Laughton, C.A.; Stevens, M.F.; Burger, A.M. Pharmacodynamics of the g-quadruplex-stabilizing telomerase inhibitor 3, 11-difluoro-6, 8, 13-trimethyl-8h-quino [4, 3, 2-kl] acridinium methosulfate (rhps4) in vitro: Activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol. Pharmacol. 2005, 68, 1551–1558. [Google Scholar]
- Salvati, E.; Leonetti, C.; Rizzo, A.; Scarsella, M.; Mottolese, M.; Galati, R.; Sperduti, I.; Stevens, M.F.; D’Incalci, M.; Blasco, M. Telomere damage induced by the g-quadruplex ligand rhps4 has an antitumor effect. J. Clin. Investig. 2007, 117, 3236–3247. [Google Scholar] [CrossRef]
- Miyazaki, T.; Pan, Y.; Joshi, K.; Purohit, D.; Hu, B.; Demir, H.; Mazumder, S.; Okabe, S.; Yamori, T.; Viapiano, M. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric g-quadruplex and inhibition of the proto-oncogene, c-myb. Clin. Cancer Res. 2012, 18, 1268–1280. [Google Scholar] [CrossRef]
- Qin, Y.; Fortin, J.S.; Tye, D.; Gleason-Guzman, M.; Brooks, T.A.; Hurley, L.H. Molecular cloning of the human platelet-derived growth factor receptor β (pdgfr-β) promoter and drug targeting of the g-quadruplex-forming region to repress pdgfr-β expression. Biochemistry 2010, 49, 4208–4219. [Google Scholar] [CrossRef]
- Sugimura, K.; Namikawa, T.; Takezaki, Y.; Kitagwa, H.; Munekage, M.; Hanazaki, K. Antitumor effects of a combined treatment with bortezomib and gemcitabine in bile duct cancer cell lines. Gan Kagaku Ryoho. Cancer Chemother. 2016, 43, 1608. [Google Scholar]
- Yapp, D.T.; Wong, M.Q.; Kyle, A.H.; Valdez, S.M.; Tso, J.; Yung, A.; Kozlowski, P.; Owen, D.A.; Buczkowski, A.K.; Chung, S.W. The differential effects of metronomic gemcitabine and antiangiogenic treatment in patient-derived xenografts of pancreatic cancer: Treatment effects on metabolism, vascular function, cell proliferation, and tumor growth. Angiogenesis 2016, 19, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; De Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Stansel, R.M.; De Lange, T.; Griffith, J.D. T-loop assembly in vitro involves binding of trf2 near the 3′ telomeric overhang. EMBO J. 2001, 20, 5532–5540. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Smogorzewska, A.; de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003, 13, 1549–1556. [Google Scholar] [CrossRef]
- Li, G.-Z.; Eller, M.S.; Hanna, K.; Gilchrest, B.A. Signaling pathway requirements for induction of senescence by telomere homolog oligonucleotides. Exp. Cell Res. 2004, 301, 189–200. [Google Scholar] [CrossRef]
- Puri, N.; Eller, M.S.; Byers, H.R.; Dykstra, S.; Kubera, J.; Gilchrest, B.A. Telomere-based DNA damage responses: A new approach to melanoma. FASEB J. 2004, 18, 1373–1381. [Google Scholar] [CrossRef]
- Yaar, M.; Eller, M.S.; Panova, I.; Kubera, J.; Wee, L.H.; Cowan, K.H.; Gilchrest, B.A. Telomeric DNA induces apoptosis and senescence of human breast carcinoma cells. Breast Cancer Res. 2007, 9, R13. [Google Scholar] [CrossRef]
- Peuscher, M.H.; Jacobs, J.J. Posttranslational control of telomere maintenance and the telomere damage response. Cell Cycle 2012, 11, 1524–1534. [Google Scholar] [CrossRef]
- Chang, W.; Dynek, J.N.; Smith, S. Trf1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003, 17, 1328–1333. [Google Scholar] [CrossRef]
- Zhong, L.; Yeh, T.-Y.J.; Hao, J.; Pourtabatabaei, N.; Mahata, S.K.; Shao, J.; Chessler, S.D.; Chi, N.-W. Nutritional energy stimulates nad+ production to promote tankyrase-mediated parsylation in insulinoma cells. PLoS ONE 2015, 10, e0122948. [Google Scholar] [CrossRef]
- Her, Y.R.; Chung, I.K. P300-mediated acetylation of trf2 is required for maintaining functional telomeres. Nucleic Acids Res. 2013, 41, 2267–2283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, L.-Y.; Han, X.; Xie, W.; Kim, H.; Yang, D.; Liu, D.; Songyang, Z. Phosphorylation of tpp1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl. Acad. Sci. USA 2013, 110, 5457–5462. [Google Scholar] [CrossRef] [PubMed]
- Ida, J.; Chan, S.K.; Glökler, J.; Lim, Y.Y.; Choong, Y.S.; Lim, T.S. G-quadruplexes as an alternative recognition element in disease-related target sensing. Molecules 2019, 24, 1079. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.K.; Meeker, A. Telomeres and telomerase in prostate cancer development and therapy. Nat. Rev. Urol. 2017, 14, 607. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).